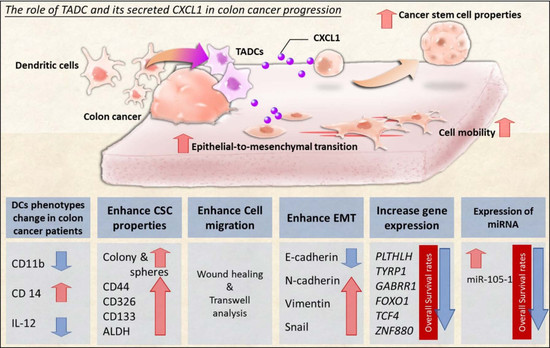
Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1
Abstract
1. Introduction
2. Results
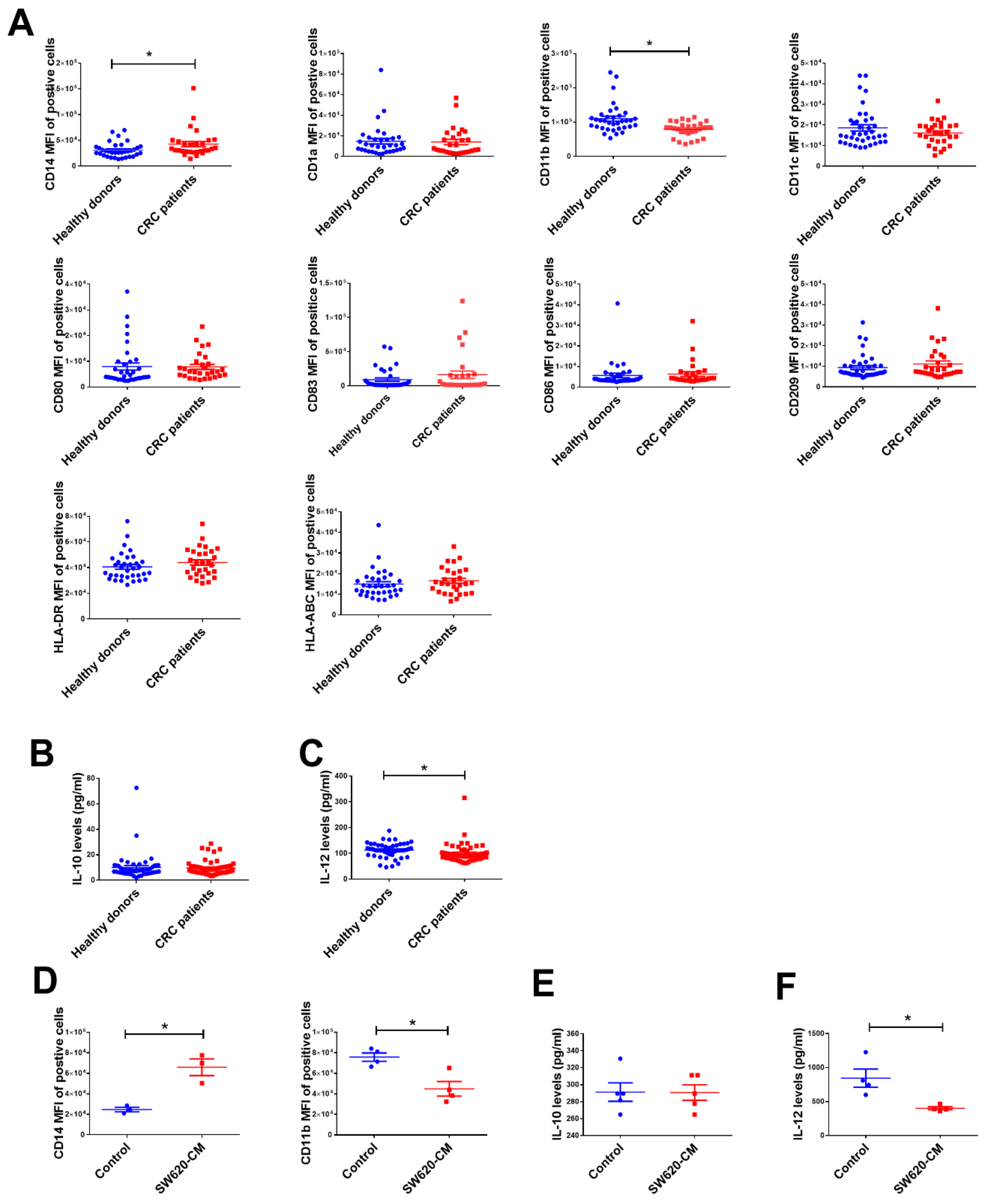
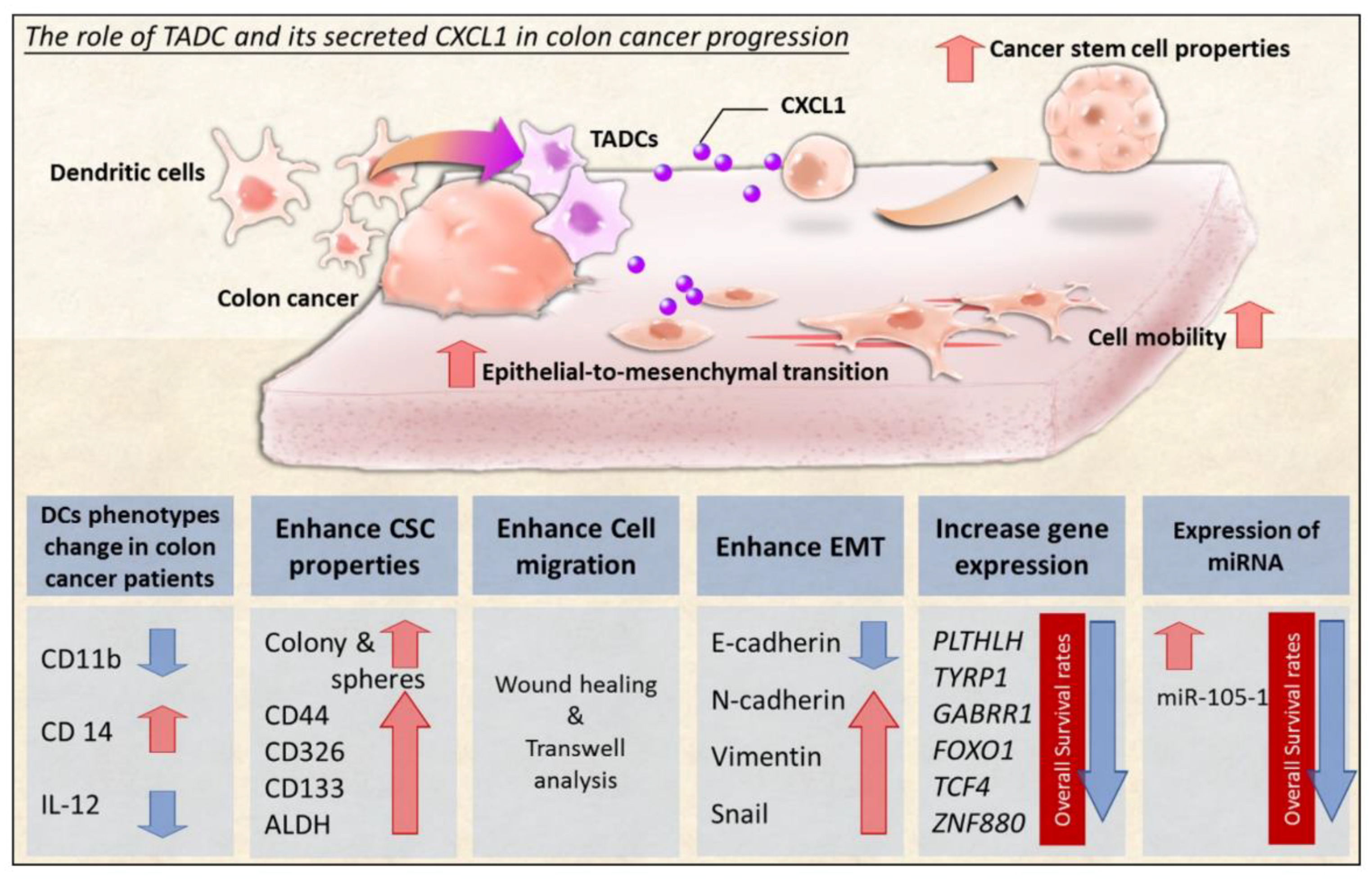
2.1. The Change in Dendritic Cell (DC) Phenotypes in Colorectal Cancer Patients
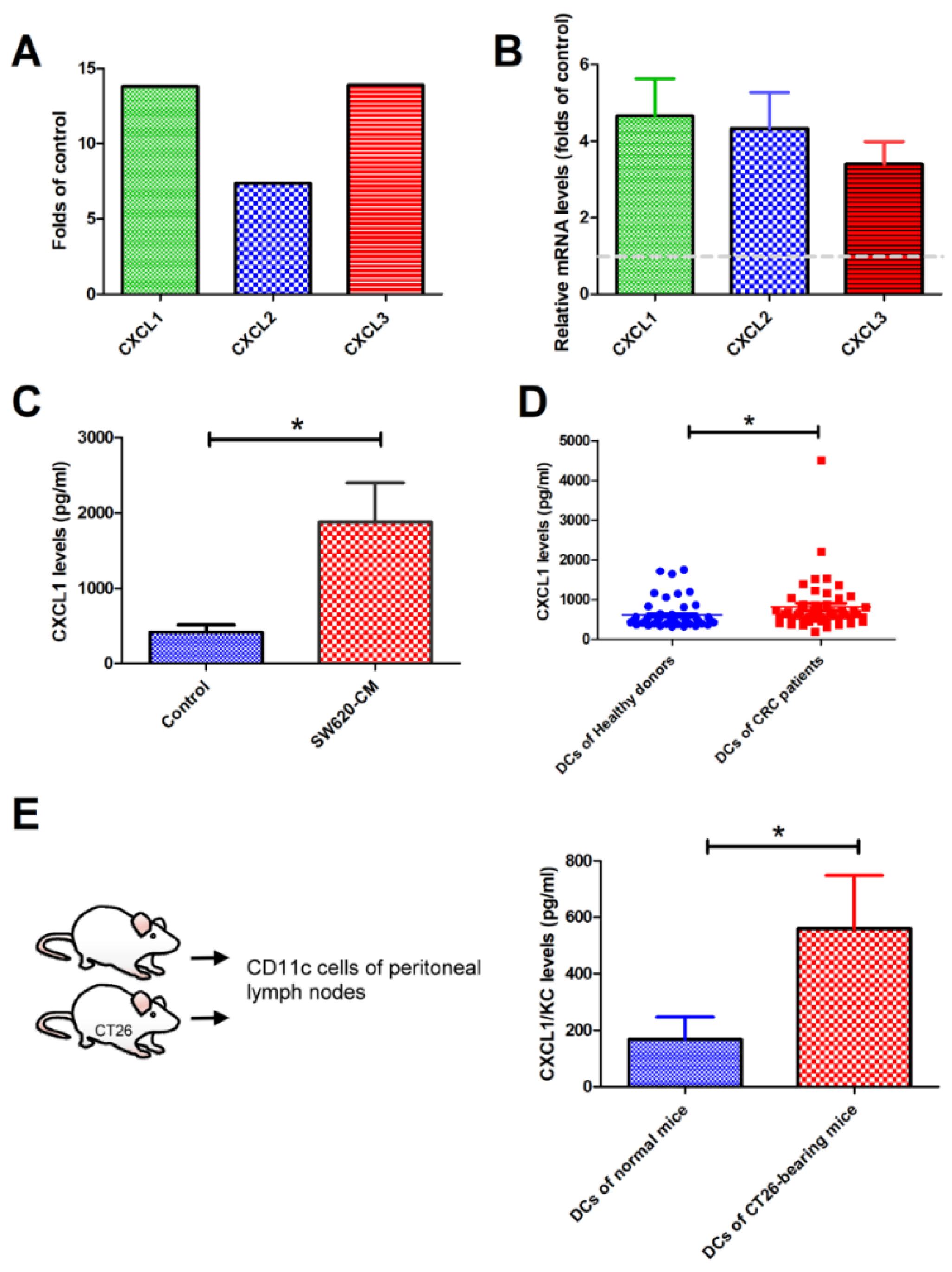
2.2. SW620 Cells Increased the Expression of CXCL1, 2, and 3 in Tumor-Associated DCs In Vitro and In Vivo
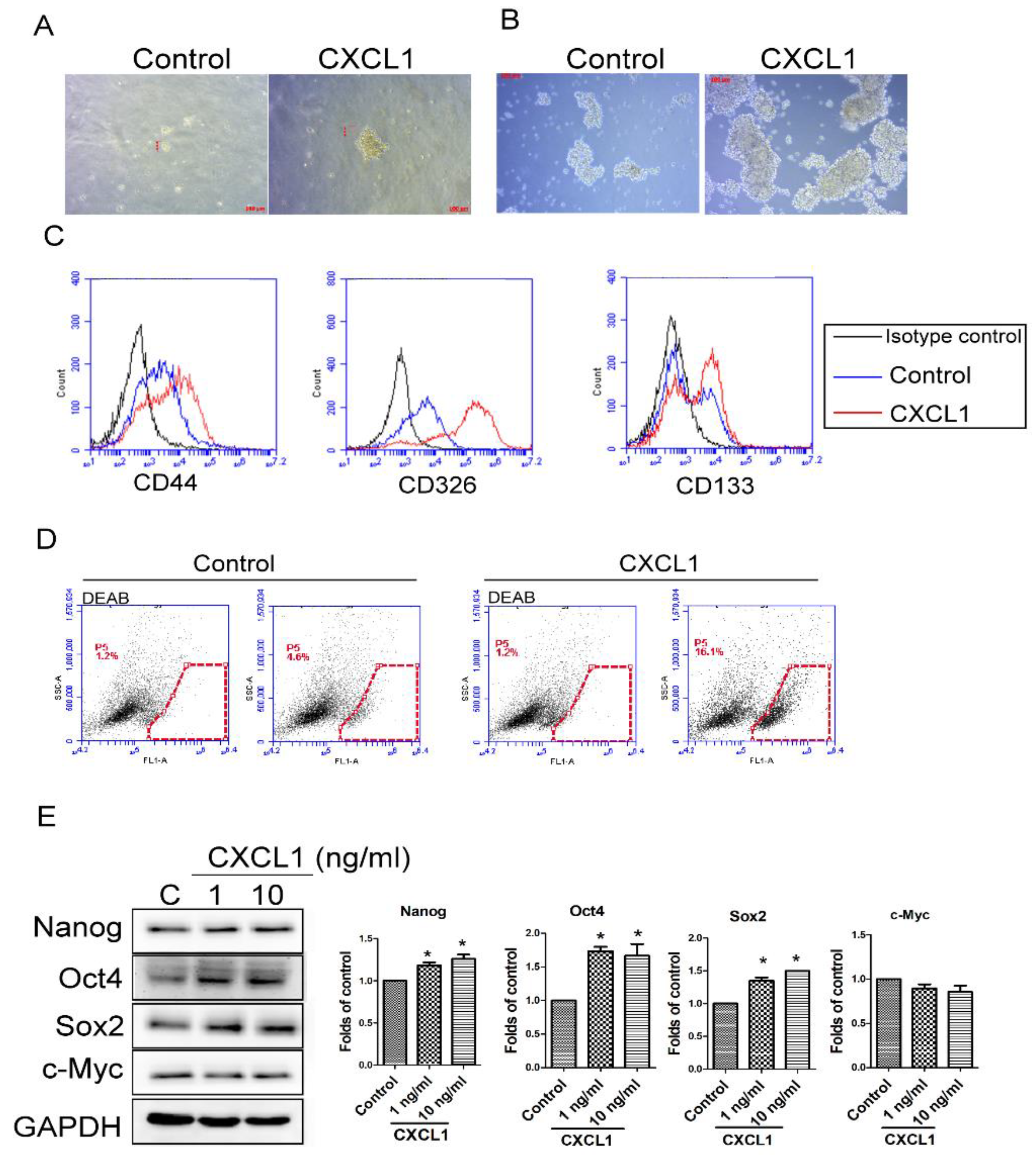
2.3. CXCL1 Increased the Cancer Stem Cell Properties of SW620 Cells
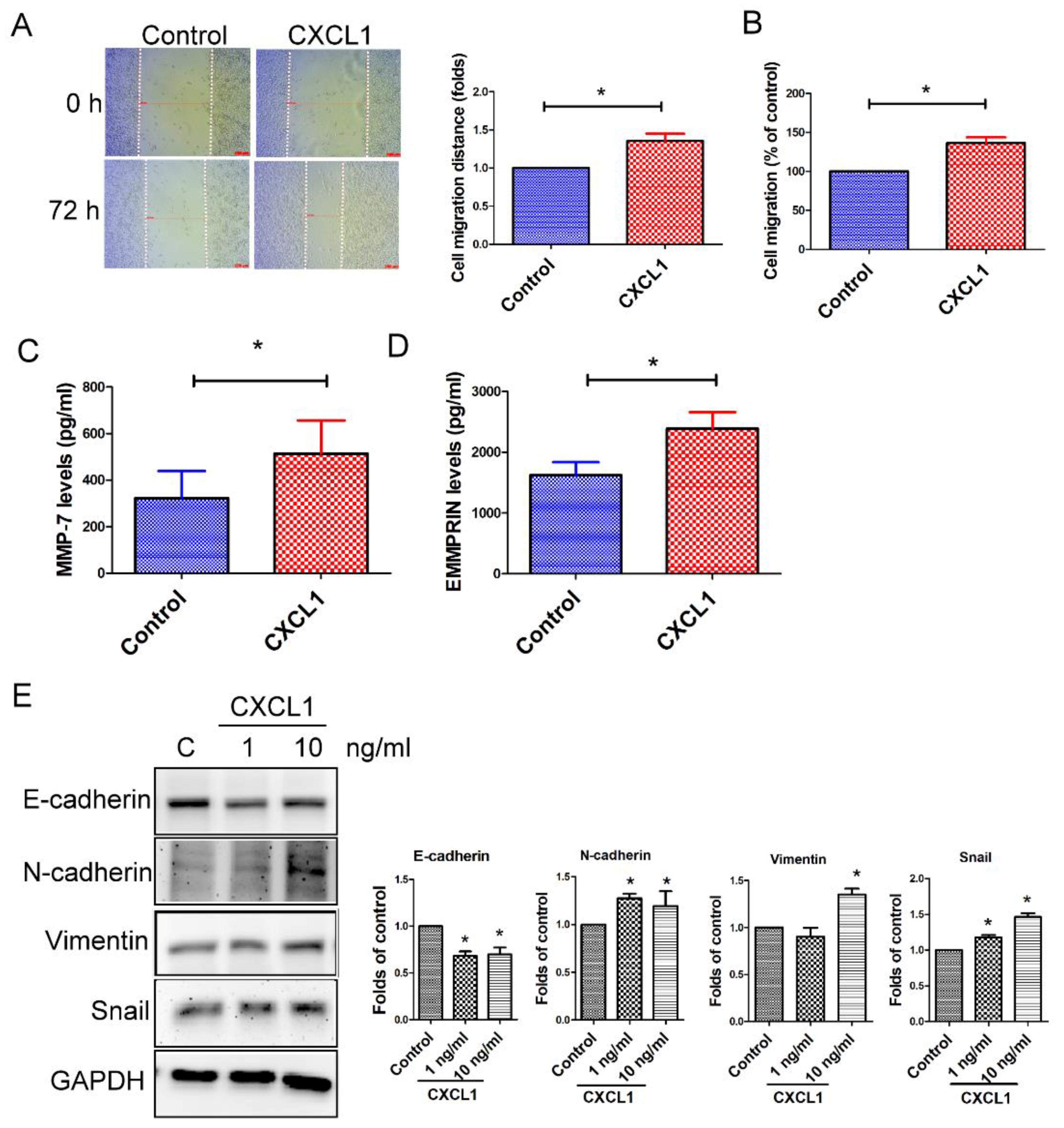
2.4. CXCL1 Enhanced Cell Migration and Epithelial-to-Mesenchymal Transition in SW620 Cells
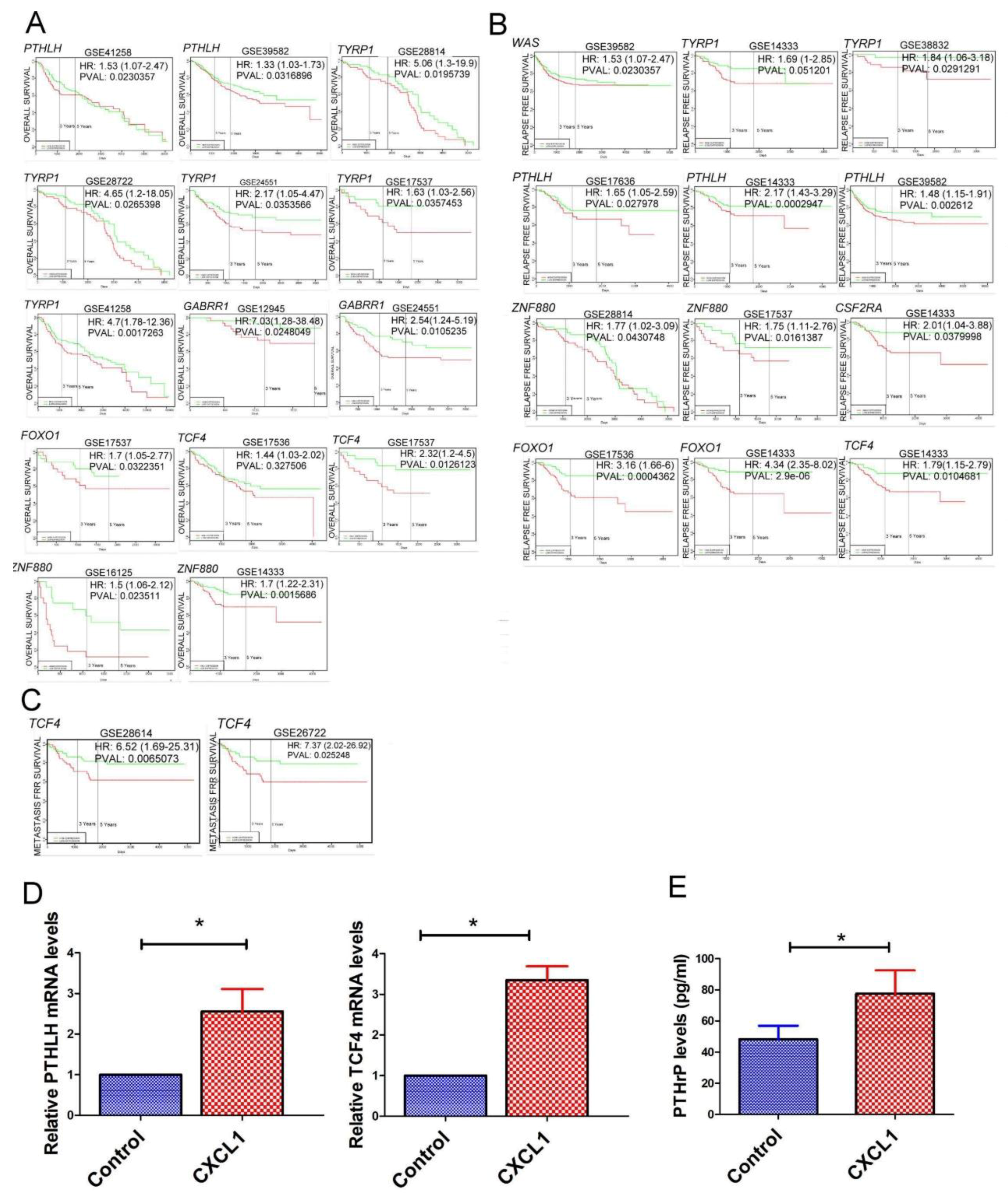
2.5. The Transcriptome of CXCL1-Treated Colon Cancer Cells
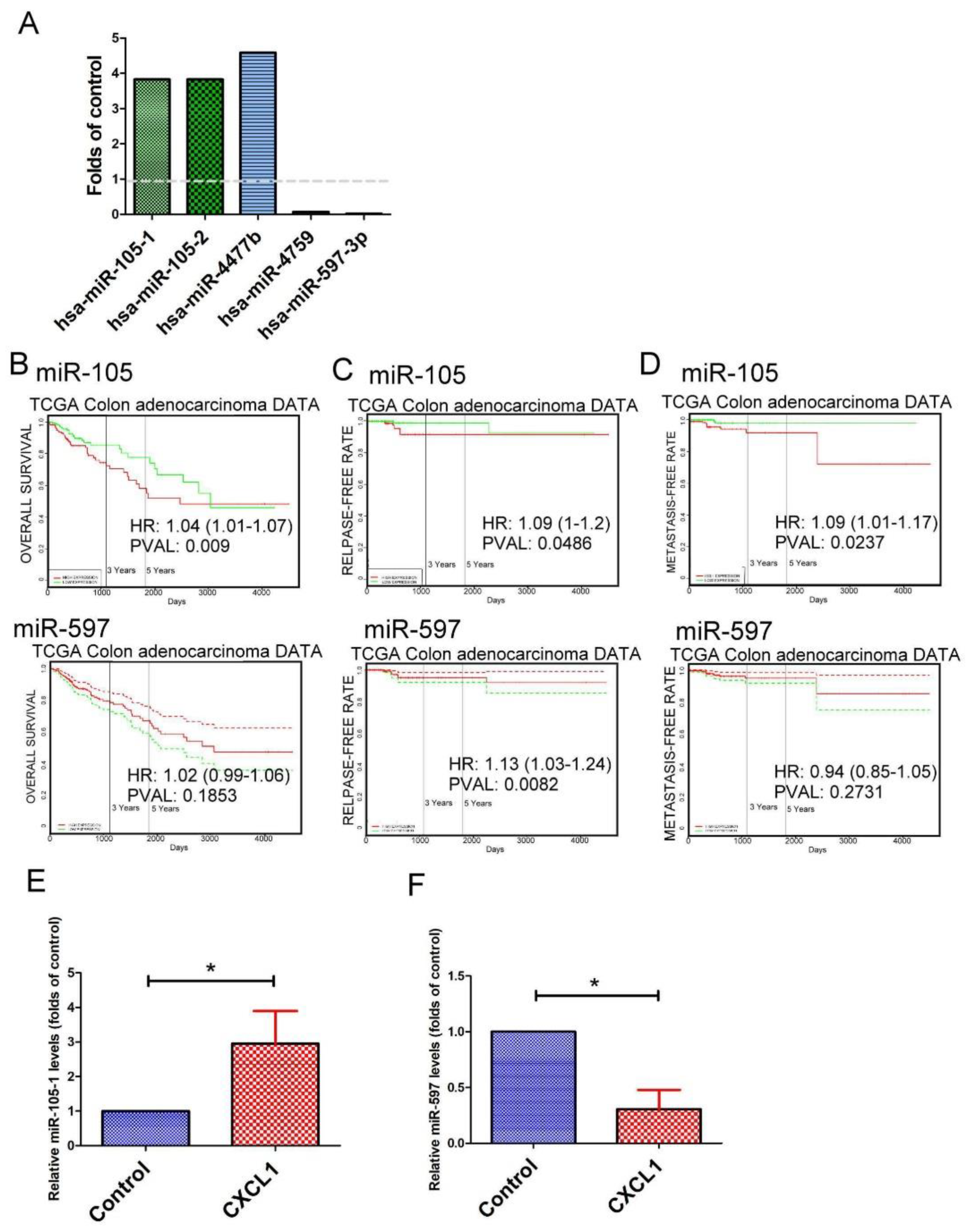
2.6. The Expression Pattern of microRNAs (miRNAs) in CXCL1-Treated Colon Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Flow Cytometry
4.3. Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
4.4. Differentiation of DCs
4.5. Animal Tumor Model
4.6. Measurement of Secreted Factors
4.7. Soft Agar Colony Formation, Tumor Spheroid and Cell Migration Analysis
4.8. Immunoblot Analysis
4.9. Next Generation Sequencing and Bioinformatics Analysis
4.10. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CM | Condition medium |
| CRC | Colorectal cancer |
| CSC | Cancer stem cell |
| CXCL1 | chemokine (C-X-C motif) ligand 1 |
| CXCR2 | chemokine (C-X-C motif) receptor 2 |
| DCs | Dendritic cells |
| EMT | Epithelial-to-mesenchymal transition |
| NGS | next generation sequencing |
| PBMCs | Peripheral blood mononuclear cells |
| TADCs | Tumor-associated dendritic cells |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Zare-Bandamiri, M.; Fararouei, M.; Zohourinia, S.; Daneshi, N.; Dianatinasab, M. Risk Factors Predicting Colorectal Cancer Recurrence Following Initial Treatment: A 5-year Cohort Study. Asian Pac. J. Cancer Prev. 2017, 18, 2465–2470. [Google Scholar] [PubMed]
- Lim, S.Y.; Yuzhalin, A.E.; Gordon-Weeks, A.N.; Muschel, R.J. Tumor-infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene 2016, 35, 5735–5745. [Google Scholar] [CrossRef] [PubMed]
- Crotti, S.; Piccoli, M.; Rizzolio, F.; Giordano, A.; Nitti, D.; Agostini, M. Extracellular Matrix and Colorectal Cancer: How Surrounding Microenvironment Affects Cancer Cell Behavior? J. Cell. Physiol. 2017, 232, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.L.; Reizis, B. Dendritic Cells: Arbiters of Immunity and Immunological Tolerance. Cold Spring Harb. Perspect. Biol. 2012, 4, 7401–7975. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-beta-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018, 48, 147–160.e7. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.; Keskinov, A.A.; Shurin, G.V.; Shurin, M.R. Tumor-derived factors modulating dendritic cell function. Cancer Immun. Immunother. 2016, 65, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Kan, J.Y.; Wu, D.C.; Yu, F.J.; Wu, C.Y.; Ho, Y.W.; Chiu, Y.J.; Jian, S.F.; Hung, J.Y.; Wang, J.Y.; Kuo, P.L. Chemokine (C-C Motif) Ligand 5 is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression Through Non-Coding RNA MALAT-1. J. Cell. Physiol. 2015, 230, 1883–1894. [Google Scholar] [CrossRef] [PubMed]
- Kenkel, J.A.; Tseng, W.W.; Davidson, M.G.; Tolentino, L.L.; Choi, O.; Bhattacharya, N.; Seeley, E.S.; Winer, D.A.; Reticker-Flynn, N.E.; Engleman, E.G. An Immunosuppressive Dendritic Cell Subset Accumulates at Secondary Sites and Promotes Metastasis in Pancreatic Cancer. Cancer Res. 2017, 77, 4158–4170. [Google Scholar] [CrossRef] [PubMed]
- Legitimo, A.; Consolini, R.; Failli, A.; Orsini, G.; Spisni, R. Dendritic cell defects in the colorectal cancer. Hum. Vaccin. Immunother. 2014, 10, 3224–3235. [Google Scholar] [CrossRef] [PubMed]
- Tran Janco, J.M.; Lamichhane, P.; Karyampudi, L.; Knutson, K.L. Tumor-Infiltrating Dendritic Cells in Cancer Pathogenesis. J. Immunol. 2015, 194, 2985–2991. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Liu, W.; Kaur, M.; Luo, Y.; Domenico, J.; Samson, J.M.; Shellman, Y.G.; Norris, D.A.; Dinarello, C.A.; Spritz, R.A.; et al. NLRP1 promotes tumor growth by enhancing inflammasome activation and suppressing apoptosis in metastatic melanoma. Oncogene 2017, 36, 3820–3830. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, X.; Xu, Q.; Ren, J.; Shen, S.; Fan, C.; Hou, Y. miR-19a promotes colitis-associated colorectal cancer by regulating tumor necrosis factor alpha-induced protein 3-NF-kappaB feedback loops. Oncogene 2017, 36, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Bowser, J.L.; Phan, L.H.; Eltzschig, H.K. The Hypoxia-Adenosine Link during Intestinal Inflammation. J. Immunol. 2018, 200, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Le Rolle, A.F.; Chiu, T.K.; Fara, M.; Shia, J.; Zeng, Z.; Weiser, M.R.; Paty, P.B.; Chiu, V.K. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. J. Trans. Med. 2015, 13, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Oladipo, O.; Conlon, S.; O’Grady, A.; Purcell, C.; Wilson, C.; Maxwell, P.J.; Johnston, P.G.; Stevenson, M.; Kay, E.W.; Wilson, R.H.; et al. The expression and prognostic impact of CXC-chemokines in stage II and III colorectal cancer epithelial and stromal tissue. Br. J. Cancer 2011, 104, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Sun, H.; Wei, J.; Cen, B.; DuBois, R.N. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017, 77, 3655–3665. [Google Scholar] [CrossRef] [PubMed]
- Roy, L.; Bobbs, A.; Sattler, R.; Kurkewich, J.L.; Dausinas, P.B.; Nallathamby, P.; Cowden Dahl, K.D. CD133 Promotes Adhesion to the Ovarian Cancer Metastatic Niche. Cancer Growth Metastasis 2018, 11, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Kadomatsu, T.; Tanoue, H.; Yugami, M.; Miyata, K.; Endo, M.; Morinaga, J.; Kobayashi, E.; Miyamoto, T.; Kurahashi, R.; et al. TET2-dependent IL-6 induction mediated by the tumor microenvironment promotes tumor metastasis in osteosarcoma. Oncogene 2018, 37, 2903–2920. [Google Scholar] [CrossRef] [PubMed]
- Piao, C.; Zhang, W.M.; Li, T.T.; Zhang, C.C.; Qiu, S.; Liu, Y.; Liu, S.; Jin, M.; Jia, L.X.; Song, W.C.; et al. Complement 5a stimulates macrophage polarization and contributes to tumor metastases of colon cancer. Exp. Cell Res. 2018, 366, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Danelli, L.; Frossi, B.; Pucillo, C.E. Mast cell/MDSC a liaison immunosuppressive for tumor microenvironment. Oncoimmunology 2015, 4, 100–123. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Valle, L.D.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, H.; Struyf, S.; Laureys, G.; Van Damme, J. The expression and role of CXC chemokines in colorectal cancer. Cytokine Growth Factor Rev. 2011, 22, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Planagumà, A.; Domènech, T.; Pont, M.; Calama, E.; García-González, V.; López, R.; Aulí, M.; López, M.; Fonquerna, S.; Ramos, I.; et al. Combined anti CXC receptors 1 and 2 therapy is a promising anti-inflammatory treatment for respiratory diseases by reducing neutrophil migration and activation. Pulm. Pharmacol. Ther. 2015, 34, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Divella, R.; Daniele, A.; DE Luca, R.; Simone, M.; Naglieri, E.; Savino, E.; Abbate, I.; Gadaleta, C.D.; Ranieri, G. Circulating Levels of VEGF and CXCL1 Are Predictive of Metastatic Organotropismin in Patients with Colorectal Cancer. Anticancer Res. 2017, 37, 4867–4871. [Google Scholar] [PubMed]
- Kasashima, H.; Yashiro, M.; Nakamae, H.; Masuda, G.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; Nakane, T.; Hino, M.; et al. Clinicopathologic significance of the CXCL1-CXCR2 axis in the tumor microenvironment of gastric carcinoma. PLoS ONE 2017, 12, 467–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Jiang, Q.; Deng, J.; Xu, F.; Chen, X.; Cheng, F.; Zhang, Y.; Yao, Y.; Xia, Z.; et al. Human Adipose-Derived Mesenchymal Stem Cell-Secreted CXCL1 and CXCL8 Facilitate Breast Tumor Growth By Promoting Angiogenesis. Stem Cells 2017, 35, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Ruiz de Porras, V.; Bystrup, S.; Martínez-Cardús, A.; Pluvinet, R.; Sumoy, L.; Howells, L.; James, M.I.; Iwuji, C.; Manzano, J.L.; Layos, L.; et al. Curcumin mediates oxaliplatin-acquired resistance reversion in colorectal cancer cell lines through modulation of CXC-Chemokine/NF-kappaB signalling pathway. Sci. Rep. 2016, 6, 246–275. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Hung, J.Y.; Tsai, E.M.; Wu, C.Y.; Ho, Y.W.; Jian, S.F.; Yen, M.C.; Chang, W.A.; Hou, M.F.; Kuo, P.L. Benzyl butyl phthalate increases the chemoresistance to doxorubicin/cyclophosphamide by increasing breast cancer-associated dendritic cell-derived CXCL1/GROalpha and S100A8/A9. Oncol. Rep. 2015, 34, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.L.; Tsai, C.F.; Chen, D.R. Peri-foci adipose-derived stem cells promote chemoresistance in breast cancer. Stem Cell Res. Ther. 2017, 8, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Esbrit, P.; Herrera, S.; Portal-Núñez, S.; Nogués, X.; Díez-Pérez, A. Parathyroid Hormone-Related Protein Analogs as Osteoporosis Therapies. Calcif. Tissue Int. 2016, 98, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Fahy, N.; Gardner, O.F.W.; Alini, M.; Stoddart, M.J. Parathyroid Hormone-Related Protein Gradients Affect the Progression of Mesenchymal Stem Cell Chondrogenesis and Hypertrophy. Tissue Eng. Part A 2018, 24, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Galindo, R.J.; Romao, I.; Valsamis, A.; Weinerman, S.; Harris, Y.T. Hypercalcemia of Malignancy and Colorectal Cancer. World J. Oncol. 2016, 7, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.M.; Lin, Y.F.; Su, C.Y.; Peng, H.Y.; Chang, Y.C.; Hsiao, J.R.; Chen, C.L.; Chang, J.Y.; Shieh, Y.S.; Hsiao, M.; et al. Parathyroid Hormone-Like Hormone is a Poor Prognosis Marker of Head and Neck Cancer and Promotes Cell Growth via RUNX2 Regulation. Sci. Rep. 2017, 7, 411–431. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Takyar, F.M.; Swan, K.; Jeong, J.; VanHouten, J.; Sullivan, C.; Dann, P.; Yu, H.; Fiaschi-Taesch, N.; Chang, W.; et al. Calcium-Sensing Receptor Promotes Breast Cancer by Stimulating Intracrine Actions of Parathyroid Hormone-Related Protein. Cancer Res. 2016, 76, 5348–5360. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.J.; Calvo, N.; de Boland, A.R.; Gentili, C. Molecular mechanisms associated with PTHrP-induced proliferation of colon cancer cells. J. Cell. Biochem. 2014, 115, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Park, S.I.; McCauley, L.K. Nuclear localization of parathyroid hormone-related peptide confers resistance to anoikis in prostate cancer cells. Endocr. Relat. Cancer 2012, 19, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Urosevic, J.; Garcia-Albéniz, X.; Planet, E.; Real, S.; Céspedes, M.V.; Guiu, M.; Fernandez, E.; Bellmunt, A.; Gawrzak, S.; Pavlovic, M.; et al. Colon cancer cells colonize the lung from established liver metastases through p38 MAPK signalling and PTHLH. Nat. Cell Biol. 2014, 16, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Huo, Z.; Song, X.; Chen, X.; Tian, X.; Wang, X. mir-106a regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol. Lett. 2018, 15, 3197–3201. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Liang, J.L.; Kuo, Y.L.; Lee, H.H.; Calkins, M.J.; Chang, H.T.; Lin, F.C.; Chen, Y.C.; Hsu, T.I.; Hsiao, M.; et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017, 19, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zhou, R.; Liu, C.; Wang, Y.; Zhan, W.; Shao, Z.; Liu, J.; Zhang, F.; Xu, L.; Zhou, X.; et al. MicroRNA-105 is involved in TNF-alpha-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017, 8, 3213–3315. [Google Scholar] [CrossRef] [PubMed]
- Goswami, C.P.; Nakshatri, H. PROGgeneV2: Enhancements on the existing database. BMC Cancer 2014, 14, 970–986. [Google Scholar] [CrossRef] [PubMed]
- Goswami, C.P.; Nakshatri, H. PROGmiR: A tool for identifying prognostic miRNA biomarkers in multiple cancers using publicly available data. J. Clin. Bioinforma 2012, 2, 23–33. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Ratio (CXCL/Control) | Log2 Ratio | p Value |
|---|---|---|---|
| WAS | 2.75575 | 1.46245 | 0.28025 |
| PTHLH | 3.09636 | 1.63057 | 0.00285 |
| TYRP1 | 3.48983 | 1.80316 | 0.0775 |
| TRIM16L | 2.21167 | 1.14514 | 0.09035 |
| TFEB | 2.5323 | 1.34045 | 0.07635 |
| CSTA | 3.14503 | 1.65307 | 0.1503 |
| AOAH | 2.95457 | 1.56295 | 0.0516 |
| GABRR1 | 2.62297 | 1.3912 | 0.03455 |
| FOXO1 | 2.7327 | 1.45032 | 0.10195 |
| SPAG17 | 2.55284 | 1.35211 | 0.31355 |
| SCNN1D | 2.31426 | 1.21055 | 0.29645 |
| DYNLRB2 | 2.77593 | 1.47297 | 0.1716 |
| RP11-231C14.4 | 3.51186 | 1.81223 | 0.00315 |
| WFDC9 | 2.10763 | 1.07562 | 0.2166 |
| TCF4 | 2.29962 | 1.20139 | 0.05605 |
| CSF2RA | 2.05608 | 1.03989 | 0.1571 |
| ZNF880 | 2.10466 | 1.07359 | 0.18095 |
| H2BFS | 3.42202 | 1.77485 | 0.13455 |
| LY6G6D | 2.55342 | 1.35243 | 0.5991 |
| CTC-435M10.3 | 2.42044 | 1.27527 | 0.36275 |
| BCL2L2-PABPN1 | 4.05028 | 2.01802 | 0.3231 |
| RP11-438J1.1 | 2.75195 | 1.46046 | 0.21695 |
| CH17-270A2.2 | 3.22723 | 1.6903 | 0.6751 |
| ABCB4 | 0.34488 | −1.5358 | 0.05905 |
| NME1-NME2 | 0.28497 | −1.8111 | 0.5181 |
| MAN2B2 | 0.484 | −1.0469 | 0.18655 |
| POU2F2 | 0.19867 | −2.3315 | 0.1448 |
| DGKG | 0.34246 | −1.546 | 0.12485 |
| HPX | 0.07198 | −3.7963 | 0.1496 |
| GBP3 | 0.21884 | −2.1921 | 0.00015 |
| GRPR | 0.45999 | −1.1203 | 0.01765 |
| KHDRBS3 | 0.25515 | −1.9706 | 0.0735 |
| ZCCHC2 | 0.47568 | −1.0719 | 0.0612 |
| CYP17A1 | 0.41877 | −1.2558 | 0.1948 |
| ALS2CR12 | 0.3871 | −1.3692 | 0.05095 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, Y.-L.; Chen, Y.-J.; Chang, W.-A.; Jian, S.-F.; Fan, H.-L.; Wang, J.-Y.; Kuo, P.-L. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. Int. J. Mol. Sci. 2018, 19, 2427. https://doi.org/10.3390/ijms19082427
Hsu Y-L, Chen Y-J, Chang W-A, Jian S-F, Fan H-L, Wang J-Y, Kuo P-L. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. International Journal of Molecular Sciences. 2018; 19(8):2427. https://doi.org/10.3390/ijms19082427
Chicago/Turabian StyleHsu, Ya-Ling, Yi-Jen Chen, Wei-An Chang, Shu-Fang Jian, Hsiao-Li Fan, Jaw-Yuan Wang, and Po-Lin Kuo. 2018. "Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1" International Journal of Molecular Sciences 19, no. 8: 2427. https://doi.org/10.3390/ijms19082427
APA StyleHsu, Y.-L., Chen, Y.-J., Chang, W.-A., Jian, S.-F., Fan, H.-L., Wang, J.-Y., & Kuo, P.-L. (2018). Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. International Journal of Molecular Sciences, 19(8), 2427. https://doi.org/10.3390/ijms19082427