Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia
Abstract
:1. Introduction
2. Results and Discussion
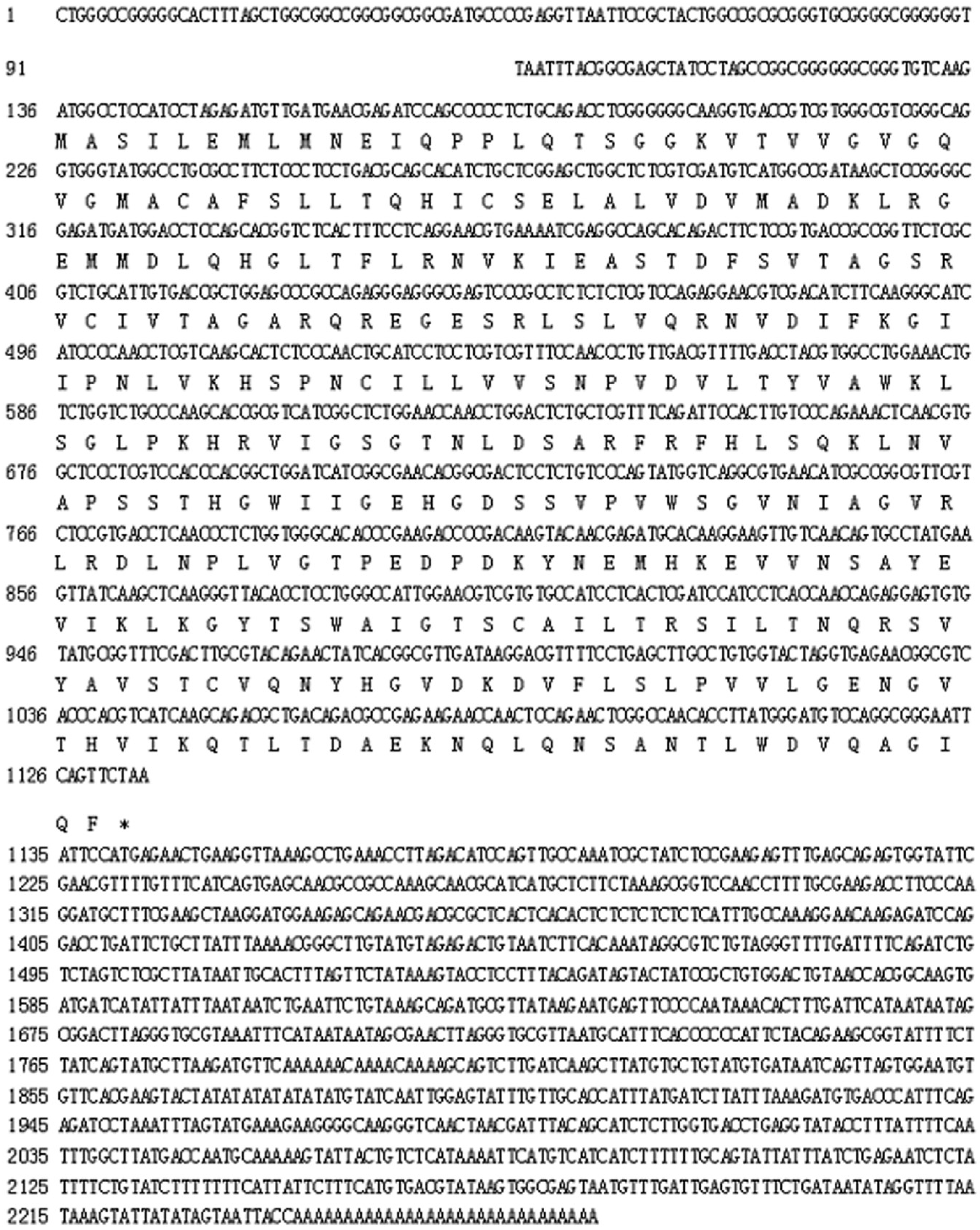
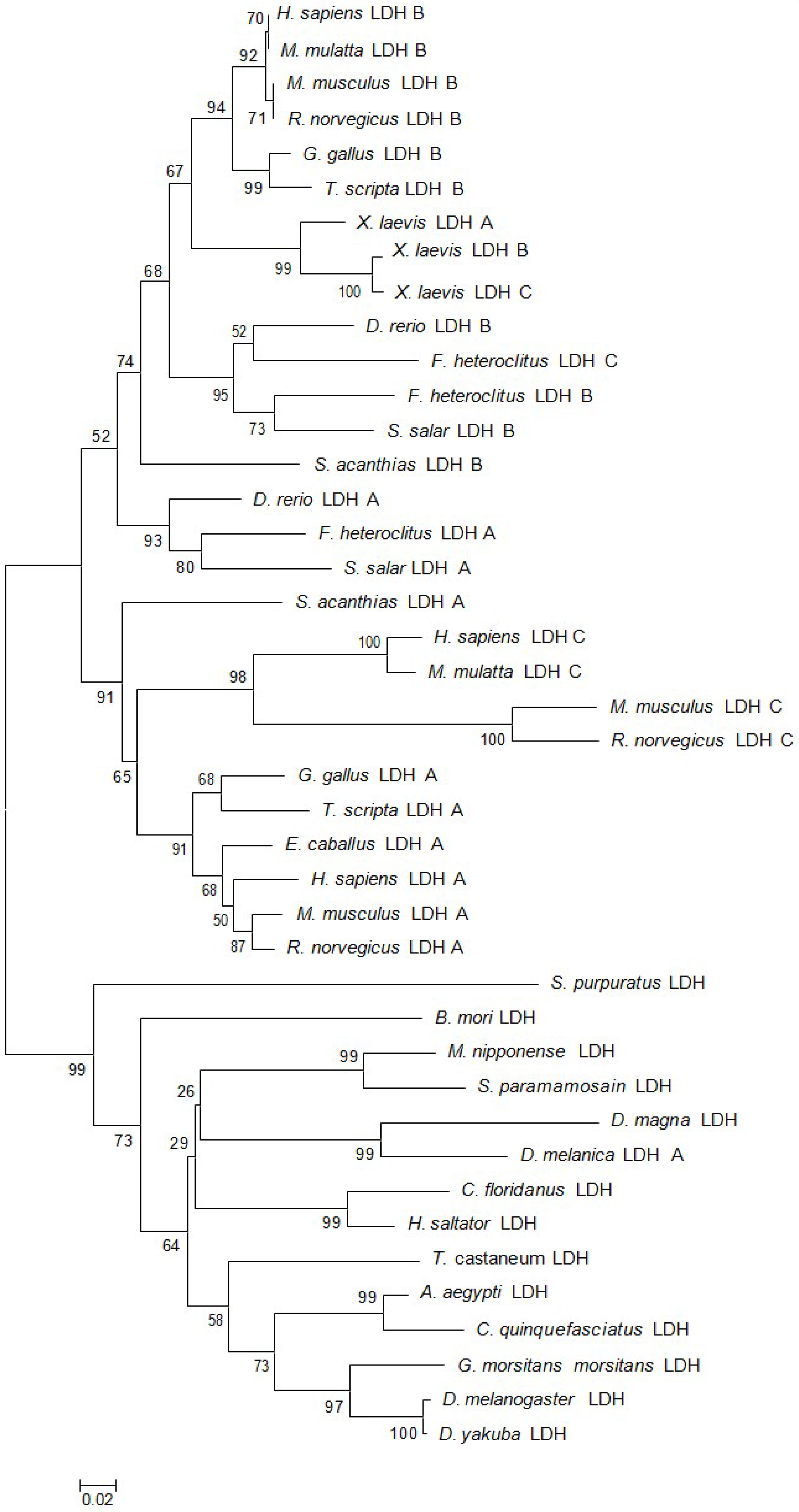
2.1. Characteristics and Phylogeny of MnLDH
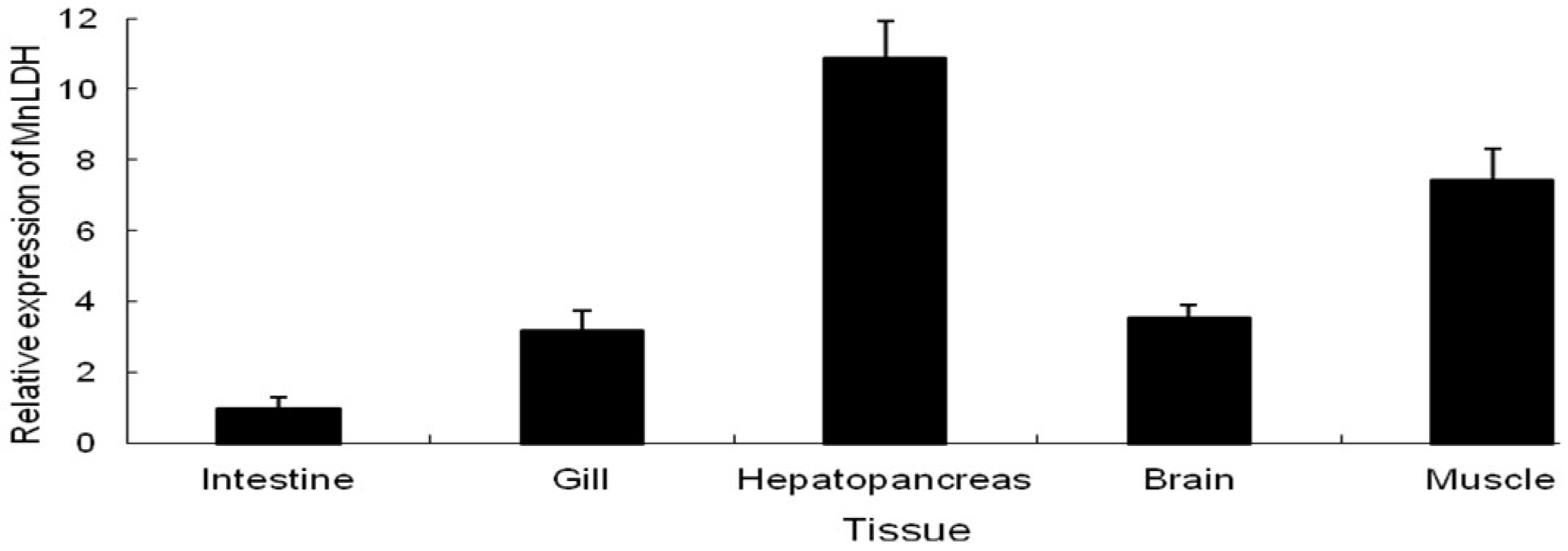
2.2. Tissue-Specific Expression of MnLDH
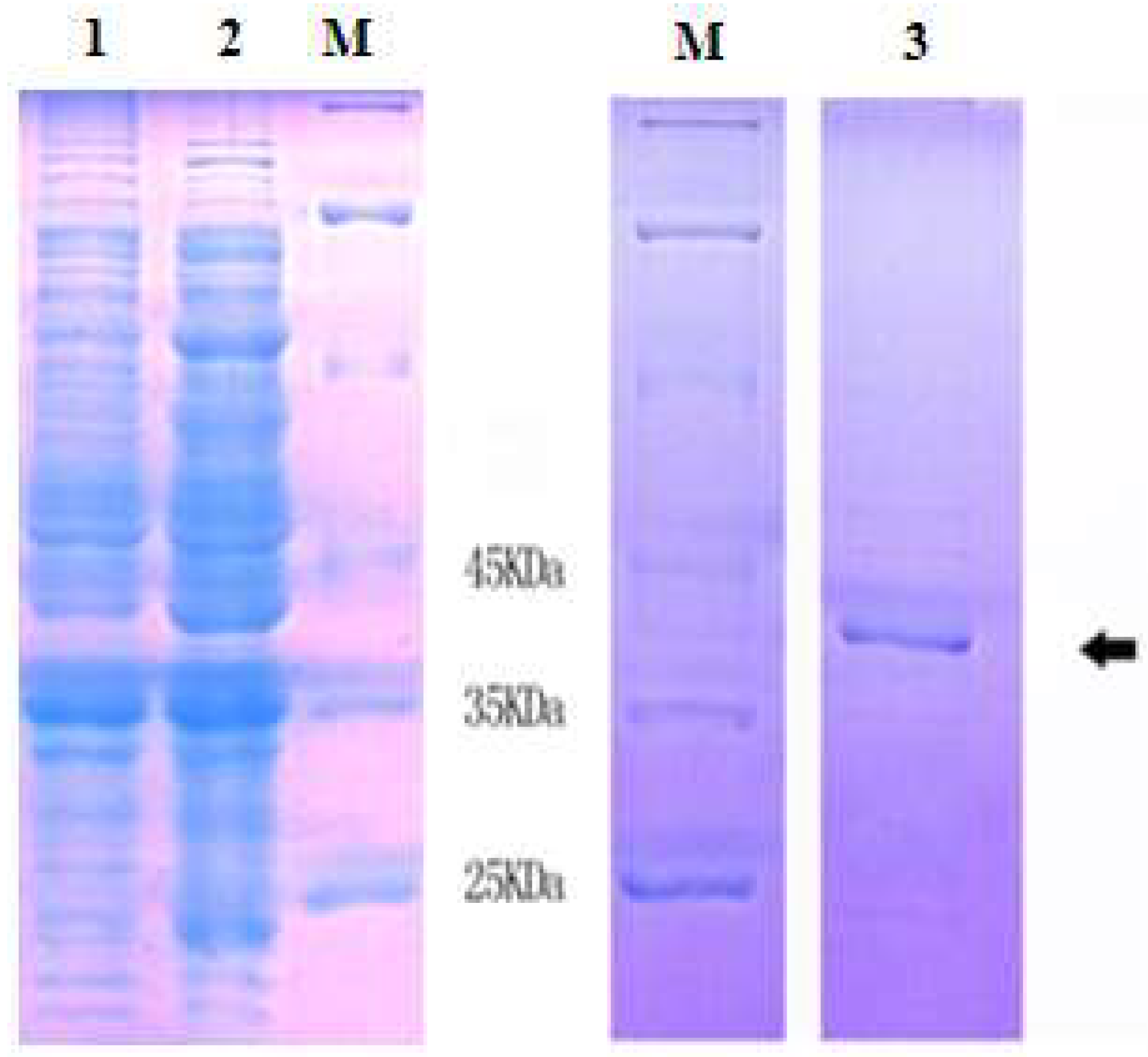
2.3 Expression and Purification of Recombinant MnLDH
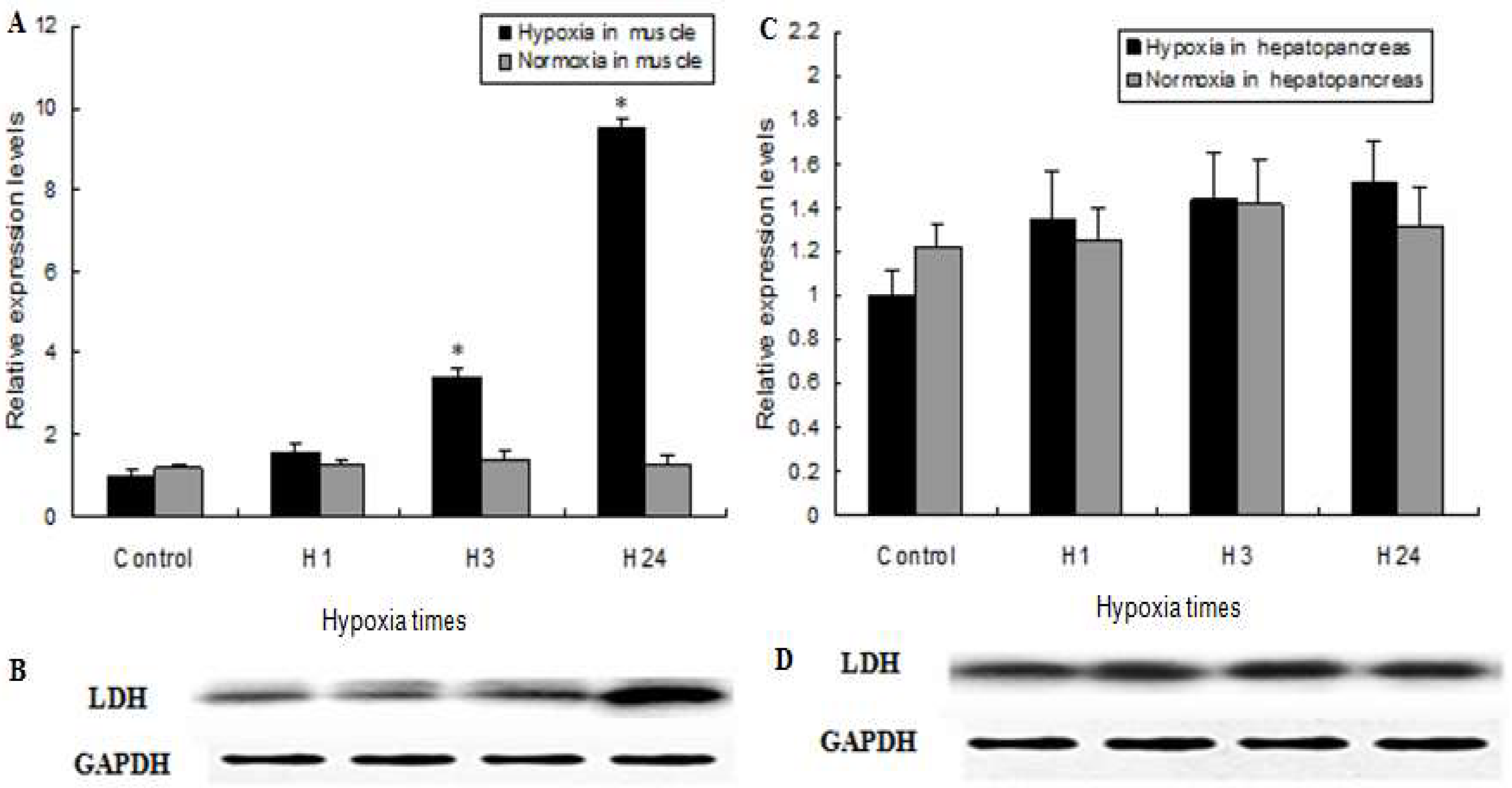
2.4. MnLDH Expression in Muscles and Hepatopancreas of Prawns during Hypoxia
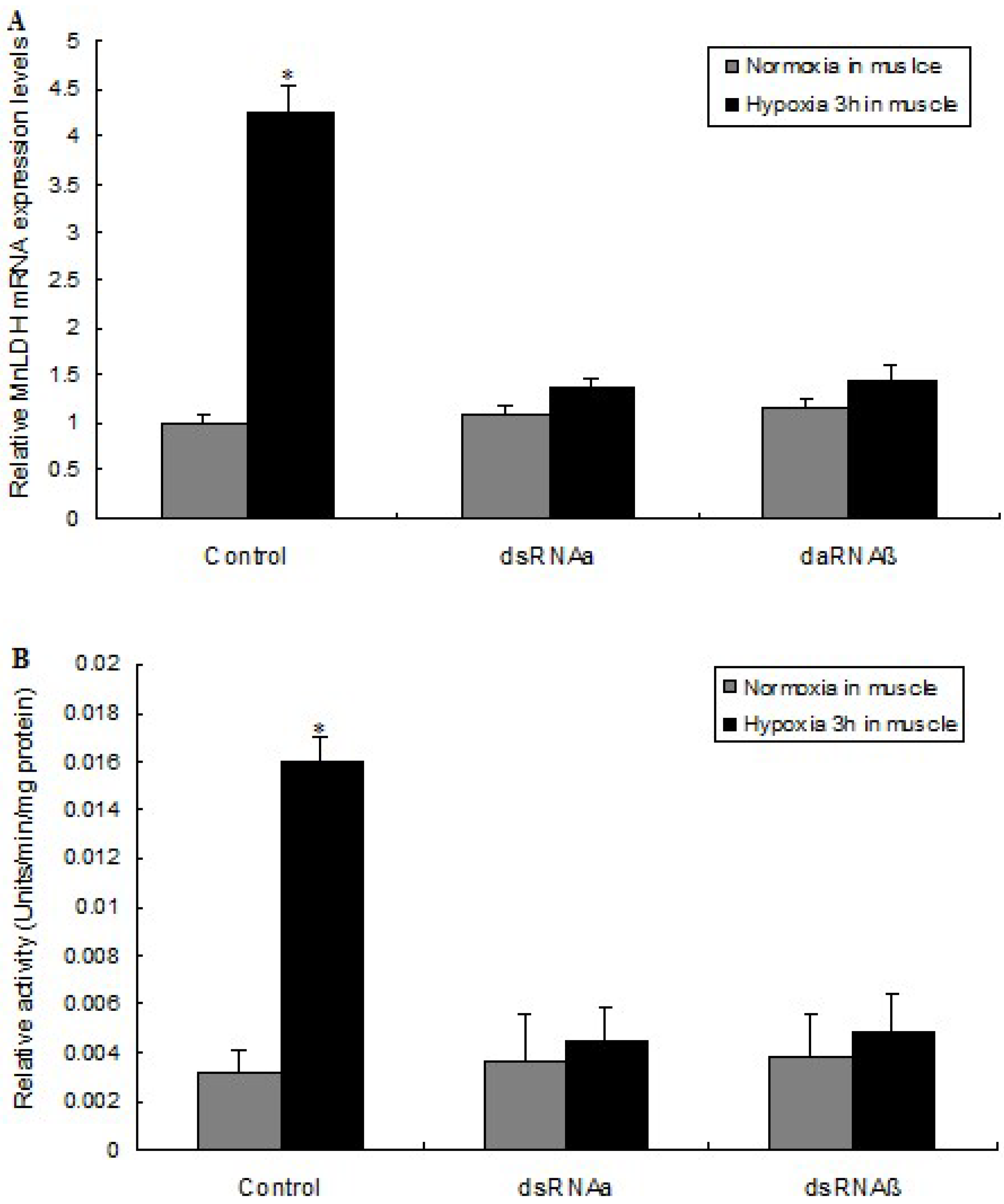
2.5. LDH mRNA Levels and Enzyme Activity Are Affected by HIF-1 Silencing
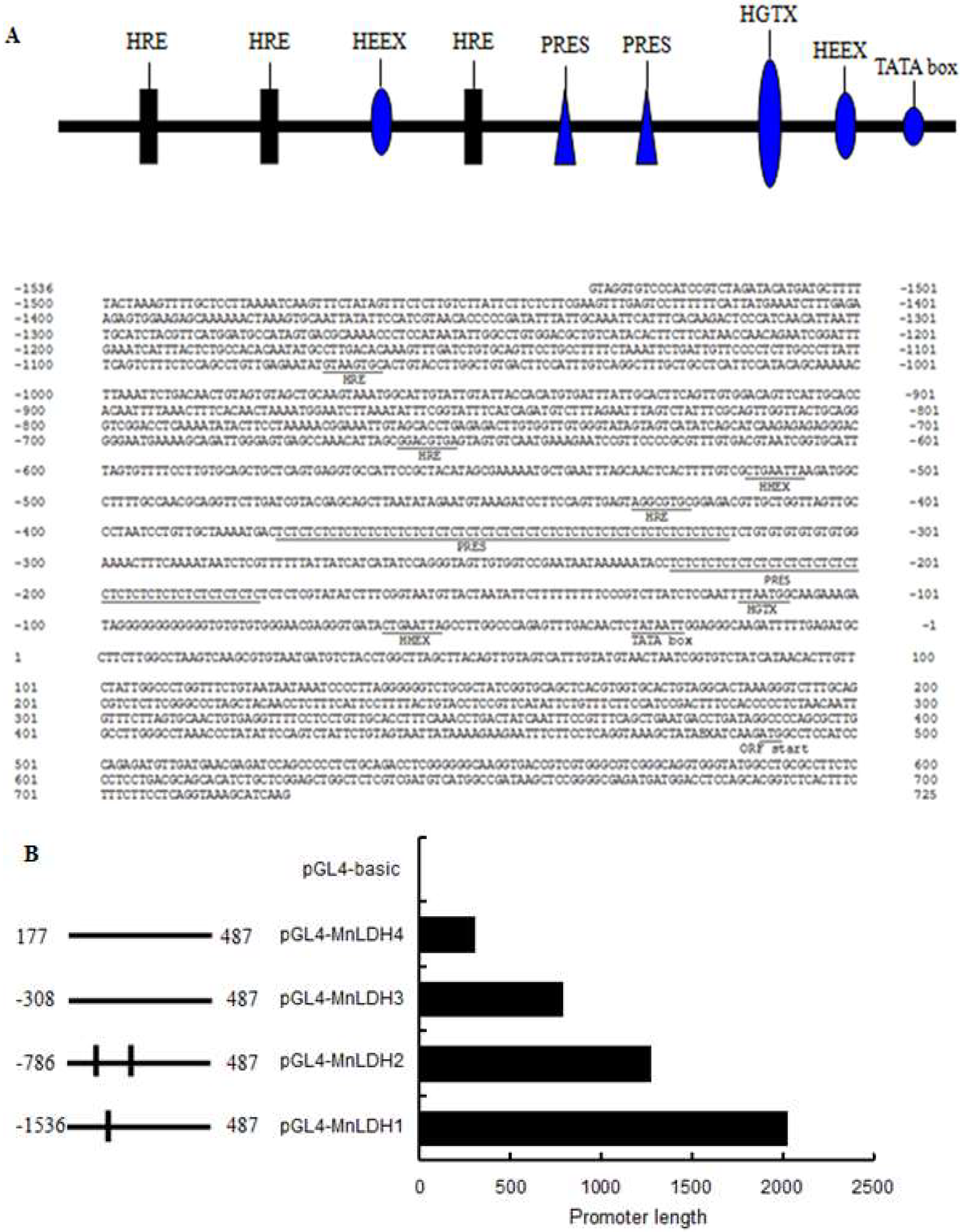
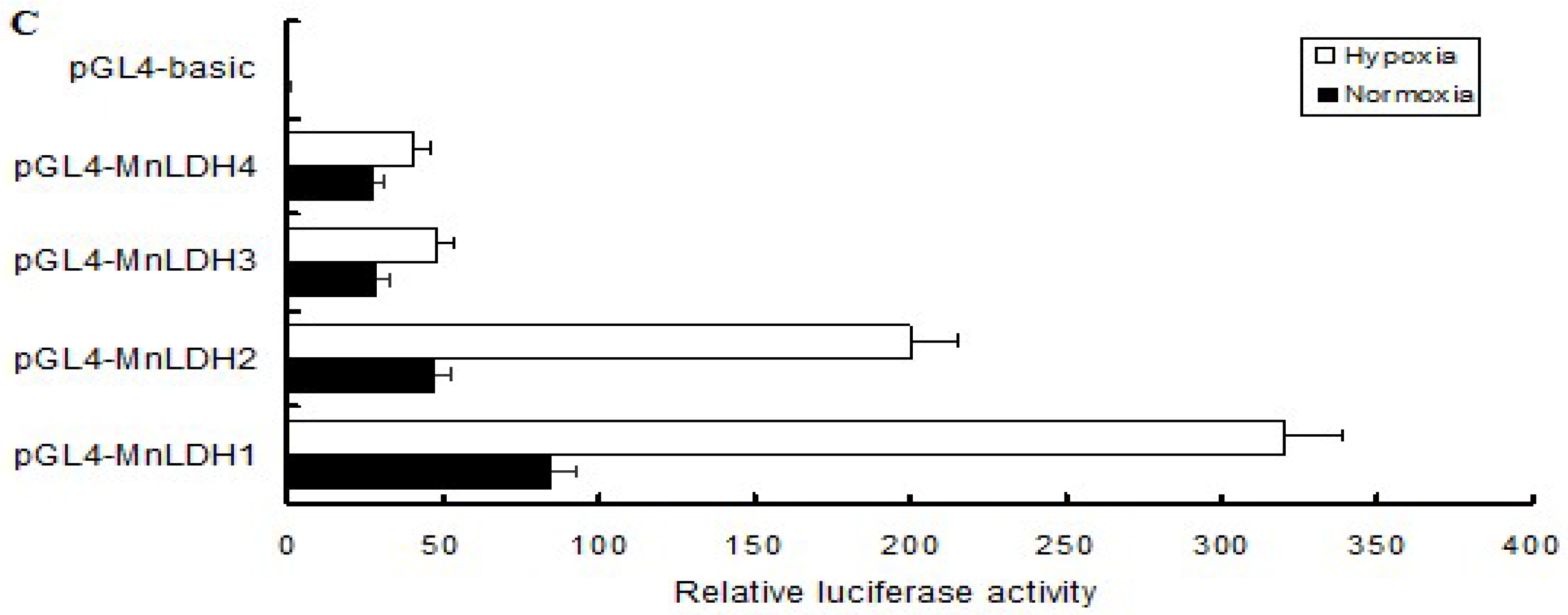
2.6. Analysis of the MnLDH promotor
3. Materials and Methods
3.1. Experimental Animals and Hypoxia Treatment
3.2. Cloning of Complete Sequence and Upstream Sequence of MnLDH
3.3. Expression and Purification of MnLDH and Antibody Production
3.4. Quantitative Real-Time (qRT)-PCR and Western Blotting Analysis of MnLDH Expression of Prawns in Response to Hypoxia
3.5. HIF-1 Silencing and Hypoxia
3.6. Enzymatic Activity Assay
3.7. Construction, Transfection and Activity Assay of Luciferase Plasmids
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| LDH | Lactate dehydrogenase |
| qRT-PCR | Quantitative real-time reverse transcription PCR |
| RACE | Rapid amplification of cDNA ends |
| HIF | Hypoxia inducible factor |
| DO | Dissolved oxygen |
| HRE | Hypoxia response elements |
| SDS | Sodium dodecyl sulfate |
| IPTG | Isopropyl-β-d-galactopyranoside |
| dsRNA | Double-stranded RNA |
| ATP | Adenosine triphosphate |
| EDTA | Ethylenediaminetetraacetic acid |
| PAGE | polyacrylamide gel electrophoresis |
References
- Soñanez-Organis, J.G.; Racotta, I.S.; Yepiz-Plascencia, G. Silencing of the Hypoxia Inducible Factor 1 -HIF-1- obliterates the effects of hipoxia on glucose and lactate concentrations in a tissue-specific manner in the white shrimp Litopenaeus vannamei. J. Exp. Mar. Biol. Ecol. 2010, 393, 51–58. [Google Scholar] [CrossRef]
- Skory, C.D. Isolation and expression of lactate dehydrogenase genes from Rhizopus oryzae. Appl. Environ. Microbiol. 2000, 66, 2343–2348. [Google Scholar] [CrossRef] [PubMed]
- Markert, C.L.; Shaklee, J.B.; Whitt, G.S. Evolution of a gene. Multiple genes for LDH isozymes p rovide a model of the evolution of gene structure, function and regulation. Science 1975, 189, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Scheid, M.J.; Awapara, J. Stereospecificity of some invertebrate lactic dehydrogenases. Comp. Biochem. Physiol. B Comp. Biochem. 1972, 43, 619–626. [Google Scholar] [CrossRef]
- Long, G.L. The stereospecific distribution and evolutionary significance of invertebrate lactate dehydrogenases. Comp. Biochem. Physiol. B Comp. Biochem. 1976, 55, 77–83. [Google Scholar] [CrossRef]
- Eichner, R.D.; Kaplan, N.O. Physical and chemical properties of lactate dehydrogenase in Homarus americanus. Arch. Biochem. Biophys. 1977, 181, 490–500. [Google Scholar] [CrossRef]
- Zietara, M.S.; Gronczewska, J.; Stachowiak, K.; Skorkowski, E.F. Lactate dehydrogenase in abdominal muscle of crayfish Orconectes limosus and shrimp Crangon crangon Decapoda: Crustacea properties and evolutionary relationship. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1996, 114, 395–401. [Google Scholar] [CrossRef]
- Mulkiewicz, E.; Zietara, M.S.; Stachowiak, K.; Skorkowski, E.F. Properties of lactate dehydrogenase from the isopod, Saduria entomon. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000, 126, 337–346. [Google Scholar] [CrossRef]
- Dendinger, J.E. Electrophoretic separation of three dehydrogenases from various tissues of the Atlantic blue crab, Callinectes sapidus. Comp. Biochem. Physiol. B Comp. Biochem. 1980, 66, 431–433. [Google Scholar] [CrossRef]
- Sundt, R.C.; Fevolden, S.E. Homogeneous genetic structure of Meganyctiphanes norvegica (Euphausiacea) in the north-east Atlantic Ocean, as interpreted from allozymic variation. Sarsia 1996, 81, 155–159. [Google Scholar] [CrossRef]
- Mulkiewicz, E.; Zietara, M.S.; Stromberg, J.O.; Skorkowski, E.F. Lactate dehydrogenase from the northern krill Meganyctiphanes norvegica: Comparison with LDH from the Antarctic krill Euphausia superb. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 128, 233–245. [Google Scholar] [CrossRef]
- Fevolden, S.E.; Schneppenheim, R. Genetic population structure of Euphausia superba DANA in the Atlantic Sector of the Southern Ocean as demonstrated by different electrophoretic techniques. Polar Biol. 1988, 9, 1–8. [Google Scholar] [CrossRef]
- Angers, A.; Pothier, F.; Sevigny, J.M.; Sainte-Marie, B. Tissue specificity and ontogeny of lactate dehydrogenase in snow crab, Chionoecetes opilio (Brachyura, Majidae). Comp. Biochem. Physiol. B Comp. Biochem. 1994, 108, 385–395. [Google Scholar] [CrossRef]
- Guan, Y.Q.; Li, L.I.; Wang, H.C.; Wang, Z.L. Effects of hypoxia on respiratory metabolism and antioxidant capability of Macrobrachium nipponense. J. Hebei Univ. 2010, 30, 301–306. [Google Scholar]
- Simon, M.C. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metab. 2006, 3, 150–151. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Guo, Z.B.; Fu, H.T.; Ge, X.P.; Zhu, J.; Gu, Z.M. Based on the metabolomic approach the energy metabolism responses of oriental river prawn Macrobrachium nipponense hepatopancreas to acute hypoxia and reoxygenation. Front. Physiol. 2018, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Guo, Z.B.; Fu, H.T.; Zhu, J.; Ge, X.P.; Wu, X.G. Hypoxia induces changes in AMP-activated protein kinase activity and energy metabolism in muscle tissue of the oriental river prawn Macrobrachium nipponense. Front. Physiol. 2018, 9, 751. [Google Scholar] [CrossRef] [PubMed]
- Almeidaval, V.M.F.; Oliveira, A.R.; Silva, M.D.N.P.D.; Ferreira-Nozawa, M.S.; Araújo, R.M.; Val, A.L.; Nozawa, S.R. Anoxia-and hypoxia-induced expression of LDH-A* in the Amazon Oscar. Astronotus crassipinis. Genet. Mol. Biol. 2011, 34, 315–322. [Google Scholar] [CrossRef]
- Soñanez-Organis, J.G.; Rodriguez-Armenta, M.; Leal-Rubio, B.; Peregrino-Uriarte, A.B.; Gómez-Jiménez, S.; Yepiz-Plascencia, G. Alternative splicing generates two lactate dehydrogenase subunits differentially expressed during hypoxia via HIF-1 in the shrimp Litopenaeus vannamei. Biochimie 2012, 94, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.L.; Constantino, H.R.; Powers, D.A. Lactate dehydrogenase-B cDNA from the teleost Fundulus heteroclitus: Evolutionary implications. Mol. Biol. 1998, 6, 369–383. [Google Scholar]
- Scislowski, P.W.D.; Biegniewska, A.; Zydowo, M. Lactate dehydrogenase from the abdomen and heart muscle of the crayfish Orconectes limosus. Comp. Biochem. Physiol. B Comp. Biochem. 1982, 7, 697–699. [Google Scholar] [CrossRef]
- Thebault, M.T. Interacting effect of temperature and pH on the catalytic properties of lactate dehydrogenase from Palaemon serratus. Mol. Physiol. 1983, 3, 331–338. [Google Scholar]
- Huang, H.W.; Liu, T.Z.; Lee, K.H.; Tu, C.F.; Lee, W.C.; Shimogiri, T.; Mannen, H.; Li, S.S.L. cDNA cloning of pig testicular lactate dehydrogenase-C, thermal stability of the expressed enzyme, and polymorphism among strains. Gene 2000, 242, 151–154. [Google Scholar] [CrossRef]
- Singh, V.; Kaushal, D.C.; Rathaur, S.; Kumar, N.; Kaushal, N.A. Cloning, overexpression, purification and characterization of Plasmodium knowlesi lactate dehydrogenase. Protein Expr. Purif. 2012, 84, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Cayenne, A.P.; Gabert, B.; Stillman, J.H. Identification of proteins interacting with lactate dehydrogenase in claw muscle of the porcelain crab Petrolisthes cinctipes. Comp. Biochem. Physiol. D Genom. Proteom. 2011, 6, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, F.; Solares, M.; Balanza, E.; Coudert, J.; Clottes, E. Expression of lactate dehydrogenase A and B genes in different tissues of rats adapted to chronic hypobaric hypoxia. J. Cell. Biochem. 2003, 89, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.J.; Rosenberg, R. Spreading dead zones and consequences for marine ecosystems. Science 2008, 321, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Soñanez-Organis, J.G.; Peregrino-Uriarte, A.B.; Sotelo-Mundo, R.R.; Forman, H.J.; Yepiz-Plascencia, G. Hexokinase from the white shrimp Litopenaeus vannamei: CDNA sequence, structural protein model and regulation via HIF-1 in response to hypoxia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Cota-Ruiz, K.; Leyva-Carrillo, L.; Peregrino-Uriarte, A.B.; Valenzuela-Soto, E.M.; Gollas-Galván, T.; Gómez-Jiménez, S.; Hernández, J.; Yepiz-Plascencia, G. Role of HIF-1 on phosphofructokinase and fructose 1, 6-bisphosphatase expression during hypoxia in the white shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2016, 198, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cota-Ruiz, K.; Peregrino-Uriarte, A.B.; Felix-Portillo, M.; Martínez-Quintana, J.A.; Yepiz-Plascencia, G. Expression of fructose 1, 6-bisphosphatase and phosphofructokinase is induced in hepatopancreas of the white shrimp Litopenaeus vannamei by hypoxia. Mar. Environ. Res. 2015, 106, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Xuan, F.J.; Fu, H.T.; Ge, X.P.; Zhu, J.; Qiao, H.; Jin, S.B.; Zhang, W.Y. Molecular characterization and mRNA expression of hypoxia inducible factor-1 and cognate inhibiting factor in Macrobrachium nipponense in response to hypoxia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 196–197, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L.; Roth, P.H.; Fang, H.M.; Wang, G.L. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994, 269, 23757–23763. [Google Scholar] [PubMed]
- Firth, J.D.; Ebert, B.L.; Ratcliffe, P.J. Hypoxic regulation of lactate dehydrogenase a interaction between hypoxia-inducible factor 1 and cAMP response elements. J. Biol. Chem. 1995, 270, 21021–21027. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.J.; Andrews, G.K.; Bittel, D.; Discher, D.J.; McCue, J.; Green, C.J.; Yanovsky, M.; Giaccia, A.; Sutherland, R.M.; Laderoute, K.R.; et al. Activation of metallithionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 1999, 59, 1315–1322. [Google Scholar] [PubMed]
- Discher, D.J.; Bishopric, N.H.; Wu, X.; Peterson, C.A.; Webster, K.A. Hypoxia regulates beta-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. J. Biol. Chem. 1998, 273, 26087–26093. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L.; Jiang, B.H.; Leung, S.W.; Passantino, R.; Concordet, J.P.; Maire, P.; Giallongo, A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 32529–32537. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Kawamoto, T.; Tanimoto, K.; Nishiyama, M.; Honda, H. Identification of Functional Hypoxia Response Elements in the Promoter Region of the DEC1 and DEC2 Genes. J. Biol. Chem. 2002, 277, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, O.; Opentanova, I.; Minchenko, D.; Ogura, T.; Esumi, H. Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase-4 gene via hypoxia-inducible factor-1α activation. FEBS Lett. 2004, 576, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.T.; Chua, M.S.; Whitlock, J.P., Jr.; Shin, Y.C.; Song, W.H.; Kim, Y.; Eom, C.Y.; An, W.G. Rodent-specific hypoxia response elements enhance PAI-1 expression through HIF-1 or HIF-2 in mouse hepatoma cells. Int. J. Oncol. 2010, 37, 1627–1638. [Google Scholar] [PubMed]
- Hisada, T.; Ayaori, M.; Ohrui, N.; Nakashima, H.; Nakaya, K.; Uto-Kondo, H.; Yakushiji, E.; Takiguchi, S.; Terao, Y.; Miyamoto, Y.; et al. Statin inhibits hypoxia-induced endothelin-1 via accelerated degradation of HIF-1α in vascular smooth muscle cells. Cardiovasc. Res. 2012, 95, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Salvi, A.; Thanabalu, T. Expression of N-WASP is regulated by HiF1α through the hypoxia response element in the N-WASP promoter. Biochem. Biophys. Rep. 2016, 9, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Xuan, F.J.; Ge, X.P.; Fu, H.T.; Zhu, J.; Zhang, S.Y. Identification of differentially expressed genes in hepatopancreas of oriental river prawn, Macrobrachium nipponense exposed to environmental hypoxia. Gene 2014, 534, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.Q.; Zhou, T.T.; Yang, S.P.; Chan, S.F. Characterization of a molt-related myostatin gene (FmMstn) from the banana shrimp Fenneropenaeus merguiensis. Gen. Comp. Endocr. 2017, 248, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Xuan, F.J.; Fu, H.T.; Zhu, J.; Ge, X.P. Molecular cloning and functional characterization of a hexokinase from the oriental river prawn. Macrobrachium nipponense in response to hypoxia. Int. J. Mol. Sci. 2017, 18, 1256. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Gu, Z.M.; Fu, H.T.; Zhu, J.; Ge, X.P.; Xuan, F.J. Molecular cloning, characterization, and expression analysis of p53 from the oriental river prawn, Macrobrachium nipponense, in response to hypoxia. Fish Shellfish Immunol. 2016, 54, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Xiong, Y.W.; Zhang, W.Y.; Fu, H.G.; Jiang, S.F.; Sun, S.M.; Bai, H.K.; Jin, S.B.; Gong, Y.S. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense and its induced expression by temperature. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 185, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.K.; Qiao, H.; Li, F.J.; Fua, H.Y.; Jiang, S.F.; Zhang, W.Y.; Yan, Y.D.; Xiong, Y.W.; Sun, S.M.; Jin, S.B.; et al. Molecular and functional characterization of the vitellogenin receptor in oriental river prawn, Macrobrachium nipponense. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2016, 194, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Babson, A.L.; Babson, S.R. Kinetic colorimetric measurement of serum lactate dehydrogenase activity. Clin. Chem. 1973, 19, 766–769. [Google Scholar] [PubMed]
- Lai, X.F.; Gao, H.; Kong, J.; Wang, Q.Y.; Meng, X.H.; Yan, B.L.; Chen, B.Y. Genomic organization and promoter characterization of FcCTL, a C-type lectin-like protein in Fenneropenaeus chinensis. Gene 2013, 518, 376–380. [Google Scholar] [CrossRef] [PubMed]

| Primer | Primer Sequence (5′-3′) |
|---|---|
| MnLDH-F1 (5′-RACE out primer) | GTTCTCGCGTCTGCATTGTG |
| MnLDH-F2 (5′-RACE in primer) | TACGTGGCCTGGAAACTGTC |
| MnLDH-R1 (3′-RACE out primer) | TTGAGGTCACGGAGACGAAC |
| MnLDH-R2 (3′-RACE in primer) | TTGGTTCCAGAGCCGATGAC |
| MnLDH-F (real-time primer) | CTGTCCCAGTATGGTCAGGC |
| MnLDH-R (real-time primer) | CCGCATACACACTCCTCTGG |
| GSP-R1 (genome walking) | GGAAAGTGAGACCGTGCT |
| GSP-R2 (genome walking) | GAGCTTATCGGCCATGAC |
| GSP-R3 (genome walking) | GGGCTGGATCTCGTTCAT |
| P1-F (promoter activity) −1536~487 | CATTTCTCTGGCCTAACTGGCCGGTACCGTAGGTGTCCCATCCGTCTAGATAC |
| P1-R (promoter activity) −1536~487 | CGAGGCCAGATCTTGATATCCTCGAGCTTGATGCTTTACCTGAGGAAG |
| P2-F (promoter activity) −786~487 | CATTTCTCTGGCCTAACTGGCCGGTACCTATACTTCCTAAAAACGGAAATTG |
| P2-R (promoter activity) −786~487 | CGAGGCCAGATCTTGATATCCTCGAGCTTGATGCTTTACCTGAGGAAG |
| P3-F (promoter activity) −308~487 | CATTTCTCTGGCCTAACTGGCCGGTACCGTGTGTGGAAAACTTTCAAAATAATC |
| P3-R (promoter activity) −308~487 | CGAGGCCAGATCTTGATATCCTCGAGCTTGATGCTTTACCTGAGGAAG |
| P4-F (promoter activity) 177~487 | CATTTCTCTGGCCTAACTGGCCGGTACCGCACTAAAGGGTCTTTGCAG |
| P4-R (promoter activity) 177~487 | CGAGGCCAGATCTTGATATCCTCGAGCTTGATGCTTTACCTGAGGAAG |
| MnpLDH CDS amplification (BamHI) | CGGGATCCATGGCCTCCATCCTAGAGATG |
| MnpLDH CDS amplification (XhoI) | CCGCTCGAGGAACTGAATTCCCGCCTGGAC |
| β-Actin F (real-time primer) | TATGCACTTCCTCATGCCATC |
| β-Actin R (real-time primer) | AGGAGGCGGCAGTGGTCAT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Fu, H.; Zhu, J.; Ge, X.; Wu, X.; Qiao, H.; Jin, S.; Zhang, W. Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia. Int. J. Mol. Sci. 2018, 19, 1990. https://doi.org/10.3390/ijms19071990
Sun S, Fu H, Zhu J, Ge X, Wu X, Qiao H, Jin S, Zhang W. Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia. International Journal of Molecular Sciences. 2018; 19(7):1990. https://doi.org/10.3390/ijms19071990
Chicago/Turabian StyleSun, Shengming, Hongtuo Fu, Jian Zhu, Xianping Ge, Xugan Wu, Hui Qiao, Shubo Jin, and Wenyi Zhang. 2018. "Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia" International Journal of Molecular Sciences 19, no. 7: 1990. https://doi.org/10.3390/ijms19071990
APA StyleSun, S., Fu, H., Zhu, J., Ge, X., Wu, X., Qiao, H., Jin, S., & Zhang, W. (2018). Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia. International Journal of Molecular Sciences, 19(7), 1990. https://doi.org/10.3390/ijms19071990









