Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis
Abstract
1. Introduction
2. Results
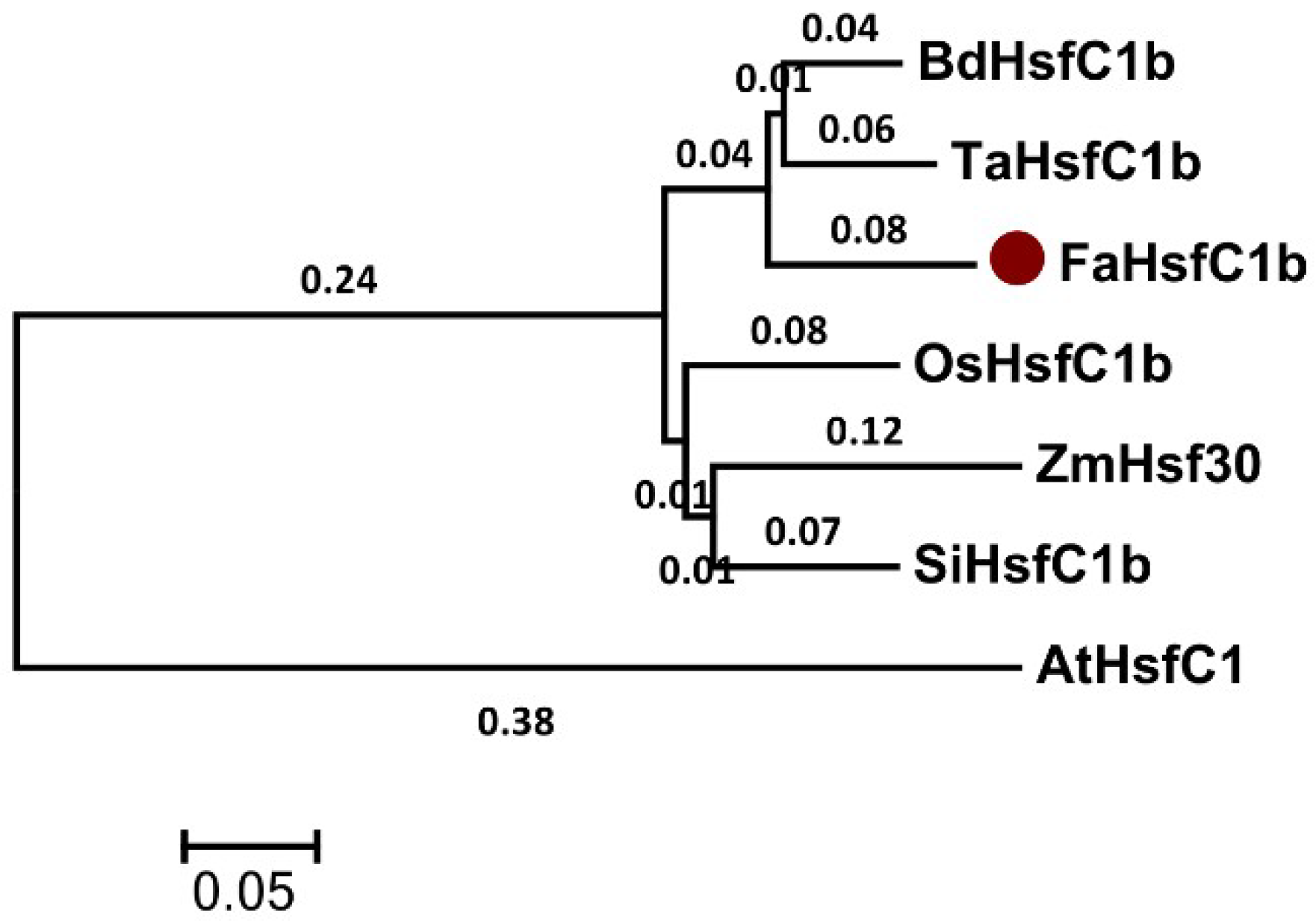
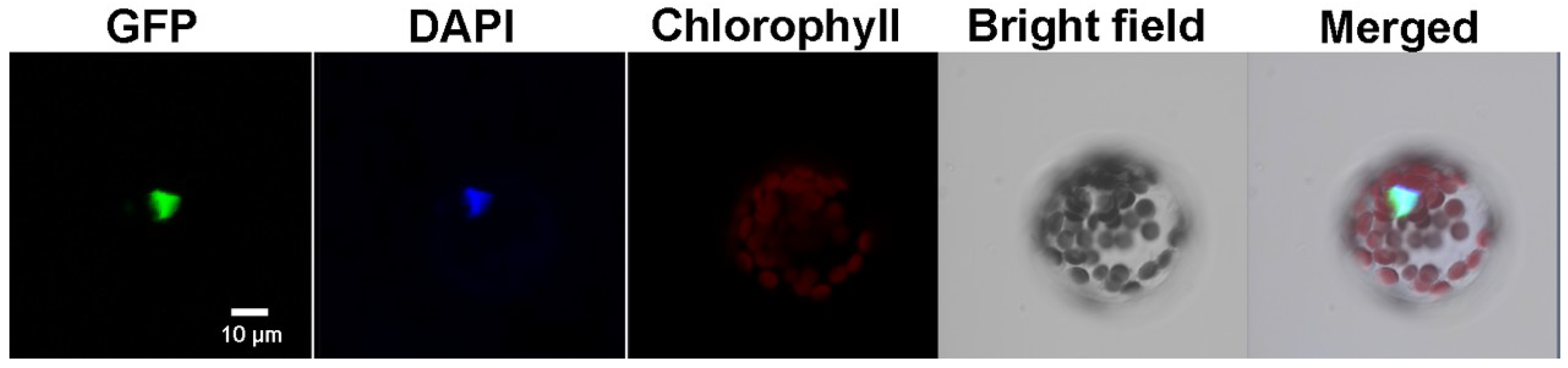
2.1. FaHsfC1b Encoded a Class C Hsf and the Gene Product is Localized in the Nucleus
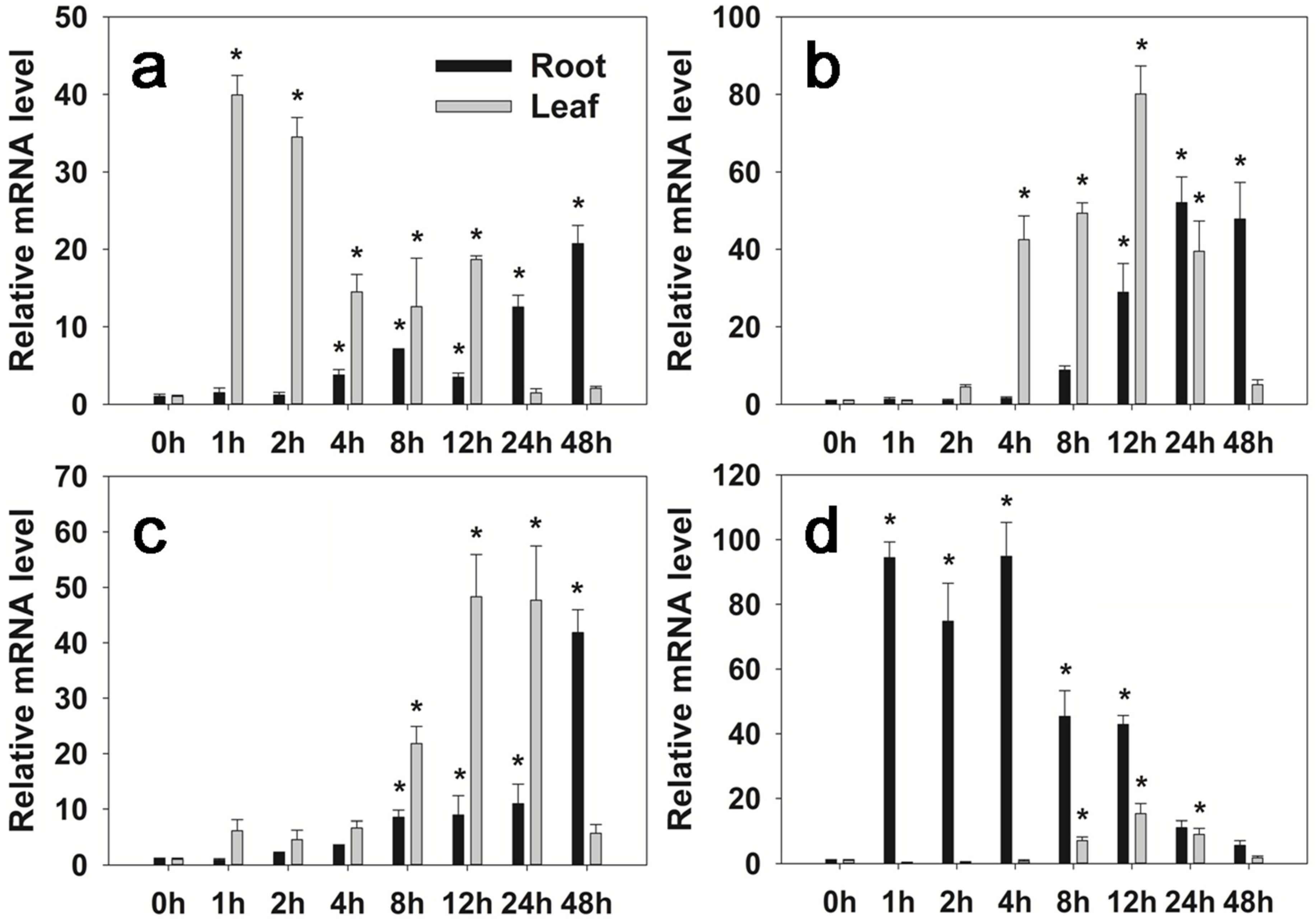
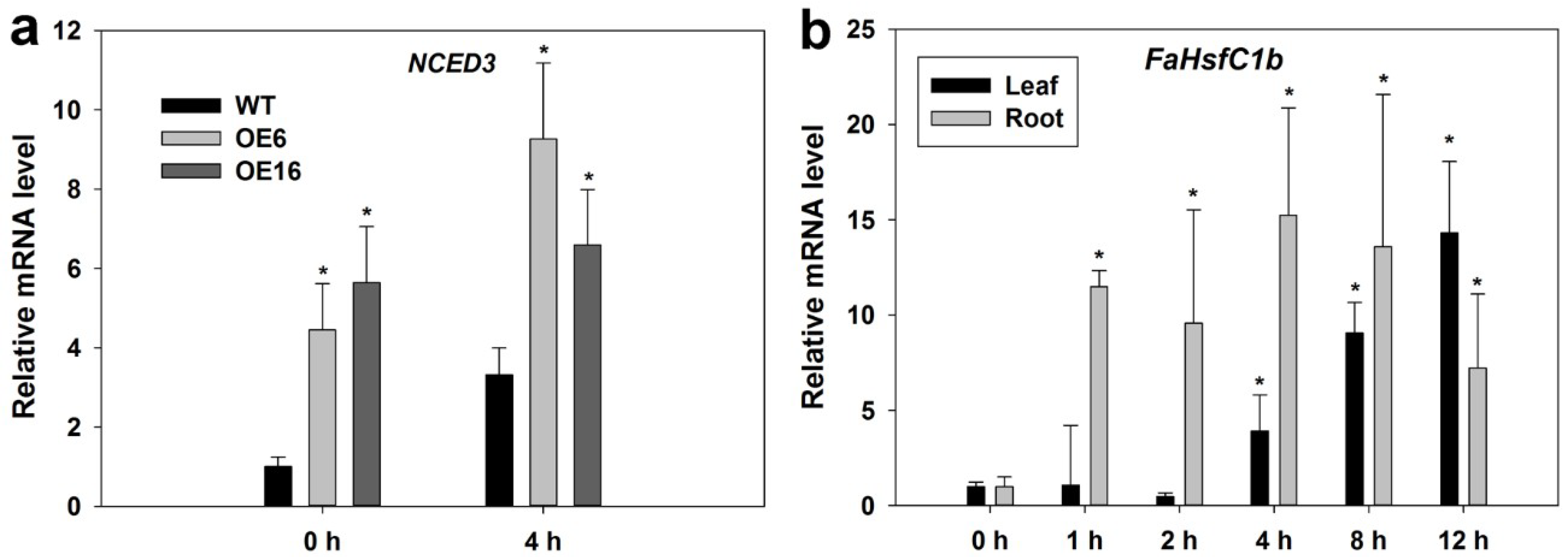
2.2. Expression Pattern of FaHsfC1b Under Different Abiotic Stresses
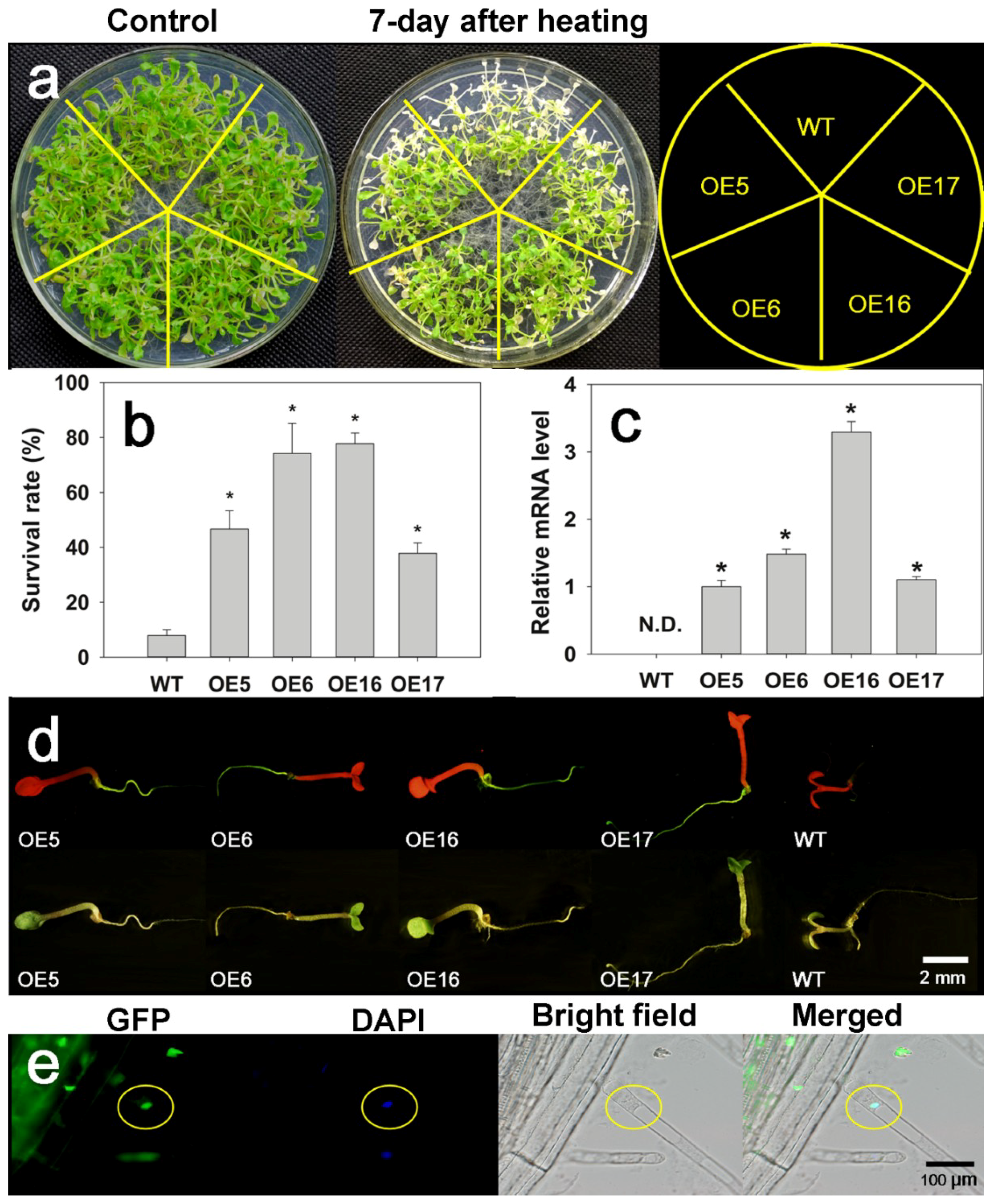
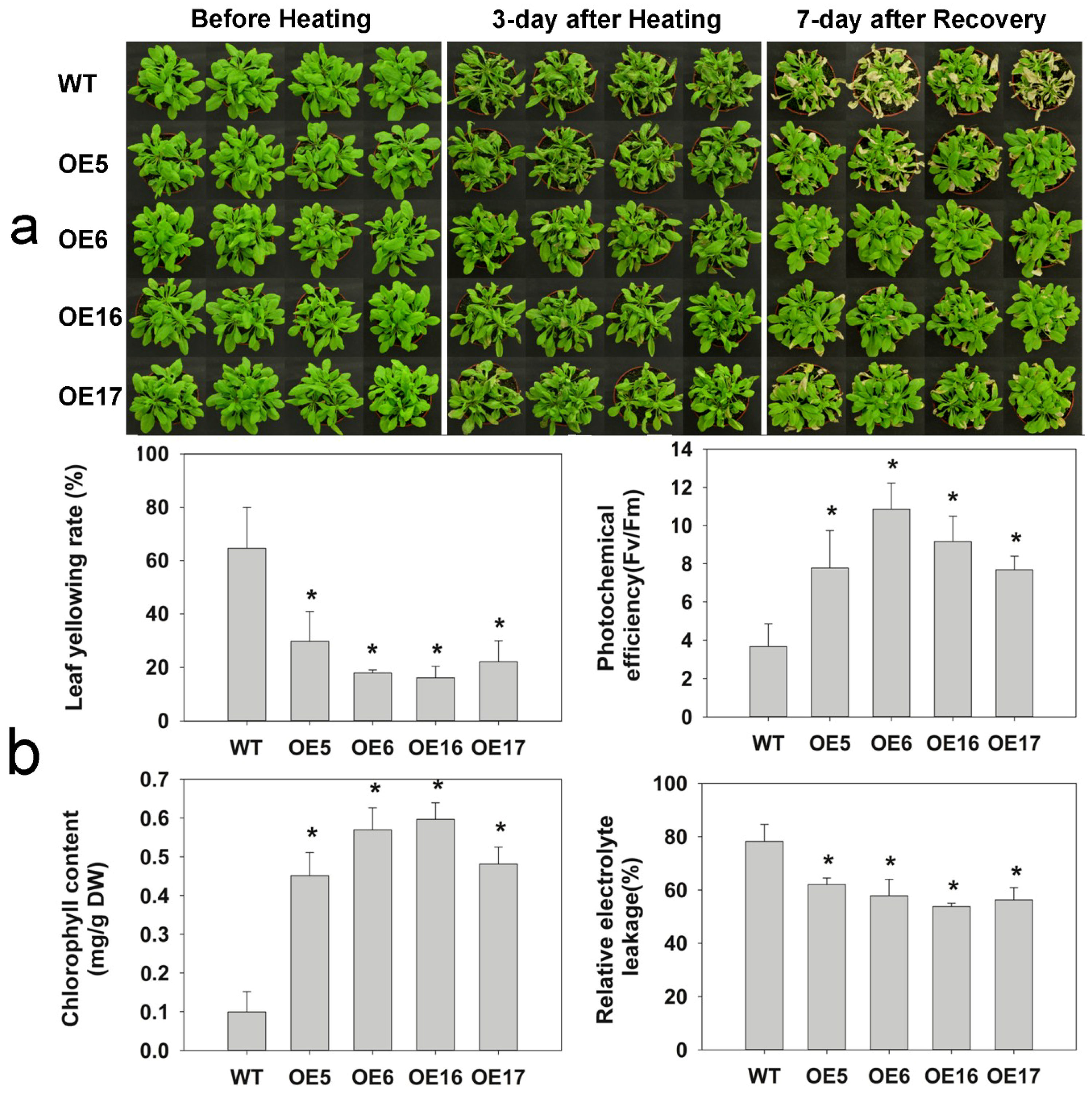
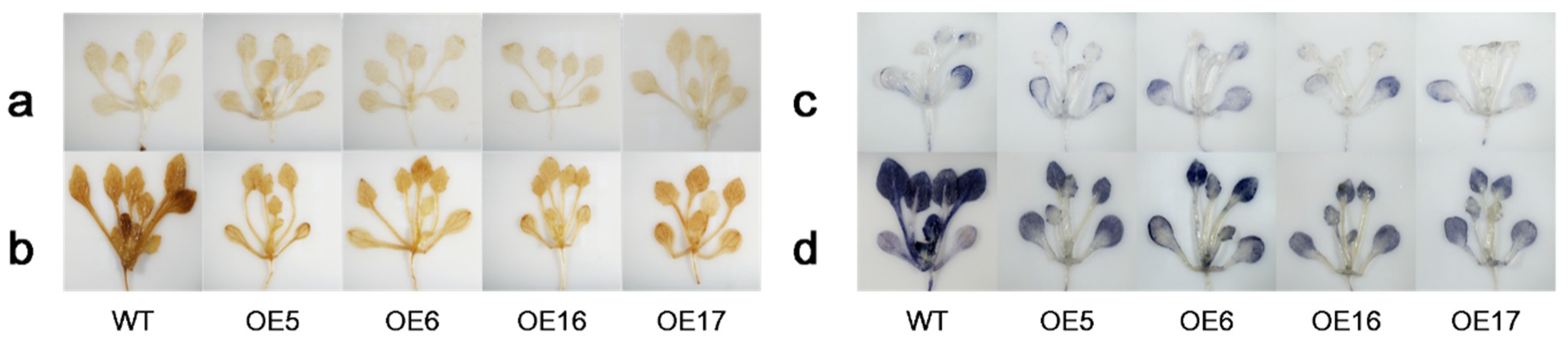
2.3. FaHsfC1b-Transgenic Arabidopsis Exhibited Improvement in Heat Tolerance
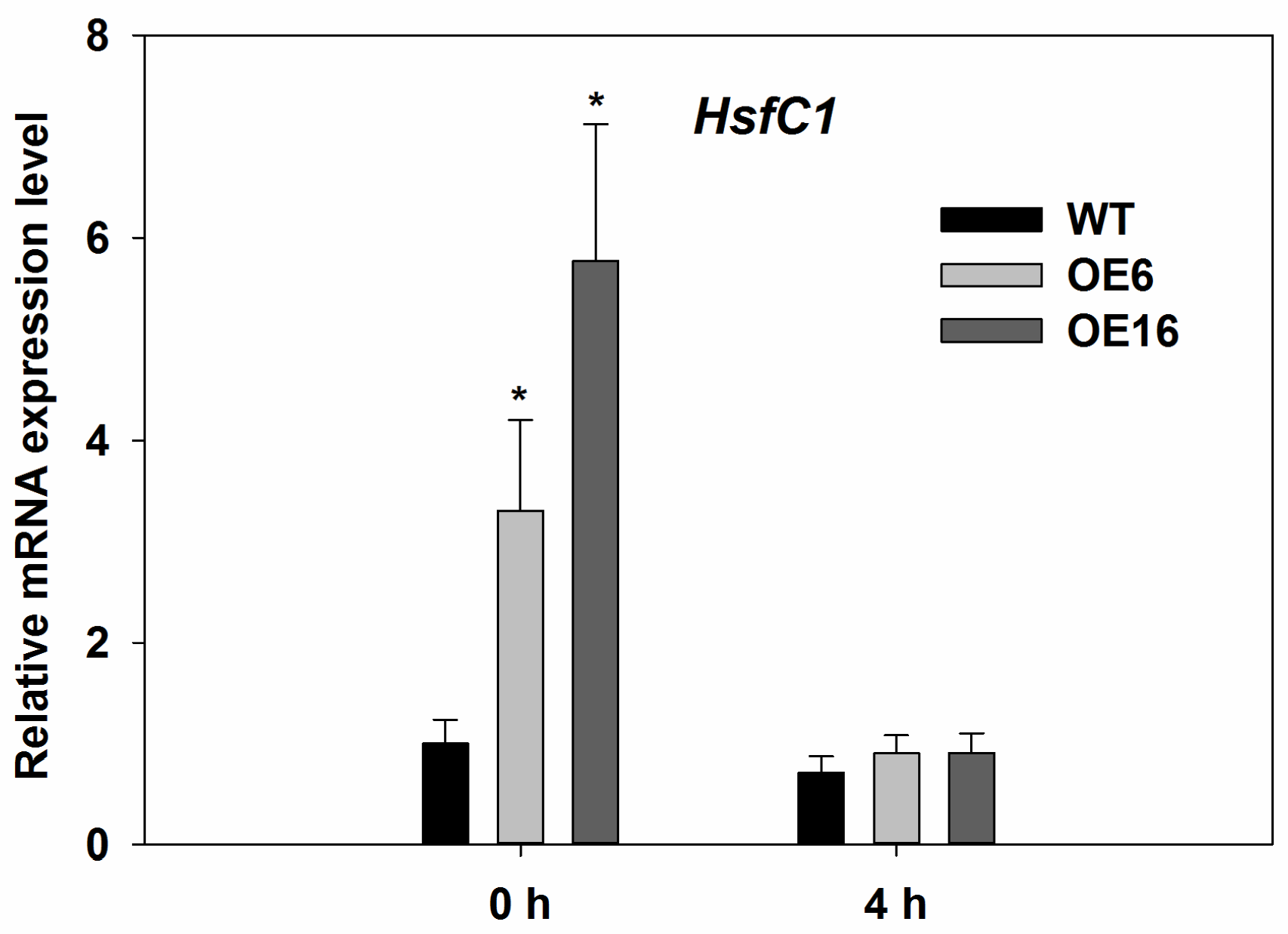
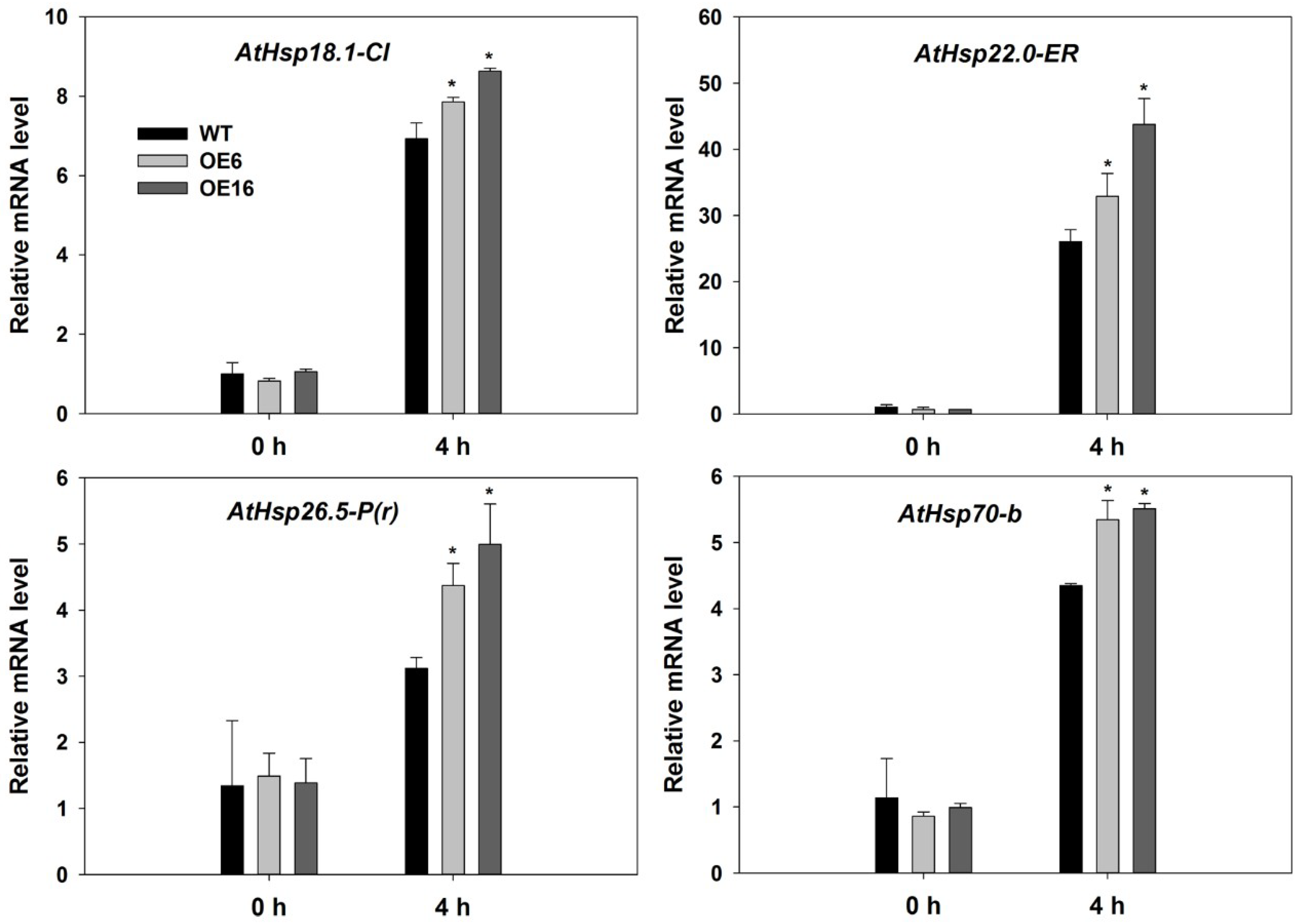
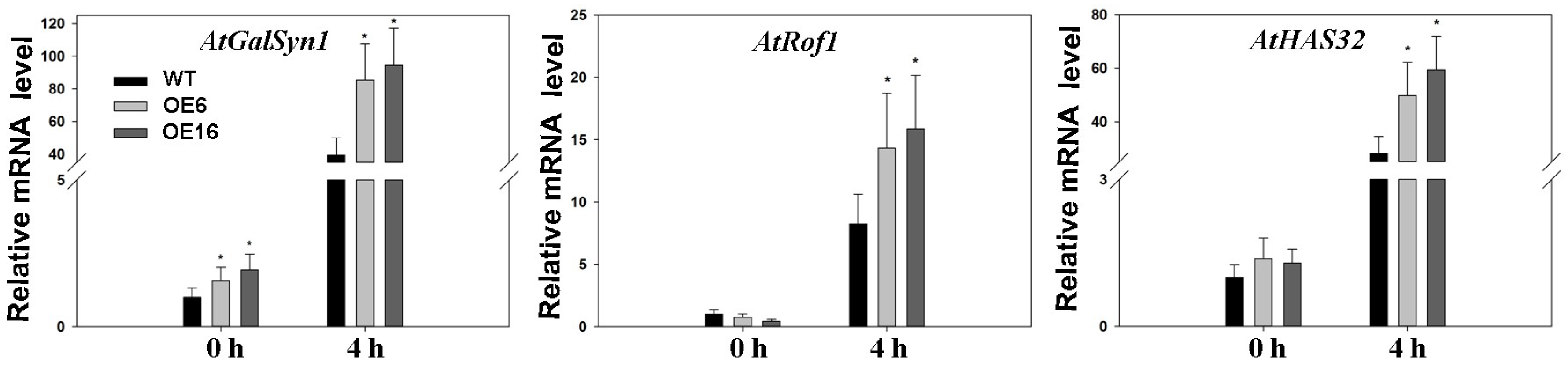
2.4. Upregulation of Endogenous Erabidopsis Hsf1C Gene, Heat Protection Protein Genes, and ABA-Related Genes Associated with Overexpression of FaHsfC1b in Response to Heat Stress
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Treatments
4.2. Isolation Fahsfc1b Gene from Tall Fescue
4.3. Subcellular Localization of Fahsfc1b Protein
4.4. Plasmid Construction and Plant Transformation
4.5. qRT-PCR Analysis
4.6. Physiological Characterization of Transgenic and Wild Type Arabidopsis Under Heat Stress
4.7. Histochemical Detection of Oxidation Resistance
4.8. Statistical Analyses
4.9. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Von, K.P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar]
- Guo, J.K.; Wu, J.; Ji, Q.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.H.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Nover, N.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.R.; Li, Y.S.; Xing, D.; Gao, C.J. Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 2009, 60, 2073–2091. [Google Scholar] [CrossRef] [PubMed]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 2010, 152, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Huang, W.L.; Yang, Z.M.; Liu, J.; Huang, B.R. Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 2016, 6, 28021. [Google Scholar] [CrossRef] [PubMed]
- Chanschaminet, K.Y.; Baniwal, S.K.; Bublak, D.; Nover, L.; Scharf, K.D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 2009, 284, 20848–20857. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Huang, W.L.; Liu, J.; Yang, Z.M.; Huang, B.R. Molecular regulation and physiological functions of a novel FaHsfA2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 2017, 15, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Zhuang, L.L.; Shi, Y.; Huang, B.R. Up-regulation of HsfA2c and HSPs by ABA contributing to improved heat tolerance in tall fescue and Arabidopsis. Int. J. Mol. Sci. 2017, 18, 1981. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P.; Sadat, S.; Drenth, J.; Mcintyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.H.; Hu, G.C.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Tao, P.; Li, B.Y.; Wang, W.H.; Yue, Z.C.; Lei, J.L.; Zhong, X.M. Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet. Mol. Res. 2015, 14, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Jiang, Q.; Lin, J.Y.; Wang, D.L.; Li, S.J.; Liu, C.R.; Sun, C.D.; Chen, K.S. Heat shock transcription factors expression during fruit development and under hot air stress in Ponkan (Citrus reticulata Blanco cv. Ponkan) fruit. Gene 2015, 559, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, M.Y.; Wang, F.; Xu, Z.S.; Huang, W.; Wang, G.L.; Ma, J.; Xiong, A.S. Heat shock factors in carrot: Genome-wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 2015, 42, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in Salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.J.; Chen, D.; Mclntyre, C.L.; Dreccer, M.F.; Zhang, Z.B.; Drenth, J.; Sundaravelpandian, K.; Chang, H.; Xue, G.P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2017, 41, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.M.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, 2012, pls011. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Kim, Y.K.; Grover, A. Rice sHsp genes: Genomic organization and expression profiling under stress and development. BMC Genom. 2009, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Haq, N.U.; Raza, S.; Luthe, D.S.; Heckathorn, S.A.; Shakeel, S.N. A dual role for the chloroplast small heat shock protein of chenopodium album including protection from both heat and metal stress. Plant Mol. Biol. Rep. 2013, 31, 398–408. [Google Scholar] [CrossRef]
- Siddique, M.; Gernhard, S.; Koskull-Döring, P.V.; Vierling, E.; Scharf, K.D. The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 2008, 13, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, L.; Sanchez-Ballesta, M.T.; Romojaro, F.; Flores, F.B. Physiological, hormonal and molecualr mechanisms regulating chilling injury in horticutural species. Postharvest technologies applied to reduce its impact. J. Sci. Food Agric. 2009, 89, 555–573. [Google Scholar] [CrossRef]
- Li, M.; Berendzen, K.W.; Friedrich, S. Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol. Biol. 2010, 73, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Schramm, F.; Ganguli, A.; Kiehlmann, E.; Englich, G.; Walch, D.; von Koskull-Döring, P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006, 60, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Charng, Y.Y.; Liu, H.C.; Liu, N.Y.; Hsu, F.C.; Ko, S.S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006, 140, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2010, 59, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Charng, Y.Y. A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol. 2014, 164, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Panikulangara, T.J.; Eggersschumacher, G.; Wunderlich, M.; Stransky, H.; Schöffl, F. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol. 2004, 136, 3148–3158. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Shekhar, S.; Singh, D.; Chakraborty, S.; Chakraborty, N. Heat Shock Proteins and Abiotic Stress Tolerance in Plants. In Regulation of Heat Shock Protein Responses; Asea, A., Kaur, P., Eds.; Heat Shock Proteins; Springer: Cham, Switzerland, 2018; Volume 13, pp. 41–69. [Google Scholar]
- Reddy, R.A.; Kumar, B.; Reddy, P.S.; Mishra, R.N.; Mahanty, S.; Kaul, T.; Nair, S.; Sopory, S.K.; Reddy, M.K. Molecular cloning and characterization of genes encoding Pennisetum glaucum ascorbate peroxidase and heat-shock factor: Interlinking oxidative and heat-stress responses. J. Plant Physiol. 2009, 166, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Schöffl, F. Interaction between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J. Exp. Bot. 2002, 53, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Hannaway, D.B.; Fransen, S.; Cropper, J.B.; Teel, M.; Chaney, T.; Griggs, T.D.; Halse, R.R.; Hart, J.M.; Cheeke, P.R.; Klinger, R.; Lane, W. Tall Fescue (Festuca arundinacea Schreb); PNW 504; Oregon State University Extension Publication: Corvallis, OR, USA, 1999. [Google Scholar]
- Perdomo, P.; Murphy, J.A.; Berkowitz, G.A. Physiological changes associated with performance of Kentucky bluegrass cultivars during summer stress. HortScience 1996, 31, 1182–1186. [Google Scholar]
- Baniwal, S.K.; Chan, K.Y.; Scharf, K.D.; Nover, L. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 2007, 282, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.F.; Wang, Y.F.; Gao, Y.; Zhou, Y.; Zhang, E.Y.; Hu, Y.; Yuan, Y.Y.; Liang, G.H.; Xu, C.W. Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol. Biol. 2014, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.; Xu, Z.S.; Feng, W.; Tan, G.F.; Li, M.Y.; Xiong, A.S. Genome-wide analysis of HSF family transcription factors and their responses to abiotic stresses in two Chinese cabbage varieties. Acta Physiol. Plant 2014, 36, 513–523. [Google Scholar]
- Wang, C.; Zhang, Q.; Shou, H.X. Identification and expression analysis of OsHsfs in rice. J. Zhejiang Univ. Sci. B 2009, 10, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Wollgiehn, R.; Neumann, D.; Nieden, U.Z.; Müsch, A.; Scharf, K.D.; Nover, L. Intracellular distribution of small heat stress proteins in cultured cells of Lycopersicon peruvianum. J. Plant Physiol. 1994, 144, 491–499. [Google Scholar] [CrossRef]
- Chen, X.H.; Lin, S.K.; Liu, Q.L.; Huang, J.; Zhang, W.F.; Lin, J.; Wang, Y.F.; Ke, Y.Q.; He, H.Q. Expression and interaction of small heat shock proteins (sHsps) in rice in response to heat stress. Biochim. Biophys. Acta 2014, 1844, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Schneider-Mergener, J.; Forreiter, C. Analysis of chaperone function and formation of hetero-oligomeric complexes of Hsp18.1 and Hsp17.7, representing two different cytoplasmic sHSP classes in Pisum sativum. J. Plant Growth Regul. 2005, 24, 226–237. [Google Scholar] [CrossRef]
- Bardel, J.; Louwagie, M.; Jaquinod, M.; Jourdain, A.; Luche, S.; Rabilloud, T.; Macherel, D.; Garin, J.; Bourguignon, J. A survey of the plant mitochondrial proteome in relation to development. Proteomics 2002, 2, 880–898. [Google Scholar] [CrossRef]
- Stupnikova, I.; Benamar, A.; Tolleter, D.; Grelet, J.; Borovskii, G.; Dorne, A.J.; Macherel, D. Pea seed mitochondria are endowed with a remarkable tolerance to extreme physiological temperatures. Plant Physiol. 2006, 140, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Li, Y.J.; Chen, S.Z. Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J. Plant Physiol. 1998, 153, 488–496. [Google Scholar] [CrossRef]
- Larkindale, J.; Huang, B.R. Effects of abscisic acid, salicylic acid, ethylene and hydrogen peroxide in thermotolerance and recovery for creeping bentgrass. Plant Growth Regul. 2005, 47, 17–28. [Google Scholar] [CrossRef]
- Zhou, W.J.; Leul, M. Uniconazole-induced tolerance of rape plants to heat stress in relation to changes in hormonal levels, enzyme activities and lipid peroxidation. Plant Growth Regul. 1999, 27, 99–104. [Google Scholar] [CrossRef]
- Bajguz, A. Brassinosteroid enhanced the level of abscisic acid in Chlorella vulgaris subjected to short-term heat stress. J. Plant Physiol. 2009, 166, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Song, L.; Wang, X.; Bi, Y. Effect of abscisic acid on heat stress tolerance in the calli from two ecotypes of Phragmites communis. Biol. Plant 2010, 54, 607–613. [Google Scholar] [CrossRef]
- Pareek, A.; Singla, S.L.; Grover, A. Protein alterations associated with salinity, desiccation, high and low temperature stresses and abscisic acid application in Lal nakanda, a drought-tolerant rice cultivar. Curr. Sci. 1998, 75, 1170–1174. [Google Scholar]
- Campbell, J.L.; Klueva, N.Y.; Zheng, H.G.; Nietosotelo, J.; Ho, T.D.; Nguyen, H.T. Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA. Biochim. Biophys. Acta 2001, 1517, 270–277. [Google Scholar] [CrossRef]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, A.L.; Chen, X.B.; Zhou, X.Y.; Gao, G.F.; Wang, W.F.; Zhang, X.W. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J. Plant Physiol. 2009, 166, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Dheeraj, M.; Yasuaki, E.; Dhruv, L.; Amanjot, S.; Hiroshi, S.; Anil, G. Binding affinities and interactions among different heat shock element types and heat shock factors in rice (Oryza sativa L.). FEBS J. 2011, 278, 3076–3085. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 357–359. [Google Scholar]
- Don, R.H.; Cox, P.T.; Wainwright, B.J.; Baker, K.; Mattick, J.S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991, 19, 4008. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.H.; Shen, S.C.; Lee, L.Y.; Lee, S.H.; Chan, M.T.; Lin, C.S. Tape-Arabidopsis Sandwich—A simpler Arabidopsis protoplast isolation method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Lee, H.; Xiong, L.; Zhu, J.K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. https://doi.org/10.3390/ijms19092702
Zhuang L, Cao W, Wang J, Yu J, Yang Z, Huang B. Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(9):2702. https://doi.org/10.3390/ijms19092702
Chicago/Turabian StyleZhuang, Lili, Wei Cao, Jian Wang, Jingjin Yu, Zhimin Yang, and Bingru Huang. 2018. "Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis" International Journal of Molecular Sciences 19, no. 9: 2702. https://doi.org/10.3390/ijms19092702
APA StyleZhuang, L., Cao, W., Wang, J., Yu, J., Yang, Z., & Huang, B. (2018). Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. International Journal of Molecular Sciences, 19(9), 2702. https://doi.org/10.3390/ijms19092702






