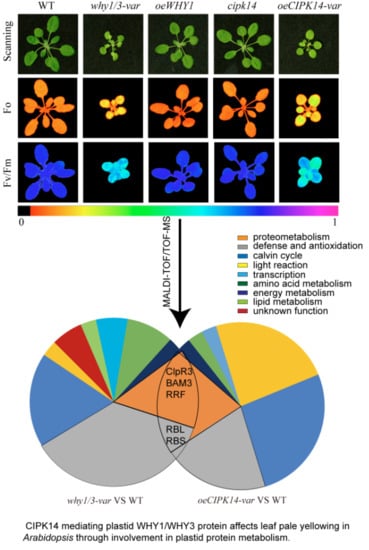
Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Proteomic Analysis of why1/3-var and oeCIPK14-var Mutants
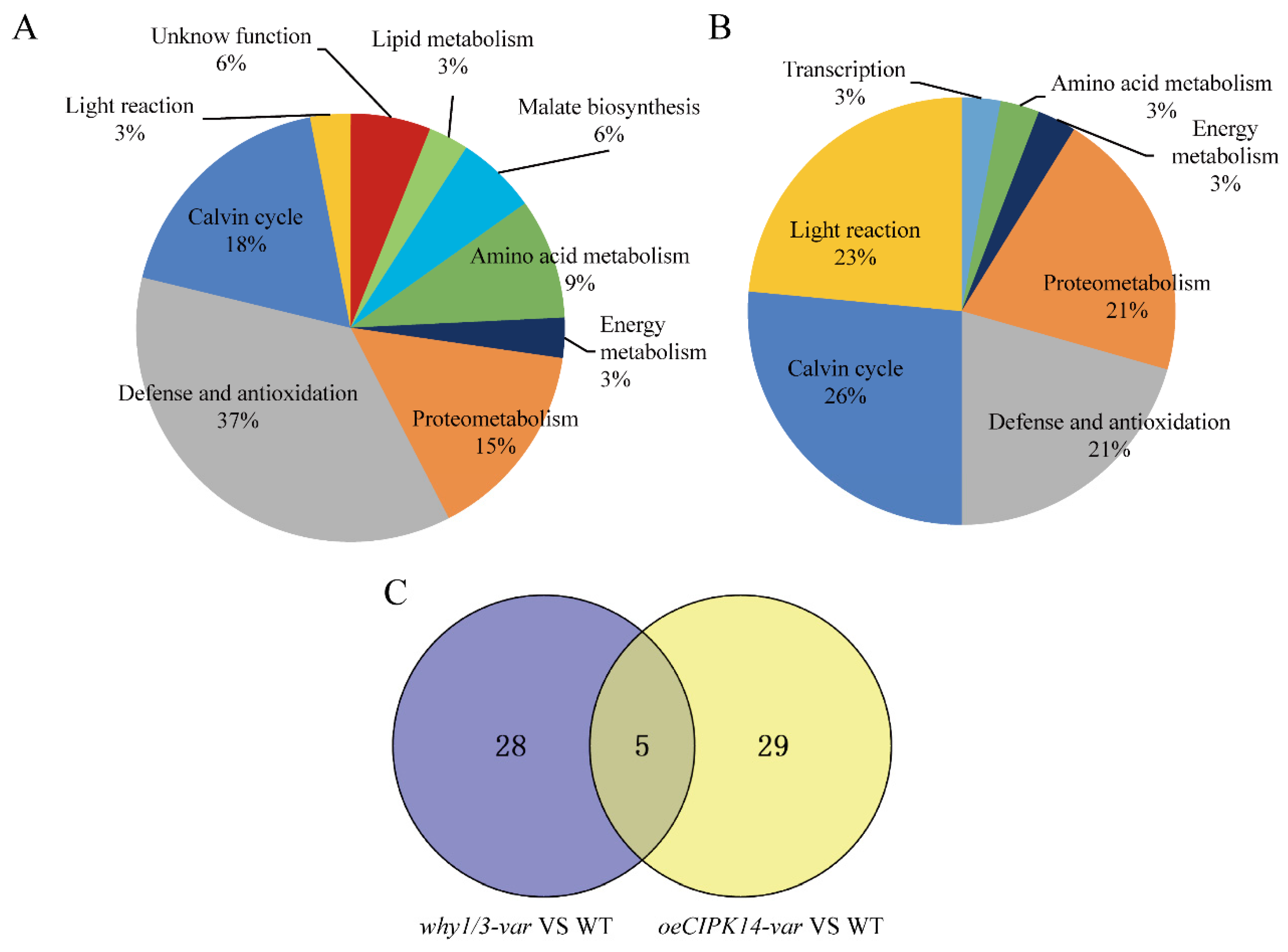
2.2. Functional Classification of Differentially Expressed Proteins
2.3. Hierarchical Clustering Analysis of Differentially Expressed Proteins
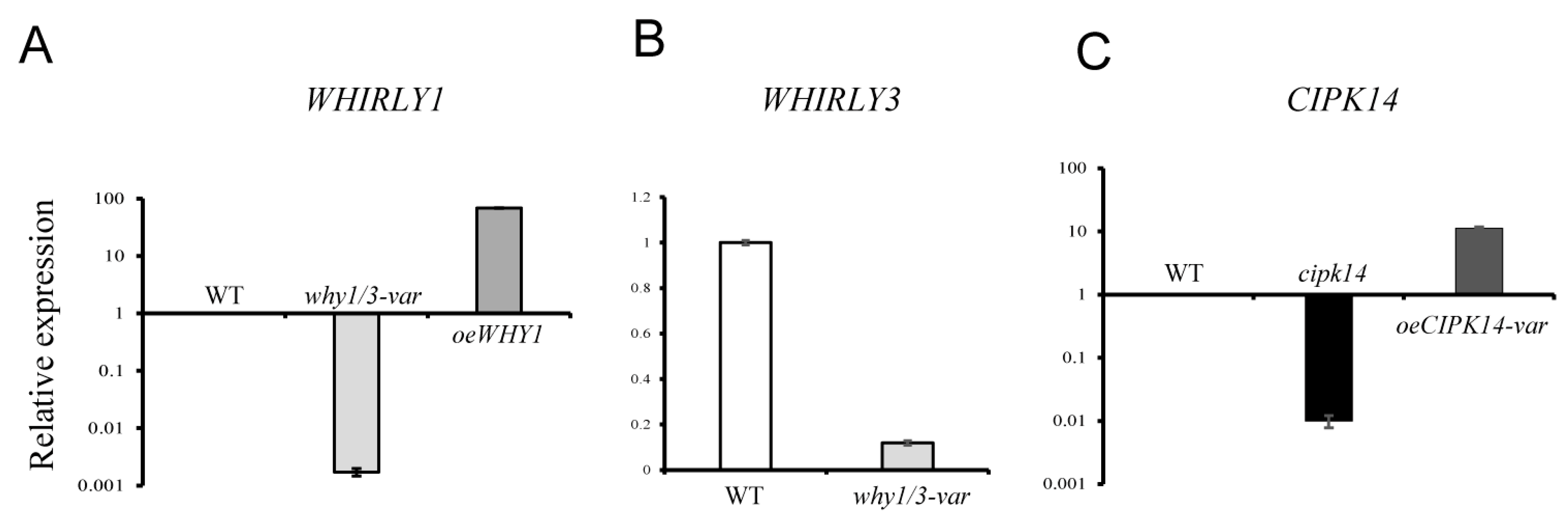
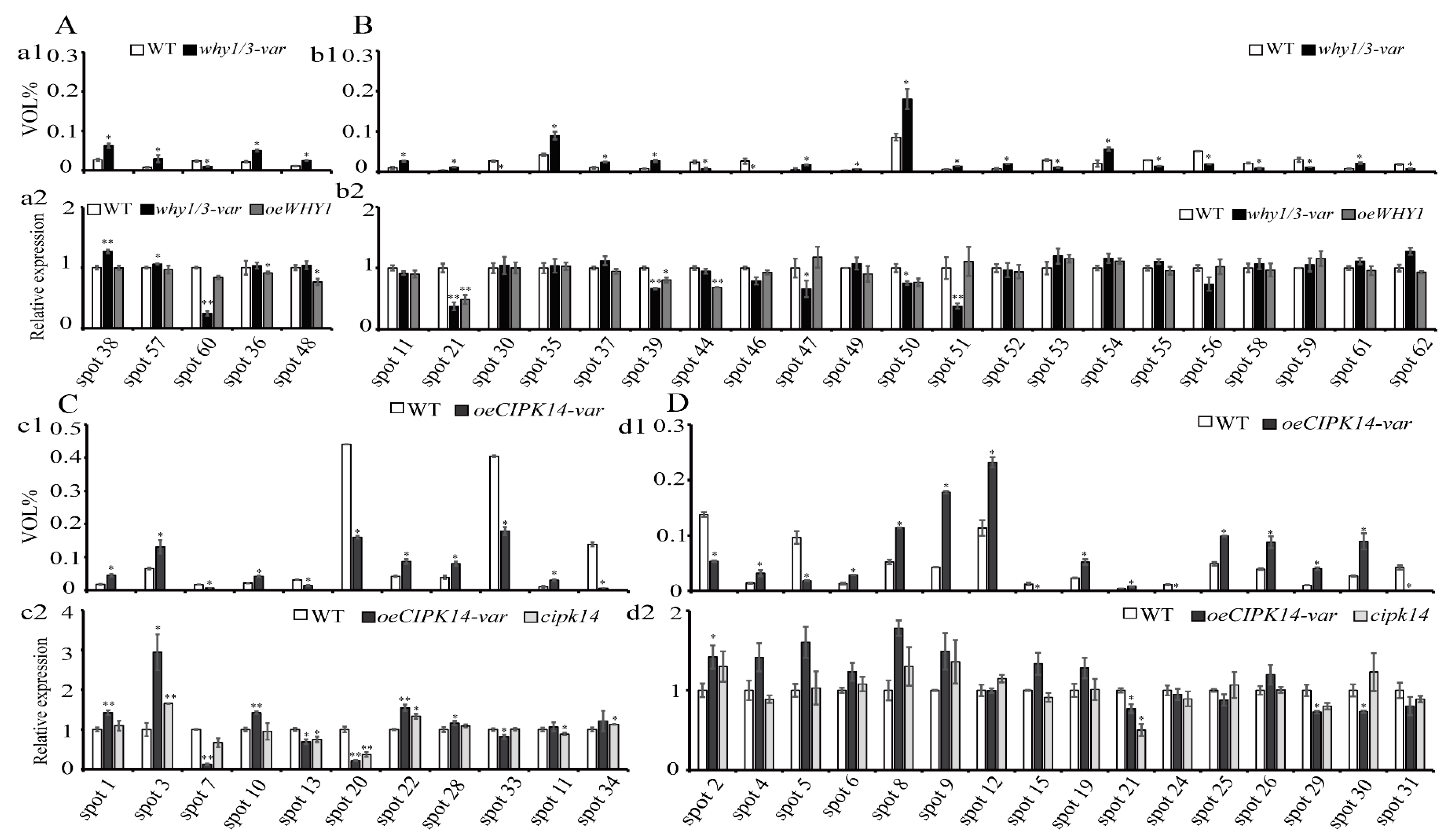
2.4. Transcriptional Level Analysis of the Encoding Genes of Differentially Expressed Proteins
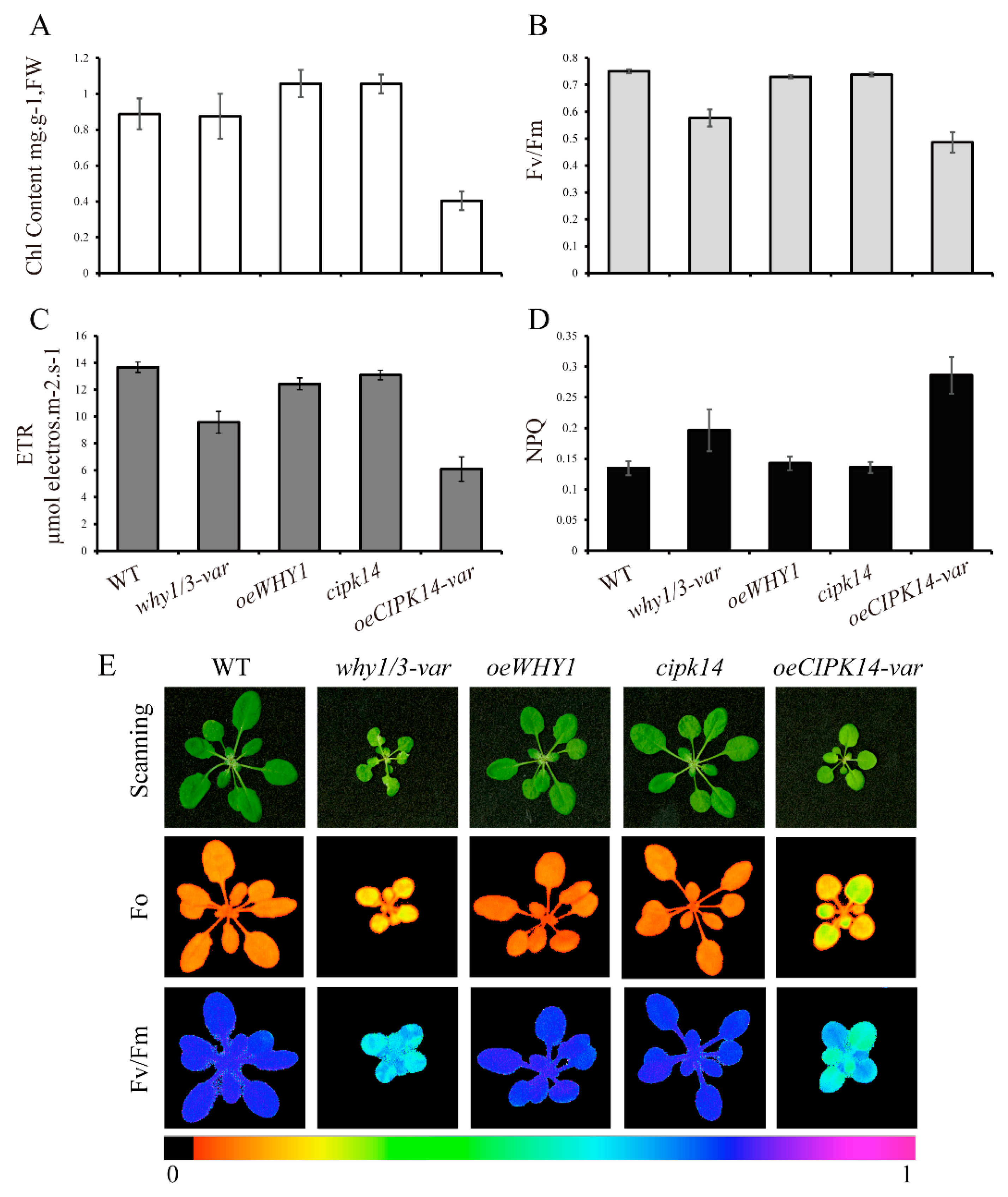
2.5. Effects of why1/3-var and oeCIPK14-var on Photosynthetic Performance
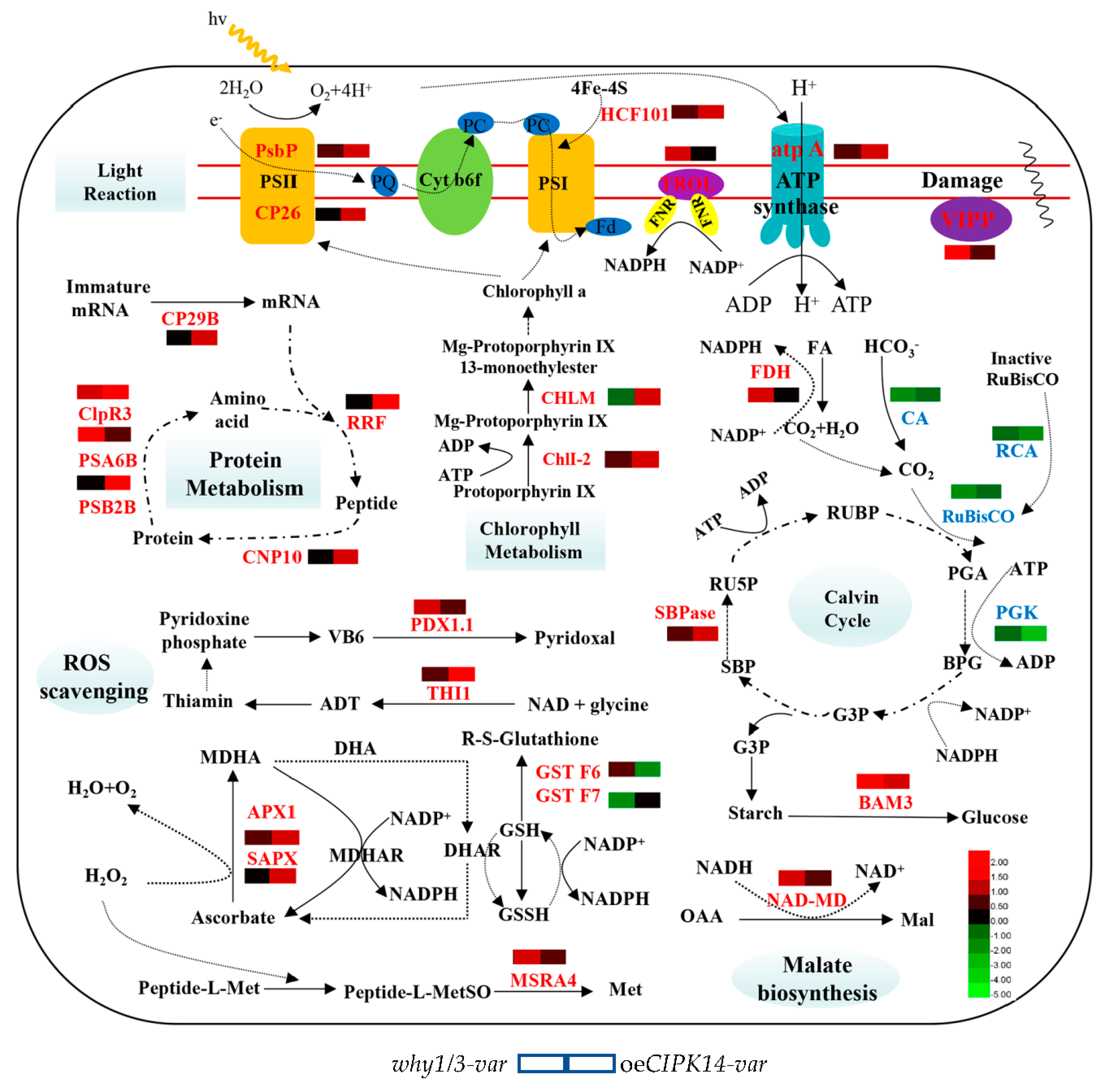
3. Discussion
3.1. Proteins Involved in Light-Dependent Reaction of Photosynthesis
3.2. Proteins Involved in the Calvin Cycle
3.3. Proteins Associated with Defense and Antioxidation
3.4. Proteins Related to Protein Metabolism
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Chlorophyll Content Measurement and Photosynthetic Parameters Analysis
4.3. Protein Extraction and 2-DE Analysis
4.4. Data Analysis In-Gel Protein Digestion and Protein Identification
4.5. RNA Extraction, Reverse Transcription, and qRT-PCR Analysis
4.6. Immunological Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ADP | adenosine diphosphate |
| ADT | adenosine diphosphate 5-(2-hydroxyethyl)-4-methylthiazole-2-carboxylic acid |
| APX1 | l-ascorbate peroxidase 1 |
| ATP | adenosine triphosphate |
| ATPA | ATP synthase alpha subunit |
| BAM3 | beta-amylase 3 |
| BPG | 1,3-diphosphoglycerate |
| CA | carbonic anhydrase |
| ChlI-2 | magnesium-chelatase subunit ChlI-2 |
| CHLM | magnesium protoporphyrin IX methyltransferase |
| CLPR3 | ATP-dependent Clp protease proteolytic subunit-related protein 3 |
| CNP10 | 10 kDa chaperonin |
| CP26 | chlorophyll a-b binding protein CP26 |
| CP29B | RNA-binding protein CP29B |
| Cyt b6f | cytochrome b6f |
| DHAR | dehydroascorbate reductase |
| FA | formate |
| Fd | ferredoxin |
| FDH | formate dehydrogenase |
| FNR | ferredoxin-NADP (H) oxidoreductase |
| G3P | glyceraldehydes-3-phosphate |
| GSH | reduced glutathione |
| GSSH | oxidized glutathione |
| GST | glutathione S-transferase |
| HCF101 | Fe-S cluster assembly factor HCF101 |
| Mal | malate |
| MD | malate dehydrogenase |
| MDHA | monodehydroascorbate |
| MDHAR | MDHA reductase |
| Met | methionine |
| MetSO | methionine sulfoxide |
| MSRA4 | peptide methionine sulfoxide reductase A4 |
| NADP+/NADPH | nicotinamide adenine dinucleotide phosphate |
| NAD+/NADH | nicotinamide adenine dinucleotide |
| OAA | oxaloacetic acid |
| PC | plastocyanin |
| PDX1.1 | pyridoxal 5′-phosphate synthase subunit PDX1.1 |
| PGA | 3-phosphoglycerate |
| PGK | phosphoglycerate kinase |
| PQ | plastoquinone |
| PSI | photosystem I |
| PSII | photosystem II |
| PSA6B | proteasome subunit alpha type-6-B |
| PSB2B | proteasome subunit beta type-2-B |
| PsbP | PsbP domain-containing protein 4 |
| RCA | ribulose-1,5-bisphosphate carboxylase/oxygenase activase |
| RRF | ribosome-recycling factor |
| Ru5P | ribulose-5-phosphate |
| RuBisCO | ribulose-1,5-bisphosphate carboxylase/oxygenase |
| THI | thiamine thiazole synthase |
| TROL | thylakoid rhodanese-like protein |
| SBP | sedoheptulose-1,7-bisphosphate |
| SBPase | sedoheptulose-1,7-bisphosphatase |
| SAPX | stromal ascorbate peroxidase |
| VB6 | vitamin B6 |
| VIPP1 | vesicle-inducing protein in plastids |
References
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Ann. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Pottier, M.; Masclaux-Daubresse, C.; Yoshimoto, K.; Thomine, S. Autophagy as a possible mechanism for micronutrient remobilization from leaves to seeds. Front. Plant Sci. 2014, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Riano-Pachon, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10 (Suppl. 1), 63–75. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.P.; Mueller-Roeber, B. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Hinderhofer, K.; Zentgraf, U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 2001, 213, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Jiang, J.; Ren, Y.; Zhao, Z. The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol. 2013, 163, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.; Kilbienski, I.; Mulisch, M.; Rodiger, A.; Schafer, A.; Krupinska, K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005, 579, 3707–3712. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, E.; Miao, Y.; Mulisch, M.; Krupinska, K. Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 2008, 147, 1800–1804. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Marechal, A.; Brisson, N. Whirly transcription factors: Defense gene regulation and beyond. Trends Plant Sci. 2005, 10, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Isemer, R.; Mulisch, M.; Schafer, A.; Kirchner, S.; Koop, H.U.; Krupinska, K. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 2012, 586, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Despres, C.; Joyeux, A.; Subramaniam, R.; Brisson, N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 2000, 12, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Subramaniam, R.; Despres, C.; Mess, J.N.; Levesque, C.; Fobert, P.R.; Dangl, J.L.; Brisson, N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 2004, 6, 229–240. [Google Scholar] [CrossRef]
- Yoo, H.H.; Kwon, C.; Lee, M.M.; Chung, I.K. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007, 49, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.Y.; Lai, C.X.; Qu, Z.; Yang, X.Y.; Qin, X.H.; Liu, G.Q. Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol. Biol. 2009, 71, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Krupinska, K.; Dahnhardt, D.; Fischer-Kilbienski, I.; Kucharewicz, W.; Scharrenberg, C.; Trosch, M.; Buck, F. Identification of WHIRLY1 as a Factor Binding to the Promoter of the Stress- and Senescence-Associated Gene HvS40. J. Plant Growth Regul. 2014, 33, 91–105. [Google Scholar] [CrossRef]
- Prikryl, J.; Watkins, K.P.; Friso, G.; van Wijk, K.J.; Barkan, A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008, 36, 5152–5165. [Google Scholar] [CrossRef] [PubMed]
- Melonek, J.; Mulisch, M.; Schmitz-Linneweber, C.; Grabowski, E.; Hensel, G.; Krupinska, K. Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 2010, 232, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmuller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Stroher, E.; Dietz, K.J. The dynamic thiol-disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiol. Plant. 2008, 133, 566–583. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Karpinska, B.; Krupinska, K. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: A hypothesis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014, 369, 20130226. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Lin, W.; Deng, B.; Ren, Y.; Miao, Y. Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 2352. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Parent, J.S.; Veronneau-Lafortune, F.; Joyeux, A.; Lang, B.F.; Brisson, N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Lepage, E.; Zampini, E.; Brisson, N. Plastid genome instability leads to reactive oxygen species production and plastid-to-nucleus retrograde signaling in Arabidopsis. Plant Physiol. 2013, 163, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Comadira, G.; Rasool, B.; Kaprinska, B.; Garcia, B.M.; Morris, J.; Verrall, S.R.; Bayer, M.; Hedley, P.E.; Hancock, R.D.; Foyer, C.H. WHIRLY1 Functions in the Control of Responses to Nitrogen Deficiency But Not Aphid Infestation in Barley. Plant Physiol. 2015, 168, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, Y.; Jiang, Y.; Wu, B.; Miao, Y. Phosphorylation of WHIRLY1 by CIPK14 Shifts Its Localization and Dual Functions in Arabidopsis. Mol. Plant 2017, 10, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Balsera, M.; Arellano, J.B.; Gutierrez, J.R.; Heredia, P.; Revuelta, J.L.; De Las Rivas, J. Structural analysis of the PsbQ protein of photosystem II by Fourier transform infrared and circular dichroic spectroscopy and by bioinformatic methods. Biochemistry 2003, 42, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999, 4, 236–240. [Google Scholar] [CrossRef]
- De Bianchi, S.; Dall’Osto, L.; Tognon, G.; Morosinotto, T.; Bassi, R. Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 2008, 20, 1012–1028. [Google Scholar] [CrossRef] [PubMed]
- Schwenkert, S.; Netz, D.J.; Frazzon, J.; Pierik, A.J.; Bill, E.; Gross, J.; Lill, R.; Meurer, J. Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 2009, 425, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Juric, S.; Hazler-Pilepic, K.; Tomasic, A.; Lepedus, H.; Jelicic, B.; Puthiyaveetil, S.; Bionda, T.; Vojta, L.; Allen, J.F.; Schleiff, E.; et al. Tethering of ferredoxin:NADP+ oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J. 2009, 60, 783–794. [Google Scholar] [PubMed]
- Kobayashi, K.; Mochizuki, N.; Yoshimura, N.; Motohashi, K.; Hisabori, T.; Masuda, T. Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 2008, 7, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Albrieux, C.; Joyard, J.; Lagrange, T.; Block, M.A. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. J. Biol. Chem. 2007, 282, 2297–2304. [Google Scholar] [CrossRef] [PubMed]
- Kieselbach, T.; Hagman, Å.; Andersson, B.; Schroder, W.P. The thylakoid lumen of chloroplasts. Isolation and characterization. J. Biol. Chem. 1998, 273, 6710–6716. [Google Scholar] [CrossRef] [PubMed]
- Granlund, I.; Storm, P.; Schubert, M.; Garcia-Cerdan, J.G.; Funk, C.; Schroder, W.P. The TL29 protein is lumen located, associated with PSII and not an ascorbate peroxidase. Plant Cell Physiol. 2009, 50, 1898–1910. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008, 46, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Portis, A.R., Jr. Rubisco activase—Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Tellez, S.; Anoman, A.D.; Flores-Tornero, M.; Toujani, W.; Alseek, S.; Fernie, A.R.; Nebauer, S.G.; Munoz-Bertomeu, J.; Segura, J.; Ros, R. Phosphoglycerate Kinases Are Co-Regulated to Adjust Metabolism and to Optimize Growth. Plant Physiol. 2018, 176, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Boisson-Dernier, A.; Israelsson-Nordstrom, M.; Bohmer, M.; Xue, S.; Ries, A.; Godoski, J.; Kuhn, J.M.; Schroeder, J.I. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010, 12, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozdemir, M.; Miller, G.; Song, L.; Kim, J.; Sodek, A.; Koussevitzky, S.; Misra, A.N.; Mittler, R.; Shintani, D. Thiamin confers enhanced tolerance to oxidative stress in Arabidopsis. Plant Physiol. 2009, 151, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Belanger, F.C.; Leustek, T.; Chu, B.; Kriz, A.L. Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol. Biol. 1995, 29, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Tambasco-Studart, M.; Titiz, O.; Raschle, T.; Forster, G.; Amrhein, N.; Fitzpatrick, T.B. Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. USA 2005, 102, 13687–13692. [Google Scholar] [CrossRef] [PubMed]
- Bilski, P.; Li, M.Y.; Ehrenshaft, M.; Daub, M.E.; Chignell, C.F. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 2000, 71, 129–134. [Google Scholar] [CrossRef]
- Maruta, T.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol. 2010, 51, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Rey, P.; Cuine, S.; Eymery, F.; Garin, J.; Court, M.; Jacquot, J.P.; Rouhier, N.; Broin, M. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J. 2005, 41, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Brot, N.; Weissbach, L.; Werth, J.; Weissbach, H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. USA 1981, 78, 2155–2158. [Google Scholar] [CrossRef] [PubMed]
- Romero, H.M.; Berlett, B.S.; Jensen, P.J.; Pell, E.J.; Tien, M. Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol. 2004, 136, 3784–3794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kato, Y.; Otters, S.; Vothknecht, U.C.; Sakamoto, W. Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 2012, 24, 3695–3707. [Google Scholar] [CrossRef] [PubMed]
- Adam, Z.; Rudella, A.; van Wijk, K.J. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 2006, 9, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Rudella, A.; Ramirez Rodriguez, V.; Zybailov, B.; Olinares, P.D.; van Wijk, K.J. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 2009, 21, 1669–1692. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef]
- Wang, L.; Ouyang, M.; Li, Q.; Zou, M.; Guo, J.; Ma, J.; Lu, C.; Zhang, L. The Arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol. Biol. 2010, 74, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Koumoto, Y.; Shimada, T.; Kondo, M.; Hara-Nishimura, I.; Nishimura, M. Chloroplasts have a novel Cpn10 in addition to Cpn20 as co-chaperonins in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 29688–29694. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, B.C.; Zhu, Y.X. Identification of proteins expressed at extremely low level in Arabidopsis leaves. Biochem. Biophys. Res. Commun. 2007, 358, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Fulton, D.C.; Stettler, M.; Mettler, T.; Vaughan, C.K.; Li, J.; Francisco, P.; Gil, M.; Reinhold, H.; Eicke, S.; Messerli, G.; et al. Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant cell 2008, 20, 1040–1058. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yang, J.; Lu, B.; Jiang, Y.; Chen, H.; Hong, Y.; Wu, B.; Miao, Y. Structure of Pigment Metabolic Pathways and Their Contributions to White Tepal Color Formation of Chinese Narcissus tazetta var. chinensis cv Jinzhanyintai. Int. J. Mol. Sci. 2017, 18, 1923. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Shu, Z.; Peng, C.L.; Lin, Z.F.; Yang, C.W.; Gu, Q. Enhanced sensitivity of Arabidopsis anthocyanin mutants to photooxidation: A study with fluorescence imaging. Funct. Plant Biol. 2008, 35, 714–724. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Deckers, P.; Swennen, R.; Panis, B. Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 2005, 5, 2497–2507. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, L.; Pan, D.; Li, J.; Tan, F.; Hoffmann-Benning, S.; Liang, W.; Chen, W. Proteomic analysis of changes in the Kandelia candel chloroplast proteins reveals pathways associated with salt tolerance. Plant Sci. 2015, 231, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Fling, S.P.; Gregerson, D.S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity tris buffer system without urea. Anal. Biochem. 1986, 155, 83–88. [Google Scholar] [CrossRef]
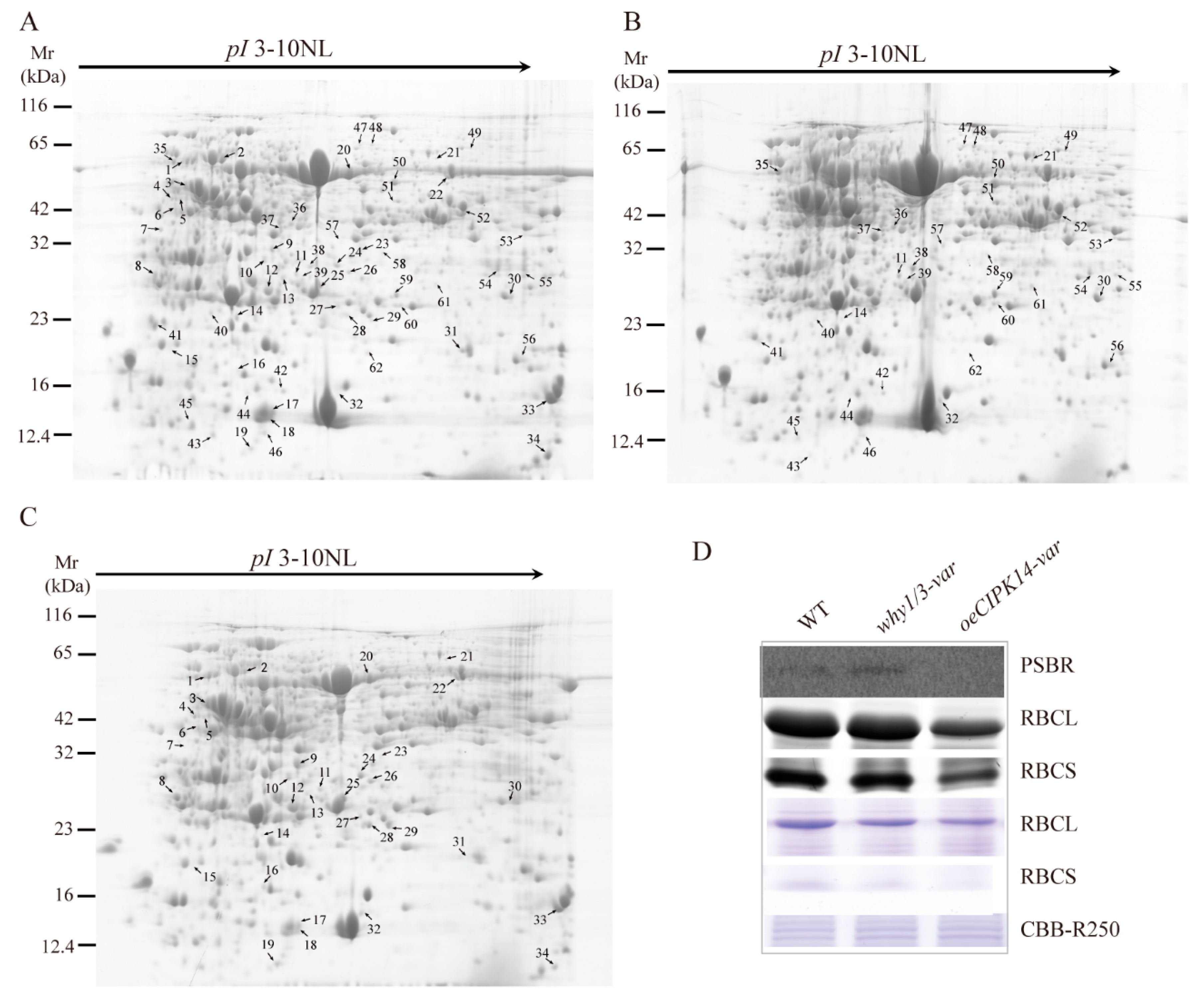

| Spot No a | Protein Name | Accession No b | Mascot Score | Matched Peptides | Theor MW (kDa)/pI c | Cov% d | Subcellular Loc e |
|---|---|---|---|---|---|---|---|
| Light reaction | |||||||
| 1 | Fe-S cluster assembly factor HCF101 | HF101_ARATH | 371 | 13 | 57.728/5.91 | 20% | plastid |
| 2 | RCA | F4IVZ7_ARATH | 499 | 18 | 48.469/7.55 | 34% | chloroplast |
| 3 | ATP synthase subunit alpha | ATPA_ARATH | 927 | 32 | 55.294/5.19 | 50% | plastid |
| 4 | Magnesium-chelatase subunit ChlI-2 | CHLI2_ARATH | 365 | 23 | 46.069/5.36 | 44% | chloroplast |
| 12 | Chlorophyll a-b binding protein CP26 | CB5_ARATH | 213 | 12 | 30.183/6 | 35% | plastid |
| 26 | Magnesium protoporphyrin IX methyltransferase | CHLM_ARATH | 653 | 23 | 33.775/7.68 | 53% | chloroplast |
| 28 | PsbP domain-containing protein 4 | PPD4_ARATH | 122 | 8 | 28.484/7.02 | 33% | plastid |
| 33 | Oxygen-evolving enhancer protein 3-2 | PSBQ2_ARATH | 464 | 16 | 24.628/9.72 | 59% | plastid |
| 34 | PSBR | A0A178WGP6_ARATH | 112 | 7 | 9.77/10.1 | 39% | plastid |
| 35 | TROL | A0A178V0X3_ARATH | 444 | 21 | 54.448/5.09 | 29% | chloroplast |
| Calvin cycle | |||||||
| 5 | Phosphoglycerate kinase 1 | PGKH1_ARATH | 642 | 24 | 50.081/5.91 | 37% | chloroplast |
| 6 | Sedoheptulose-1,7-bisphosphatase | S17P_ARATH | 354 | 18 | 42.388/6.17 | 28% | chloroplast |
| 14 | Ribulose bisphosphate carboxylase large chain (Fragment) | A0A142I795_ARATH | 292 | 12 | 51.833/6.17 | 9% | chloroplast |
| 17 | Ribulose bisphosphate carboxylase small chain | A0A178UL15_ARATH | 326 | 14 | 20.33/7.59 | 45% | chloroplast |
| 18 | Ribulose bisphosphate carboxylase small chain 1A | RBS1A_ARATH | 369 | 15 | 20.203/7.59 | 45% | chloroplast |
| 23 | Ribulose bisphosphate carboxylase large chain (Fragment) | A0A142I795_ARATH | 292 | 12 | 51.833/6.17 | 18% | plastid |
| 27 | Ribulose bisphosphate carboxylase large chain | RBL_ARATH | 740 | 31 | 52.922/5.88 | 42% | chloroplast |
| 32 | Ribulose bisphosphate carboxylase small chain | A0A178UL15_ARATH | 326 | 14 | 20.33/7.59 | 45% | chloroplast |
| 40 | Ribulose bisphosphate carboxylase large chain (Fragment) | A0A142I795_ARATH | 292 | 12 | 51.833/6.17 | 26% | chloroplast |
| 43 | Ribulose bisphosphate carboxylase small chain 1A | RBS1A_ARATH | 362 | 13 | 20.203/7.79 | 49% | chloroplast |
| 45 | Ribulose bisphosphate carboxylase small chain 1B | RBS1B_ARATH | 348 | 14 | 18.506/8.22 | 53% | chloroplast |
| 59 | Beta carbonic anhydrase 1 | BCA1_ARATH | 307 | 15 | 37.426/5.74 | 48% | plastid/ cytomembrane |
| Defense and antioxidation | |||||||
| 7 | Glucan endo-1,3-beta-glucosidase, acidic isoform | E13A_ARATH | 568 | 16 | 37.316/4.85 | 33% | secretion |
| 9 | Thiamine thiazole synthase | THI4_ARATH | 183 | 10 | 36.641/5.82 | 28% | plastid |
| 10 | SAPX | A0A178V0Q5_ARATH | 524 | 24 | 40.446/8.31 | 47% | chloroplast |
| 16 | Glycine-rich RNA-binding protein 7 | RBG7_ARATH | 167 | 9 | 16.88/5.85 | 51% | cytoplasm/nucleus |
| 20 | Glutathione S-transferase F6 | GSTF6_ARATH | 355 | 15 | 23.471/5.8 | 49% | cytoplasm |
| 24 | V-type proton ATPase subunit E1 | VATE1_ARATH | 328 | 30 | 26.044/6.04 | 75% | vacuole |
| 25 | l-ascorbate peroxidase 1 | F4HU93_ARATH | 389 | 16 | 27.503/5.85 | 52% | cytoplasm |
| 37 | Pyridoxal 5′-phosphate synthase subunit PDX1.1 | PDX11_ARATH | 423 | 23 | 32.841/5.75 | 37% | cytoplasm |
| 38 | Thioredoxin-like protein CDSP32 | CDSP_ARATH | 371 | 20 | 33.663/8.65 | 35% | chloroplast |
| 44 | GRP7 | A0A178VQY8_ARATH | 315 | 9 | 16.937/5.85 | 37% | cytoplasm/nucleus |
| 46 | V-type proton ATPase subunit G1 | VATG1_ARATH | 304 | 10 | 12.389/5.77 | 70% | vacuole |
| 51 | Formate dehydrogenase | FDH_ARATH | 354 | 18 | 42.383/7.12 | 36% | chloroplast/mitochondria |
| 52 | 12-oxophytodienoate reductase 3 | OPR3_ARATH | 782 | 24 | 42.664/7.71 | 54% | peroxysome |
| 54 | Thylakoid lumenal 29 kDa protein | TL29_ARATH | 475 | 24 | 37.911/8.59 | 53% | plastid |
| 55 | Remorin | REMO_ARATH | 427 | 26 | 20.955/8.63 | 65% | plasmalemma |
| 57 | VIPP1 | A0A178W0D3_ARATH | 412 | 23 | 28.895/5.9 | 67% | plastid |
| 60 | Glutathione S-transferase F7 | GSTF7_ARATH | 552 | 18 | 23.583/6.14 | 52% | cytoplasm |
| 61 | Peptide methionine sulfoxide reductase A4 | MSRA4_ARATH | 236 | 12 | 38.626/8.96 | 26% | plastid |
| Amino acid metabolism | |||||||
| 22 | Serine hydroxymethyltransferase 4 | GLYC4_ARATH | 504 | 22 | 51.685/6.8 | 46% | cytoplasm |
| 48 | Asparagine synthetase [glutamine-hydrolyzing] 2 | ASNS2_ARATH | 235 | 18 | 64.989/6.01 | 27% | cytoplasm/plasmodesmata |
| 50 | Glutamate-glyoxylate aminotransferase 1 | GGT1_ARATH | 478 | 20 | 53.267/6.49 | 37% | peroxysome |
| 58 | Acetylglutamate kinase | NAGK_ARATH | 424 | 14 | 36.572/9.04 | 36% | plastid |
| Proteometabolism | |||||||
| 11 | ATP-dependent Clp protease proteolytic subunit-related protein 3 | CLPR3_ARATH | 597 | 23 | 36.284/8.64 | 41% | chloroplast |
| 13 | Proteasome subunit alpha type-6-A | PSA6A_ARATH | 926 | 25 | 27.277/5.6 | 58% | cytoplasm/nucleus |
| 15 | 30S ribosomal protein S6 alpha | RR6_ARATH | 119 | 8 | 22.746/5.92 | 26% | plastid |
| 19 | CPN10 | O80504_ARATH | 567 | 10 | 15.04/8.75 | 45% | chloroplast |
| 29 | Proteasome subunit beta type-2-B | PSB2B_ARATH | 715 | 22 | 21.97/6.21 | 70% | cytoplasm/nucleus |
| 30 | Ribosome-recycling factor | RRFC_ARATH | 497 | 18 | 30.403/9.46 | 44% | plastid |
| 31 | Peptidyl-prolyl cis-trans isomerase CYP18-3 | CP18C_ARATH | 307 | 11 | 18.361/7.68 | 34% | cytoplasm |
| 39 | Proteasome subunit alpha type-6-B | PSA6B_ARATH | 312 | 19 | 27.333/5.75 | 56% | cytoplasm/nucleus |
| 56 | Peptidyl-prolyl cis-trans isomerase CYP19-1 | CP19A_ARATH | 406 | 14 | 18.48/8.65 | 43% | cytoplasm |
| 62 | Nascent polypeptide-associated complex subunit beta | A0A178W6R8_ARATH | 332 | 14 | 16.935/5.50 | 45% | cytoplasm/nucleus |
| Energy metabolism | |||||||
| 21 | Beta-amylase 3 | BAM3_ARATH | 510 | 24 | 61.314/6.59 | 41% | chloroplast |
| Malate biosynthesis | |||||||
| 36 | Malate dehydrogenase | MDHP_ARATH | 334 | 12 | 42.379/8.66 | 23% | chloroplast |
| 47 | NADP-dependent malic enzyme 2 | MAOP2_ARATH | 323 | 19 | 64.372/6.01 | 25% | cytoplasm |
| 49 | l-galactono-1,4-lactone dehydrogenase | GLDH_ARATH | 759 | 35 | 68.513/8.7 | 38% | mitochondria |
| Lipid metabolism | |||||||
| 53 | GDSL esterase/lipase At1g29670 | GDL15_ARATH | 841 | 19 | 39.847/8.85 | 44% | secretion |
| Transcription | |||||||
| 8 | RNA-binding protein CP29B | CP29B_ARATH | 602 | 16 | 30.699/5.06 | 52% | plastid |
| Unknow function | |||||||
| 41 | At1g13930/F16A14.27 | Q9XI93_ARATH | 310 | 14 | 16.154/4.82 | 87% | chloroplast/plasmalemma |
| 42 | Kinesin-like calmodulin-binding protein | KCBP_ARATH | 54 | 35 | 143.359/6.69 | 21% | cytoplasm |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, Z.; Wang, W.; Yu, X.; Lin, W.; Miao, Y. Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2231. https://doi.org/10.3390/ijms19082231
Guan Z, Wang W, Yu X, Lin W, Miao Y. Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(8):2231. https://doi.org/10.3390/ijms19082231
Chicago/Turabian StyleGuan, Zhe, Wanzhen Wang, Xingle Yu, Wenfang Lin, and Ying Miao. 2018. "Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis" International Journal of Molecular Sciences 19, no. 8: 2231. https://doi.org/10.3390/ijms19082231
APA StyleGuan, Z., Wang, W., Yu, X., Lin, W., & Miao, Y. (2018). Comparative Proteomic Analysis of Coregulation of CIPK14 and WHIRLY1/3 Mediated Pale Yellowing of Leaves in Arabidopsis. International Journal of Molecular Sciences, 19(8), 2231. https://doi.org/10.3390/ijms19082231