FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. FOXP3 Increases UBC9 mRNA and Protein Levels
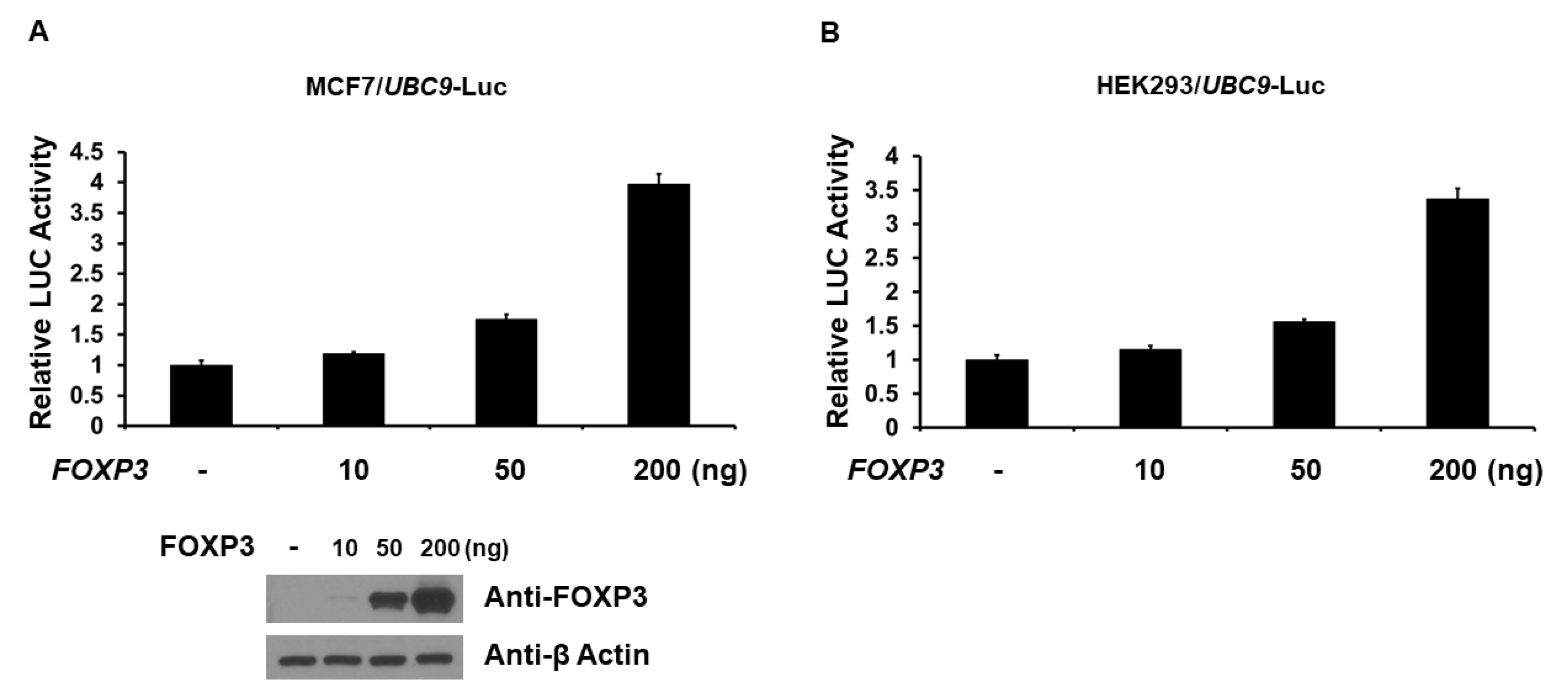
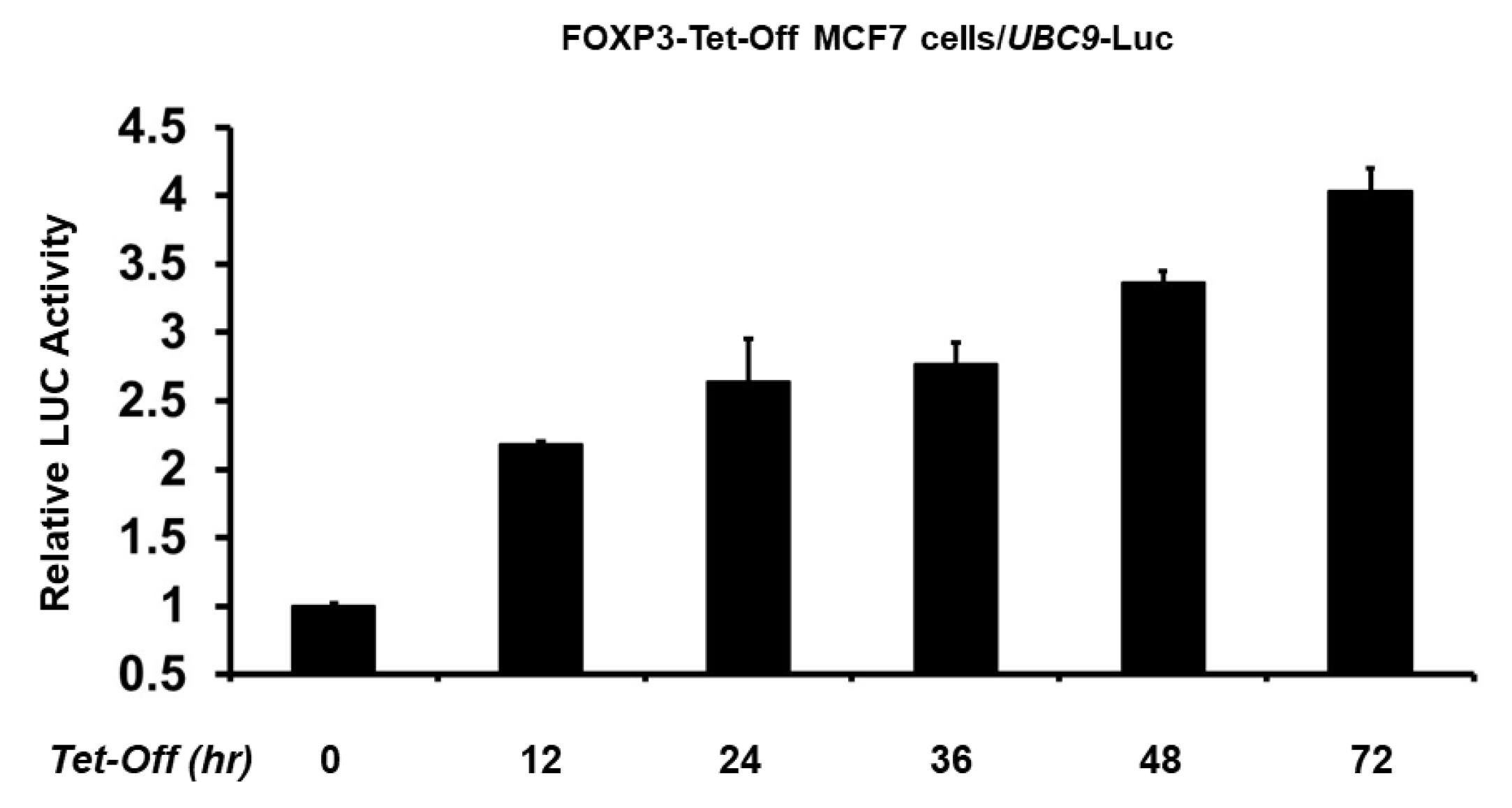
2.2. FOXP3 Is an Activator of the UBC9 Promoter
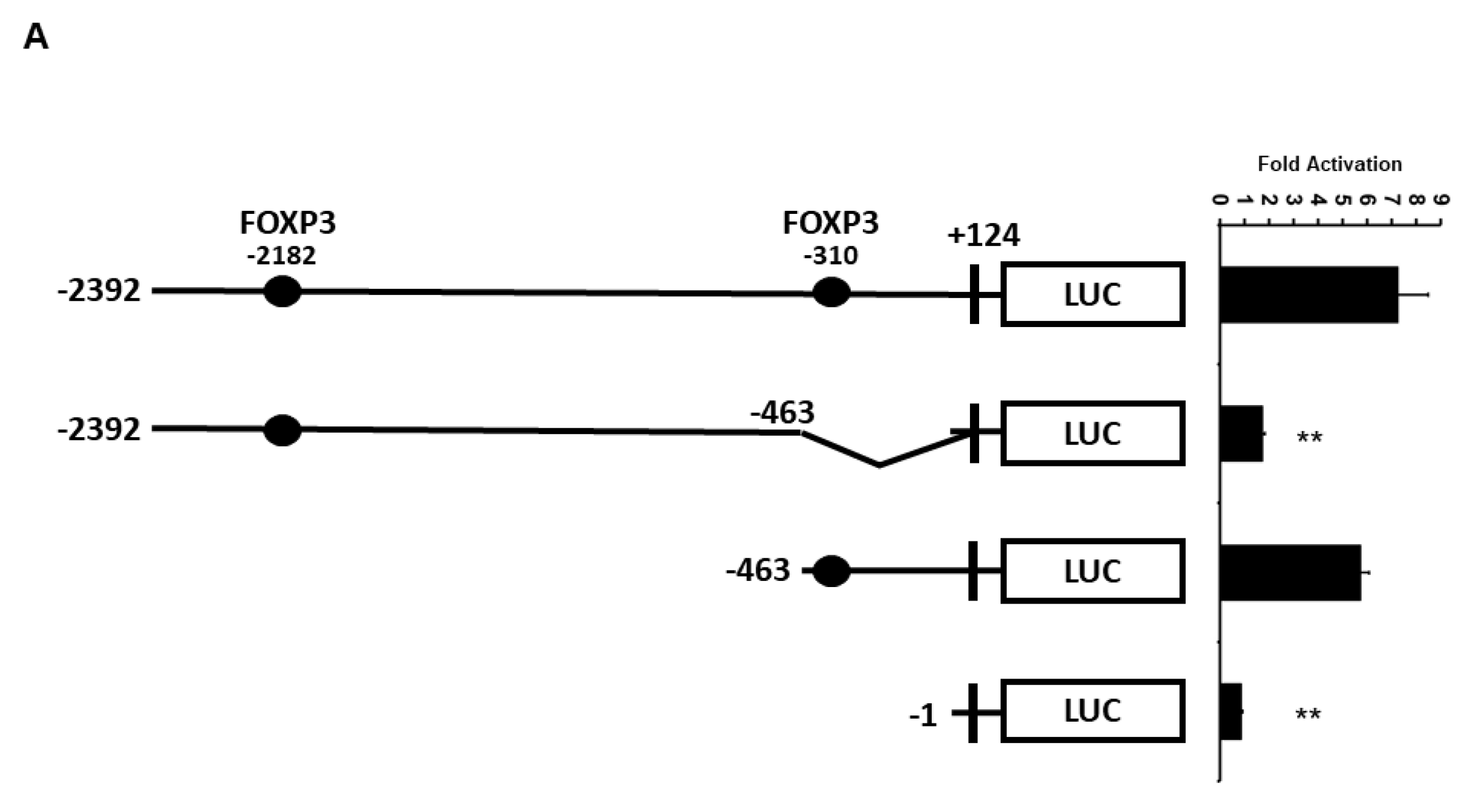
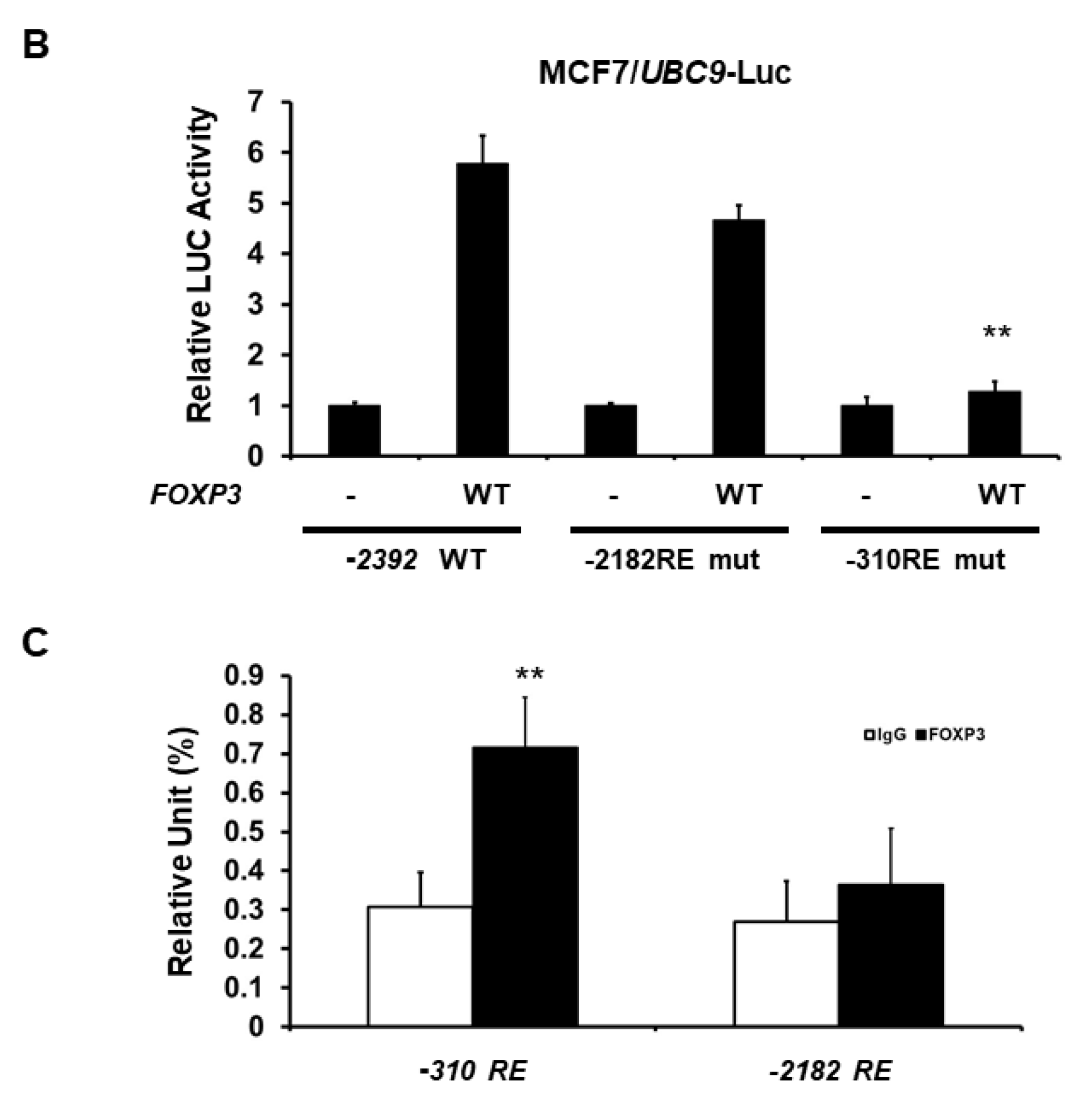
2.3. Minimal UBC9 Promoter Region Responsive to FOXP3 Activation
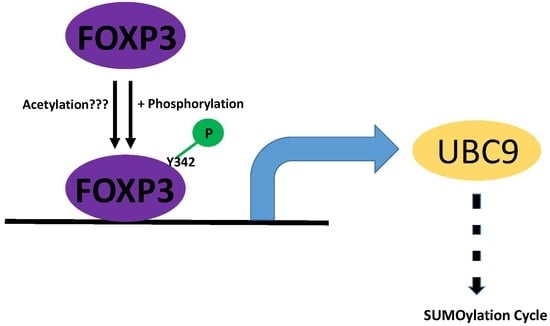
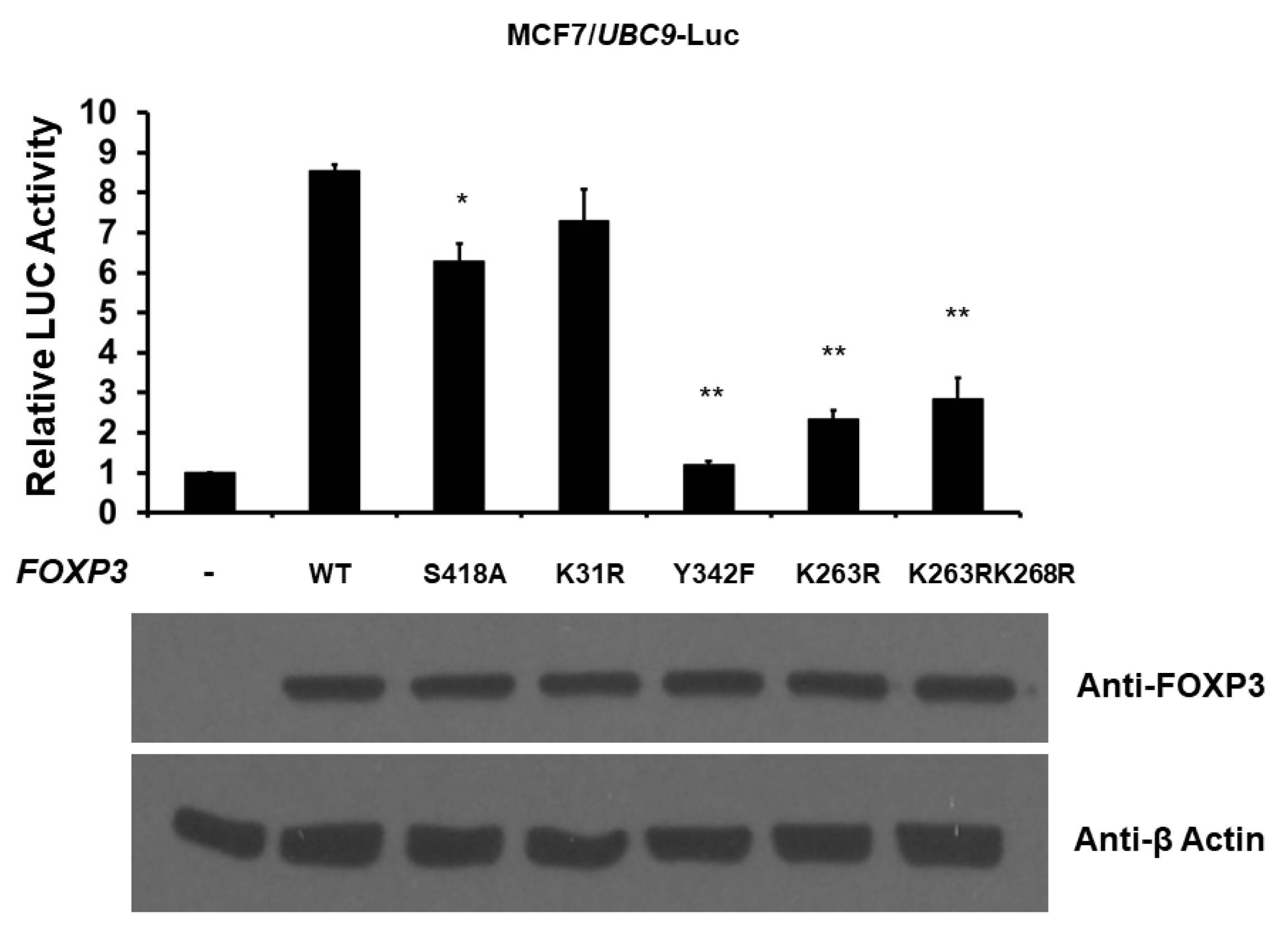
2.4. Post-Translational Modifications of FOXP3 Are Involved for FOXP3-Mediated UBC9 Transcriptional Activity
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. DNA Constructs
4.3. Cell Culture and Transfection
4.4. UBC9 Luciferase Reporter Assay
4.5. RT-PCR and Real-Time ChIP
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| FOXP3 | Forkhead Box Protein P3 |
| UBC9 | SUMO-conjugating enzyme UBC9 |
| SUMO | small ubiquitin-like modifier |
| SUMOylation | a post-translational modification process involving SUMO addition |
| BRCA1 | breast cancer type 1 susceptibility protein |
| CD44 | CD44 antigen |
| HER2/ERBB2 | receptor tyrosine-protein kinase erbB2 |
| SKP2 | S-phase kinase-associated protein 2 |
| MCF7 | a breast cancer cell line |
| LATS2 | serine/threonine-protein kinase LATS2 |
| LCK | Lymphocyte-specific protein tyrosine kinase |
References
- Chatila, T.A.; Blaeser, F.; Ho, N.; Lederman, H.M.; Voulgaropoulos, C.; Helms, C.; Bowcock, A.M. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J. Clin. Investig. 2000, 106, R75–R81. [Google Scholar] [CrossRef] [PubMed]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Wildin, R.S.; Ramsdell, F.; Peake, J.; Faravelli, F.; Casanova, J.L.; Buist, N.; Levy-Lahad, E.; Mazzella, M.; Goulet, O.; Perroni, L.; et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001, 27, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Barbi, J.; Pan, F. The regulation of immune tolerance by FOXP3. Nat. Rev. Immunol. 2017, 17, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Khattri, R.; Cox, T.; Yasayko, S.A.; Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003, 4, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Ladoire, S.; Mignot, G.; Apetoh, L.; Ghiringhelli, F. Human FOXP3 and cancer. Oncogene 2010, 29, 4121–4129. [Google Scholar] [CrossRef] [PubMed]
- Redpath, M.; Xu, B.; van Kempen, L.C.; Spatz, A. The dual role of the X-linked FoxP3 gene in human cancers. Mol. Oncol. 2011, 5, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.L.; Ruan, L.W. Association between FOXP3 promoter polymorphisms and cancer risk: A meta-analysis. Oncol. Lett. 2014, 8, 2795–2799. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Wang, L.; Morrison, C.; Chang, X.; Zhang, H.; Li, W.; Liu, Y.; Wang, Y.; Liu, X.; Chan, M.W.; et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell 2007, 129, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Katoh, H.; Wang, L.; Yu, X.; Du, Z.; Yan, X.; Zheng, P.; Liu, Y. FOXP3 regulates sensitivity of cancer cells to irradiation by transcriptional repression of BRCA1. Cancer Res. 2013, 73, 2170–2180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, Y.; Hao, Q.; Wang, S.; Li, H.; Li, J.; Gao, Y.; Li, M.; Li, W.; Xue, X.; et al. FOXP3 suppresses breast cancer metastasis through downregulation of CD44. Int. J. Cancer 2015, 137, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Liu, R.; Zhang, H.; Chang, X.; Liu, Y.; Wang, L.; Zheng, P.; Liu, Y. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J. Clin. Investig. 2007, 117, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, C.; Chen, D.; Yang, W.H.; Liu, X.; Liu, C.G.; Dugas, C.M.; Tang, F.; Zheng, P.; Liu, Y.; et al. FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015, 75, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Yi, B.; Wei, S.; Yang, W.H.; Hart, K.M.; Chauhan, P.; Zhang, W.; Mao, X.; Liu, X.; Liu, C.G.; et al. FOXP3-miR-146-NF-κB axis and therapy for precancerous lesions in prostate. Cancer Res. 2015, 75, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, Y.; Wang, M.; Li, Z.; Zhao, Z.; Wang, R.X.; Wu, R.; Yuan, Z.; Cui, R.; Jiao, K.; et al. MicroRNA-155, induced by FOXP3 through transcriptional repression of BRCA1, is associated with tumor initiation in human breast cancer. Oncotarget 2017, 8, 41451–41464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K.; et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Wang, A.; Ma, X.; Demarque, M.; Jin, W.; Xin, H.; Dejean, A.; Dong, C. Protein SUMOylation is required for regulatory t cell expansion and function. Cell Rep. 2016, 16, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Ding, X.; Demarque, M.; Liu, X.; Pan, D.; Xin, H.; Zhong, B.; Wang, X.; Dejean, A.; Jin, W.; et al. Ubc9 is required for positive selection and late-stage maturation of thymocytes. J. Immunol. 2017, 198, 3461–3470. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Ji, W.K.; Hu, X.H.; Hu, W.F.; Tang, X.C.; Huang, Z.X.; Li, L.; Liu, M.; Xiang, S.H.; Wu, E.; et al. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, 5574–5579. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.Y.; Jang, H.; Lee, J.H.; Huh, J.Y.; Choi, S.; Chung, J.; Kim, J.B. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol. Cell. Biol. 2014, 34, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.M.; Brennan, V.C.; Gutierrez, N.M.; Wang, X.; Wang, L.; Yang, W.H. SUMOylation of ATF3 alters its transcriptional activity on regulation of TP53 gene. J. Cell. Biochem. 2013, 114, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Nepveu-Traversy, M.É.; Berthoux, L. The conserved sumoylation consensus site in TRIM5α modulates its immune activation functions. Virus Res. 2014, 184, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Taylor-Jaffe, K.M.; Nordin, K.M.; Prasad, M.S.; Lander, R.M.; LaBonne, C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell Biol. 2012, 198, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Bao, H.; Pan, Y.; Yin, M.; Liu, Y.; Wu, S.; Li, H. SUMOylation alterations are associated with multidrug resistance in hepatocellular carcinoma. Mol. Med. Rep. 2014, 9, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.M.; Liu, R.; Wang, L.; Nascimento, L.; Brennan, V.C.; Yang, W.H. SUMOylation of FOXM1B alters its transcriptional activity on regulation of miR-200 family and JNK1 in MCF7 human breast cancer cells. Int. J. Mol. Sci. 2014, 15, 10233–10251. [Google Scholar] [CrossRef] [PubMed]
- Meredith, L.J.; Wang, C.M.; Nascimento, L.; Liu, R.; Wang, L.; Yang, W.H. The key regulator for language and speech development, FOXP2, is a novel substrate for SUMOylation. J. Cell. Biochem. 2016, 117, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Zheng, Y.; Li, R.; Guo, T.B.; He, D.; Fang, L.; Liu, X.; Xiao, L.; Chen, X.; Wan, B.; et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat. Med. 2013, 19, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, K.; Morita, A.; Kim, N.S.; Yanagihara, I. Phosphorylation of FOXP3 by LCK downregulates MMP9 expression and represses cell invasion. PLoS ONE 2013, 8, e77099. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.S.; Lim, H.W.; Wu, J.; Schnölzer, M.; Verdin, E.; Ott, M. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J. Immunol. 2012, 188, 2712–2721. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, L.; Chen, G.; Katoh, H.; Chen, C.; Liu, Y.; Zheng, P. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009, 69, 2252–2259. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, L.; Katoh, H.; Liu, R.; Zheng, P.; Liu, Y. Identification of a tumor suppressor relay between the FOXP3 and the Hippo pathways in breast and prostate cancers. Cancer Res. 2011, 71, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Estruch, S.B.; Graham, S.A.; Deriziotis, P.; Fisher, S.E. The language-related transcription factor FOXP2 is post-translationally modified with small ubiquitin-like modifiers. Sci. Rep. 2016, 6, 20911. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-M.; Yang, W.H.; Liu, R.; Wang, L.; Yang, W.-H. FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2036. https://doi.org/10.3390/ijms19072036
Wang C-M, Yang WH, Liu R, Wang L, Yang W-H. FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells. International Journal of Molecular Sciences. 2018; 19(7):2036. https://doi.org/10.3390/ijms19072036
Chicago/Turabian StyleWang, Chiung-Min, William H. Yang, Runhua Liu, Lizhong Wang, and Wei-Hsiung Yang. 2018. "FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells" International Journal of Molecular Sciences 19, no. 7: 2036. https://doi.org/10.3390/ijms19072036
APA StyleWang, C.-M., Yang, W. H., Liu, R., Wang, L., & Yang, W.-H. (2018). FOXP3 Activates SUMO-Conjugating UBC9 Gene in MCF7 Breast Cancer Cells. International Journal of Molecular Sciences, 19(7), 2036. https://doi.org/10.3390/ijms19072036