In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators
Abstract
1. Introduction
2. Results
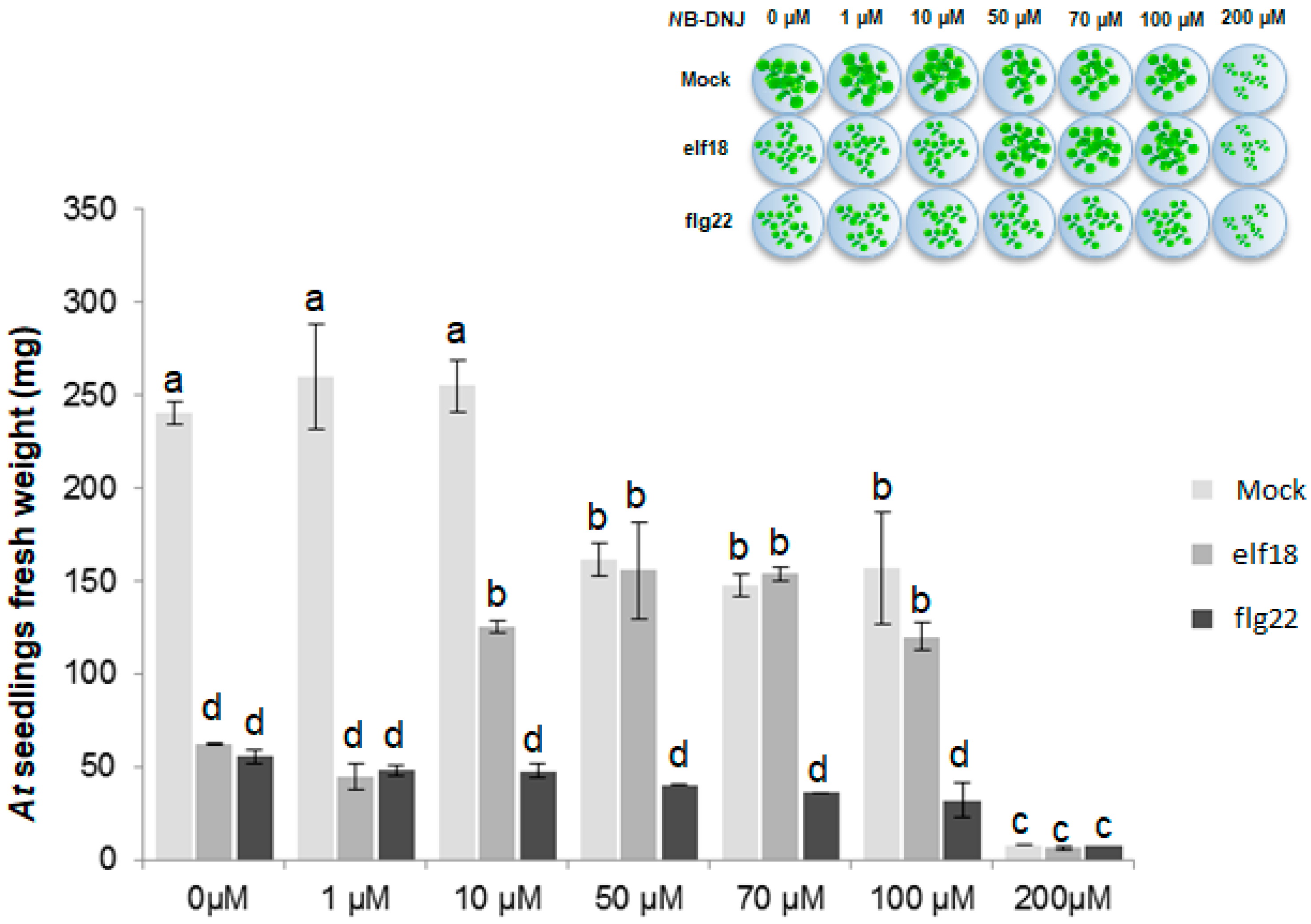
2.1. The Iminosugar NB-DNJ Is Toxic to Arabidopsis thaliana Plants at Concentrations above 200 µM
2.2. NB-DNJ Doses Higher than 50 µM Render Arabidopsis thaliana Plants Insensitive to the elf18 Peptide
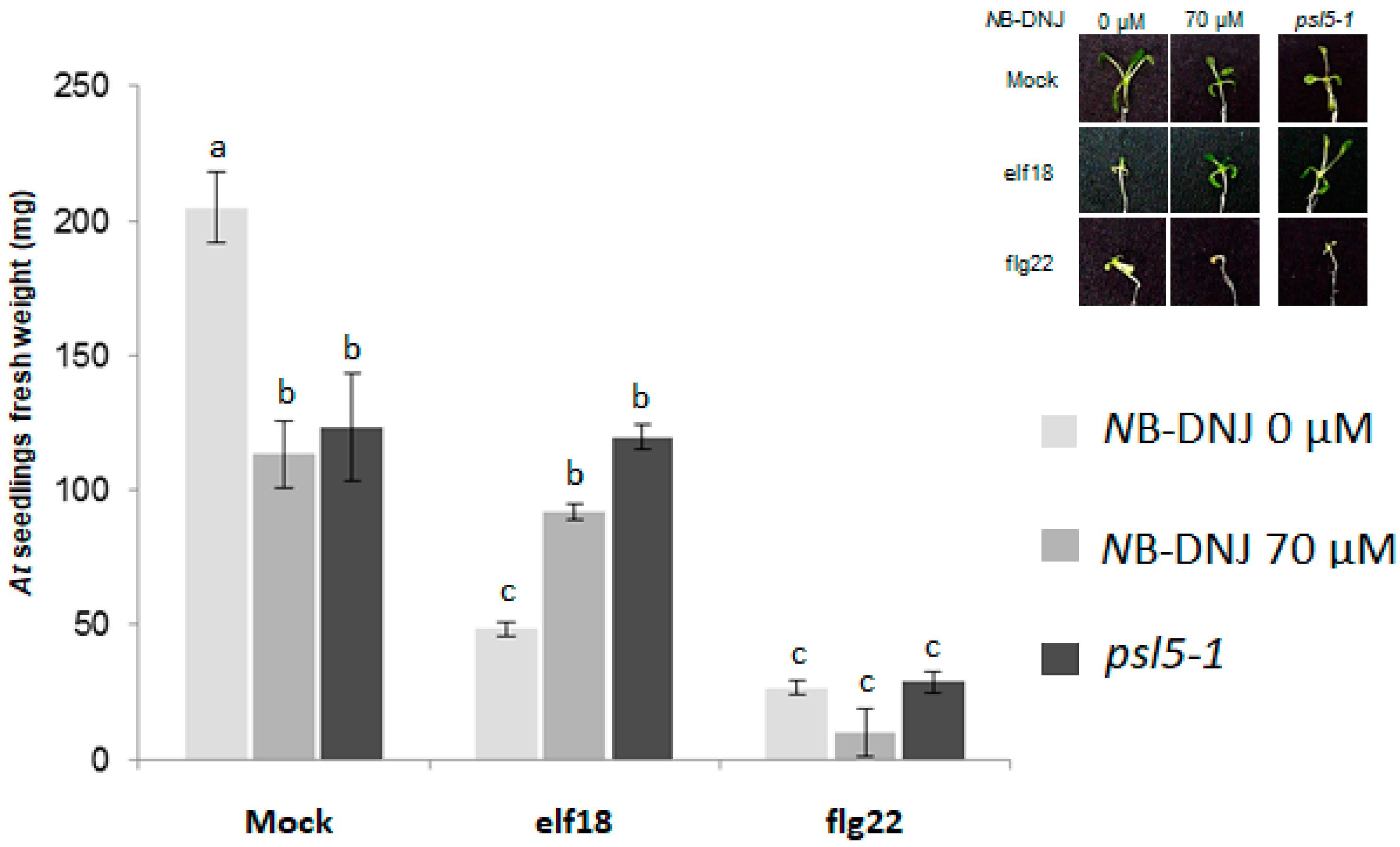
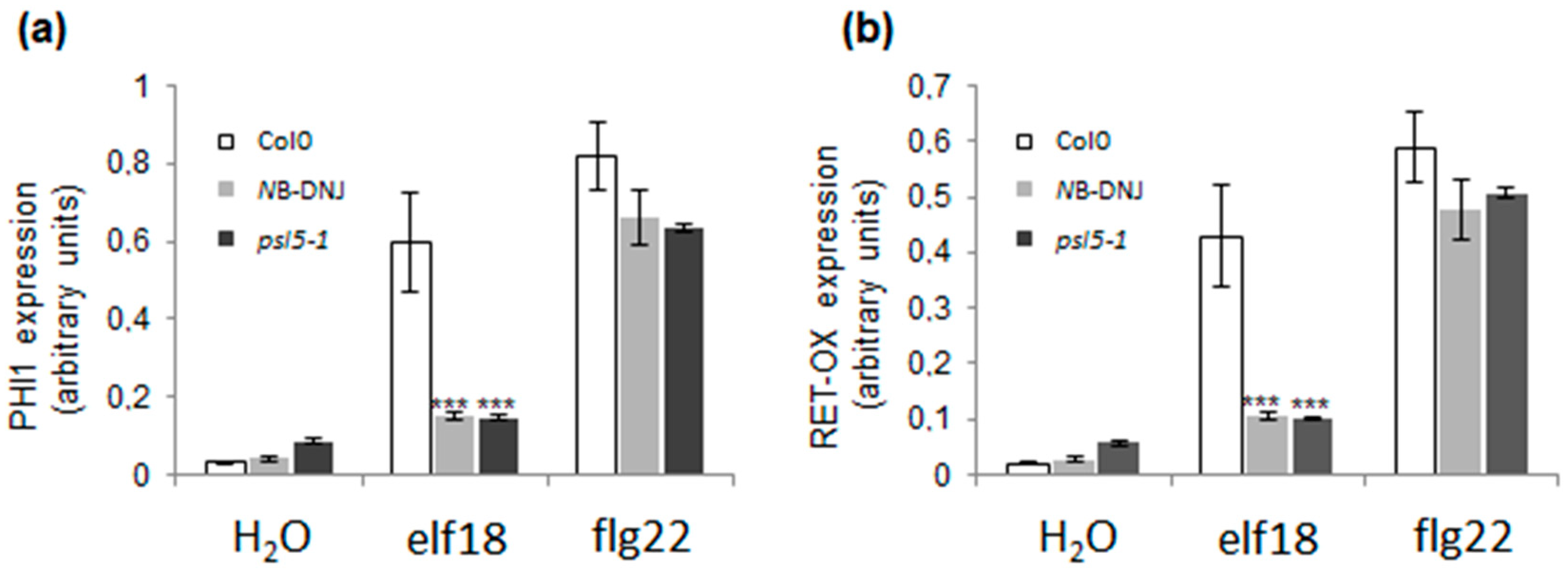
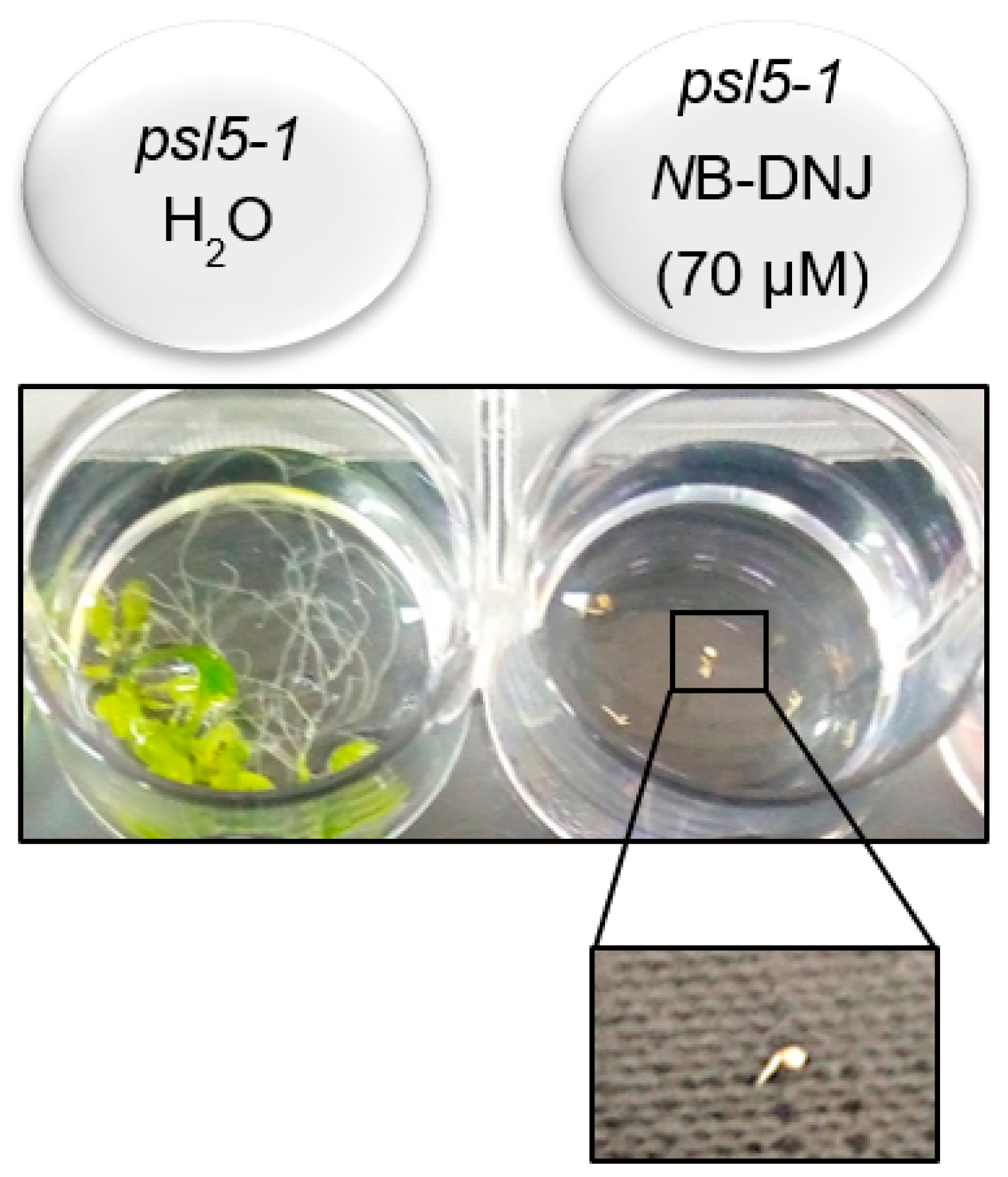
2.3. Treatment with 70 µM NB-DNJ Is Lethal to the At psl5-1 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions and Treatments
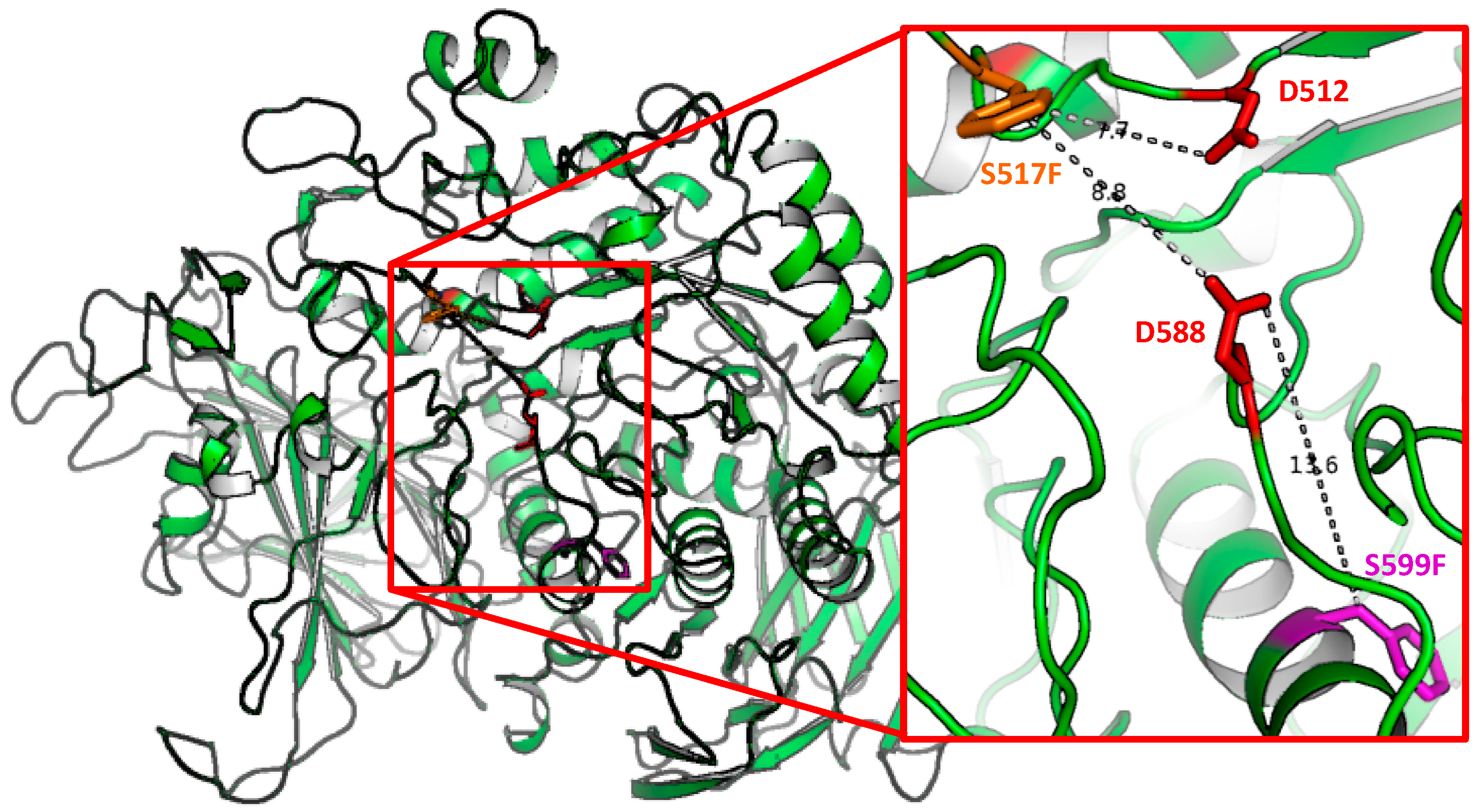
4.2. Homology Modelling
4.3. Gene Transcription Analysis
4.4. Statistical Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gidalevitz, T.; Stevens, F.; Argon, Y. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 2013, 1833, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Alonzi, D.S.; Scott, K.A.; Dwek, R.A.; Zitzmann, N. Iminosugar antivirals: The therapeutic sweet spot. Biochem. Soc. Trans. 2017, 45, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, M.; Crispin, M.; Scanlan, C.N.; Zitzmann, N.; Dwek, R.A. Emerging principles for the therapeutic exploitation of glycosylation. Science 2014, 343, 1235681. [Google Scholar] [CrossRef] [PubMed]
- Butters, T.D.; Mellor, H.R.; Narita, K.; Dwek, R.A.; Platt, F.M. Small-molecule therapeutics for the treatment of glycolipid lysosomal storage disorders. Philos. Trans. R Soc. Lond. B Biol. Sci. 2003, 358, 927–945. [Google Scholar] [CrossRef] [PubMed]
- Roversi, P.; Marti, L.; Caputo, A.T.; Alonzi, D.S.; Hill, J.C.; Dent, K.C.; Kumar, A.; Levasseur, M.D.; Lia, A.; Waksman, T.; et al. Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint. Proc. Natl. Acad. Sci. USA 2017, 114, 8544–8549. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; de Domenico, S.; Maffia, M.; Piro, G.; Sansebastiano, G.-P. Transgenic Plants as Low-Cost Platform for Chemotherapeutic Drugs Screening. IJMS 2015, 16, 2174–2186. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Durantel, D.; Macovei, A.; Zitzmann, N.; Zoulim, F.; Dwek, R.A.; Branza-Nichita, N. Treatment of hepatitis B virus-infected cells with alpha-glucosidase inhibitors results in production of virions with altered molecular composition and infectivity. Antivir. Res. 2007, 76, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, E.; Whitfield, T.; Kallis, S.; Dwek, R.A.; Zitzmann, N.; Pietschmann, T.; Bartenschlager, R. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 2007, 46, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Pollock, S.; Dwek, R.A.; Burton, D.R.; Zitzmann, N. N-Butyldeoxynojirimycin is a broadly effective anti-HIV therapy significantly enhanced by targeted liposome delivery. AIDS 2008, 22, 1961–1969. [Google Scholar] [CrossRef] [PubMed]
- Warfield, K.L.; Plummer, E.M.; Sayce, A.C.; Alonzi, D.S.; Tang, W.; Tyrrell, B.E.; Hill, M.L.; Caputo, A.T.; Killingbeck, S.S.; Beatty, P.R.; et al. Inhibition of endoplasmic reticulum glucosidases is required for in vitro and in vivo dengue antiviral activity by the iminosugar UV-4. Antiviral Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, B.E.; Sayce, A.C.; Warfield, K.L.; Miller, J.L.; Zitzmann, N. Iminosugars: Promising therapeutics for influenza infection. Crit. Rev. Microbiol. 2017, 43, 521–545. [Google Scholar] [CrossRef] [PubMed]
- Sayce, A.C.; Alonzi, D.S.; Killingbeck, S.S.; Tyrrell, B.E.; Hill, M.L.; Caputo, A.T.; Iwaki, R.; Kinami, K.; Ide, D.; Kiappes, J.L.; et al. Iminosugars Inhibit Dengue Virus Production via Inhibition of ER Alpha-Glucosidases-Not Glycolipid Processing Enzymes. PLoS Negl. Trop. Dis. 2016, 10, e0004524. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.F.; Pritchard, K. In vitro screening for drug repositioning. J. Biomol. Screen. 2015, 20, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D.G. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Häweker, H.H.A.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Tintor, N.; Mentzel, T.; Kombrink, E.; Boller, T.; Robatzek, S.; Schulze-Lefert, P.; Saijo, Y. Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl. Acad. Sci. USA 2009, 106, 22522–22527. [Google Scholar] [CrossRef] [PubMed]
- Caputo, A.T.; Alonzi, D.S.; Marti, L.; Reca, I.-B.; Kiappes, J.L.; Struwe, W.B.; Cross, A.; Basu, S.; Lowe, E.D.; Darlot, B.; et al. Structures of mammalian ER α-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals. Proc. Natl. Acad. Sci. USA 2016, 113, E4630–E4638. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, G.B.; Butters, T.D.; Dwek, R.A.; Platt, F.M. Effects of the imino sugar N-butyldeoxynojirimycin on the N-glycosylation of recombinant gp120. J. Biol. Chem. 1993, 268, 570–576. [Google Scholar] [PubMed]
- Platt, F.M.; Karlsson, G.B.; Jacob, G.S. Modulation of cell-surface transferrin receptor by the imino sugar N-butyldeoxynojirimycin. Eur. J. Biochem. 1992, 208, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Gravino, M.; Savatin, D.V.; Macone, A.; De Lorenzo, G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015, 84, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Silverblatt-Buser, E.W.; Frick, M.A.; Rabeler, C.; Kaplinsky, N.J. Genetic Interactions Between BOB1 and Multiple 26S Proteasome Subunits Suggest a Role for Proteostasis in Regulating Arabidopsis Development. G3 (Bethesda) 2018, 8, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.; Galli, C.; Vanoni, O.; Arnold, S.M.; Kaufman, R.J. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mole. Cell 2005, 20, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Herrera, F.; Moreno, A.A.; Tapia, R.; Reyes, F.; Araya, M.; D’Alessio, C.; Parodi, A.; Orellana, A. The UDP-glucose: Glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol. 2015, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Fanchiotti, S.; Fernandez, F.; D’Alessio, C.; Parodi, A.J. The UDP-Glc: Glycoprotein glucosyltransferase is essential for Schizosaccharomyces pombe viability under conditions of extreme endoplasmic reticulum stress. J. Cell Biol. 1998, 143, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Artal-Sanz, M.; de Jong, L.; Tavernarakis, N. Caenorhabditis elegans: A versatile platform for drug discovery. Biotechnol. J. 2006, 1, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Aiyar, A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 2000, 132, 221–241. [Google Scholar] [PubMed]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.-Y.; Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008, 426, 145–159. [Google Scholar] [PubMed]
- DeLano, W.L. The case for open-source software in drug discovery. Drug Discov. Today 2005, 10, 213–217. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Lange, I.G.; Daxenberger, A.; Meyer, H.H. Tissue-specific expression pattern of estrogen receptors (ER): Quantification of ER alpha and ER beta mRNA with real-time RT-PCR. APMIS 2001, 109, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Galletti, R.; Vairo, D.; Cervone, F.; De Lorenzo, G. Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol. Plant Microbe Interact. 2006, 19, 931–936. [Google Scholar] [CrossRef] [PubMed]
| GENE | AG CODE | FORWARD PRIMER (5′-3′) | REVERSE PRIMER (5′-3′) |
|---|---|---|---|
| UBQ5 | AT3G62250 | GGAATCGACGCTTCATCTCG | ATGAAAGTCCCAGCTCCACA |
| PHI1 | AT1G35140 | TTGGTTTAGACGGGATGGTG | ACTCCAGTACAAGCCGATCC |
| RET-OX | AT1G26380 | AGGTTCTCGAACCCTAACAACA | GCACAGACGACACGTAAGAAAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marti, L.; Lia, A.; Reca, I.-B.; Roversi, P.; Santino, A.; Zitzmann, N. In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators. Int. J. Mol. Sci. 2018, 19, 2135. https://doi.org/10.3390/ijms19072135
Marti L, Lia A, Reca I-B, Roversi P, Santino A, Zitzmann N. In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators. International Journal of Molecular Sciences. 2018; 19(7):2135. https://doi.org/10.3390/ijms19072135
Chicago/Turabian StyleMarti, Lucia, Andrea Lia, Ida-Barbara Reca, Pietro Roversi, Angelo Santino, and Nicole Zitzmann. 2018. "In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators" International Journal of Molecular Sciences 19, no. 7: 2135. https://doi.org/10.3390/ijms19072135
APA StyleMarti, L., Lia, A., Reca, I.-B., Roversi, P., Santino, A., & Zitzmann, N. (2018). In Planta Preliminary Screening of ER Glycoprotein Folding Quality Control (ERQC) Modulators. International Journal of Molecular Sciences, 19(7), 2135. https://doi.org/10.3390/ijms19072135






