Identification and Functional Analysis of the psaD Promoter of Chlorella vulgaris Using Heterologous Model Strains
Abstract
:1. Introduction
2. Results and Discussion
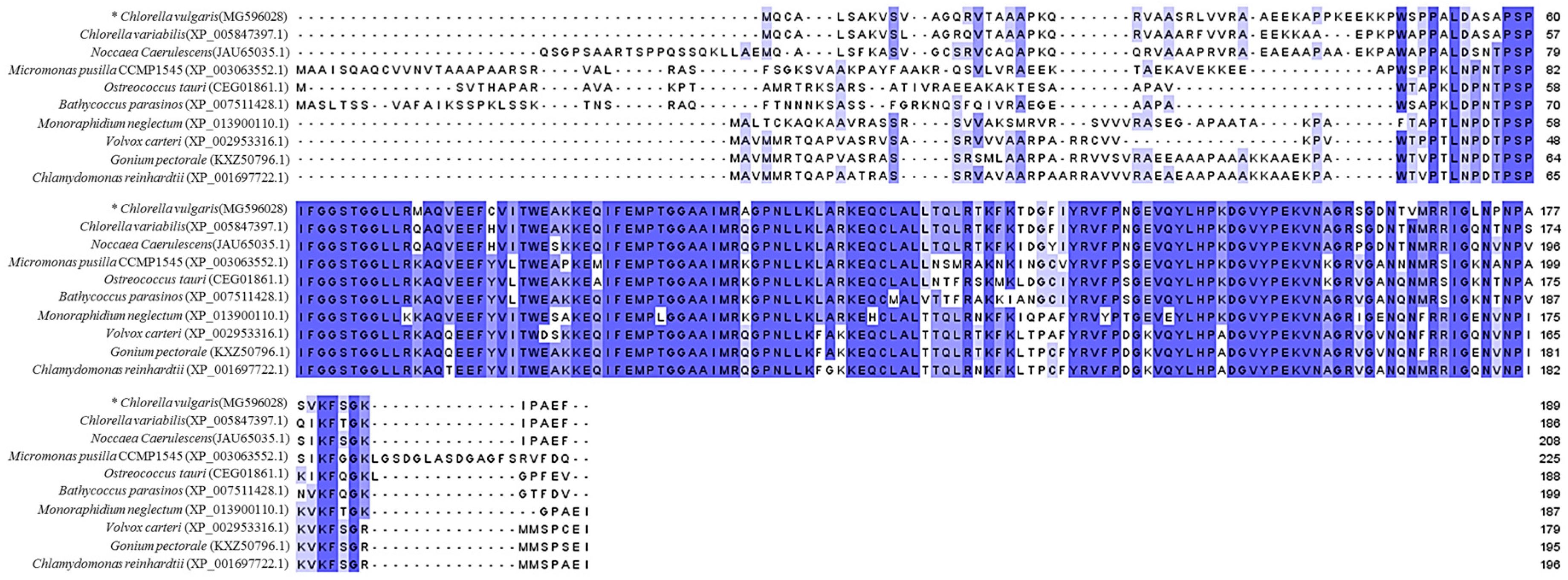
2.1. Isolation and Computational Analysis of CvpsaD Gene and the 5′ Upstream Region
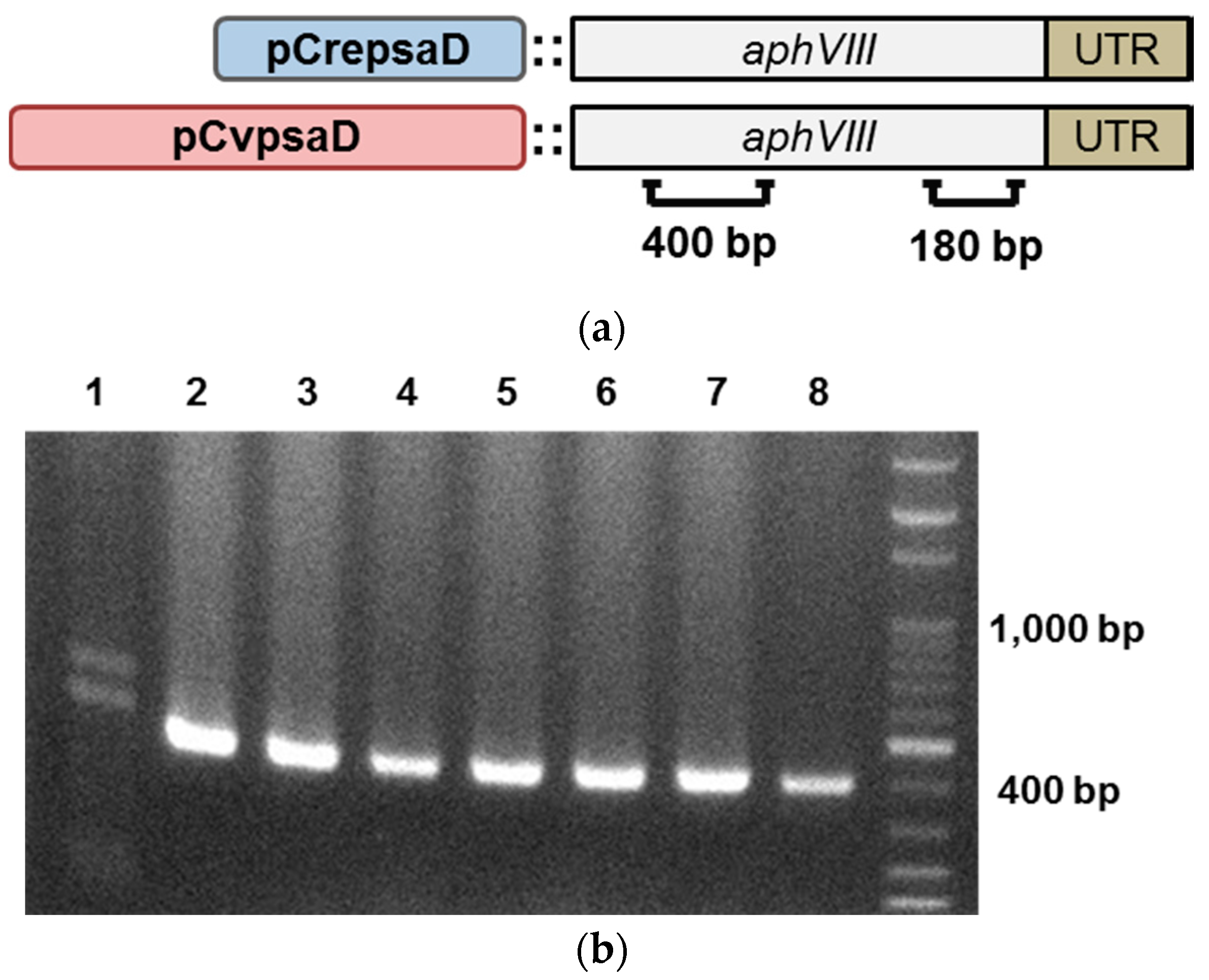
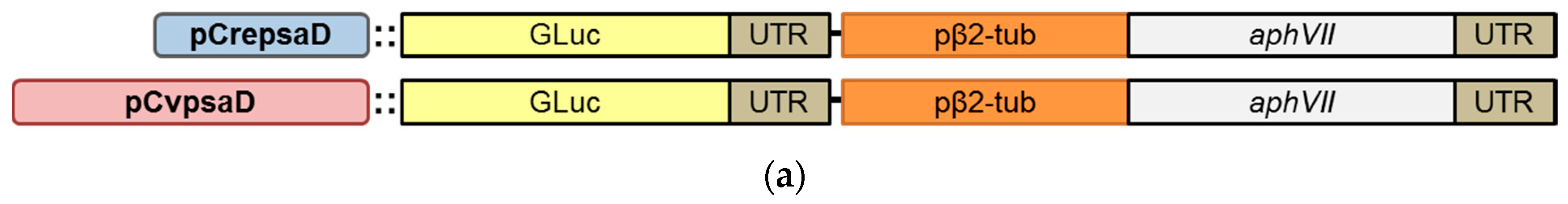
2.2. Construction of the CvpsaD Promoter Cassettes and Validation of Its Function in C. reinhardtii
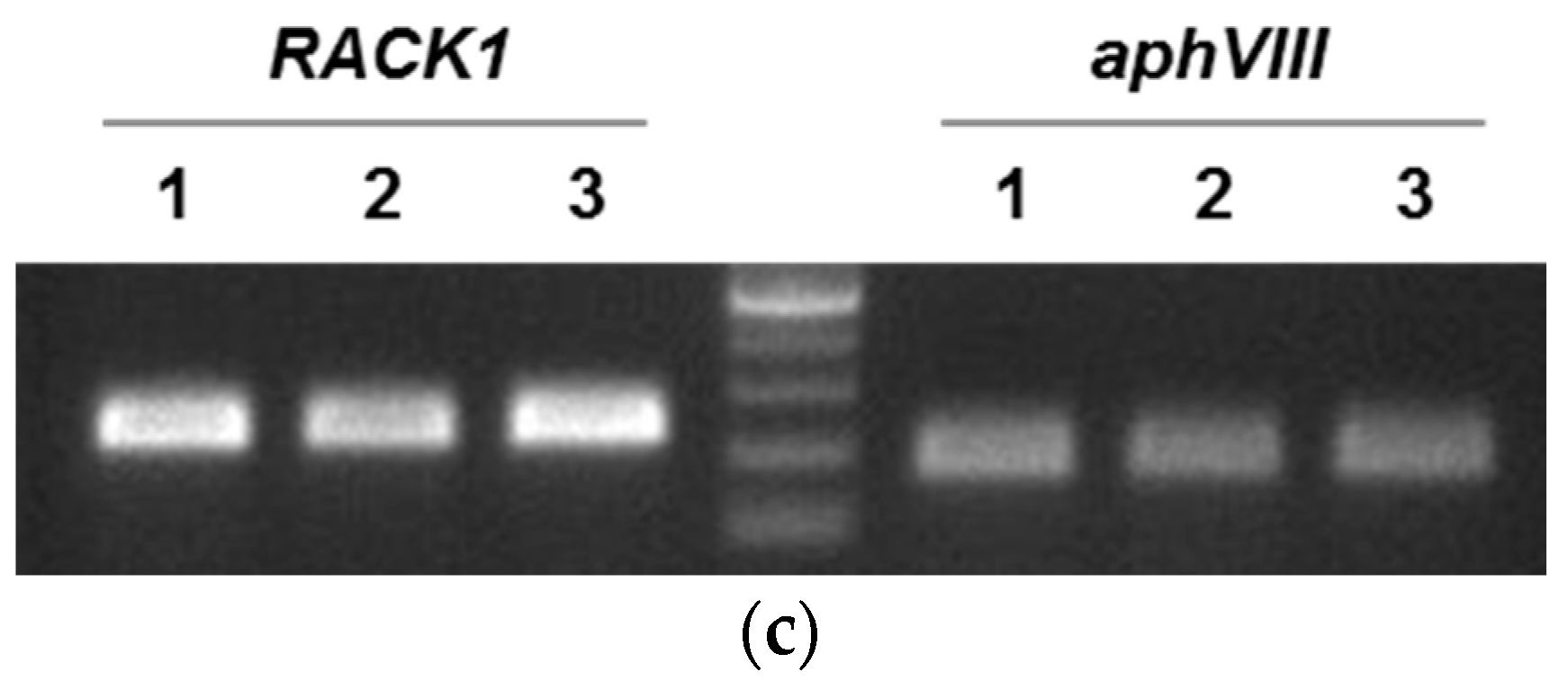
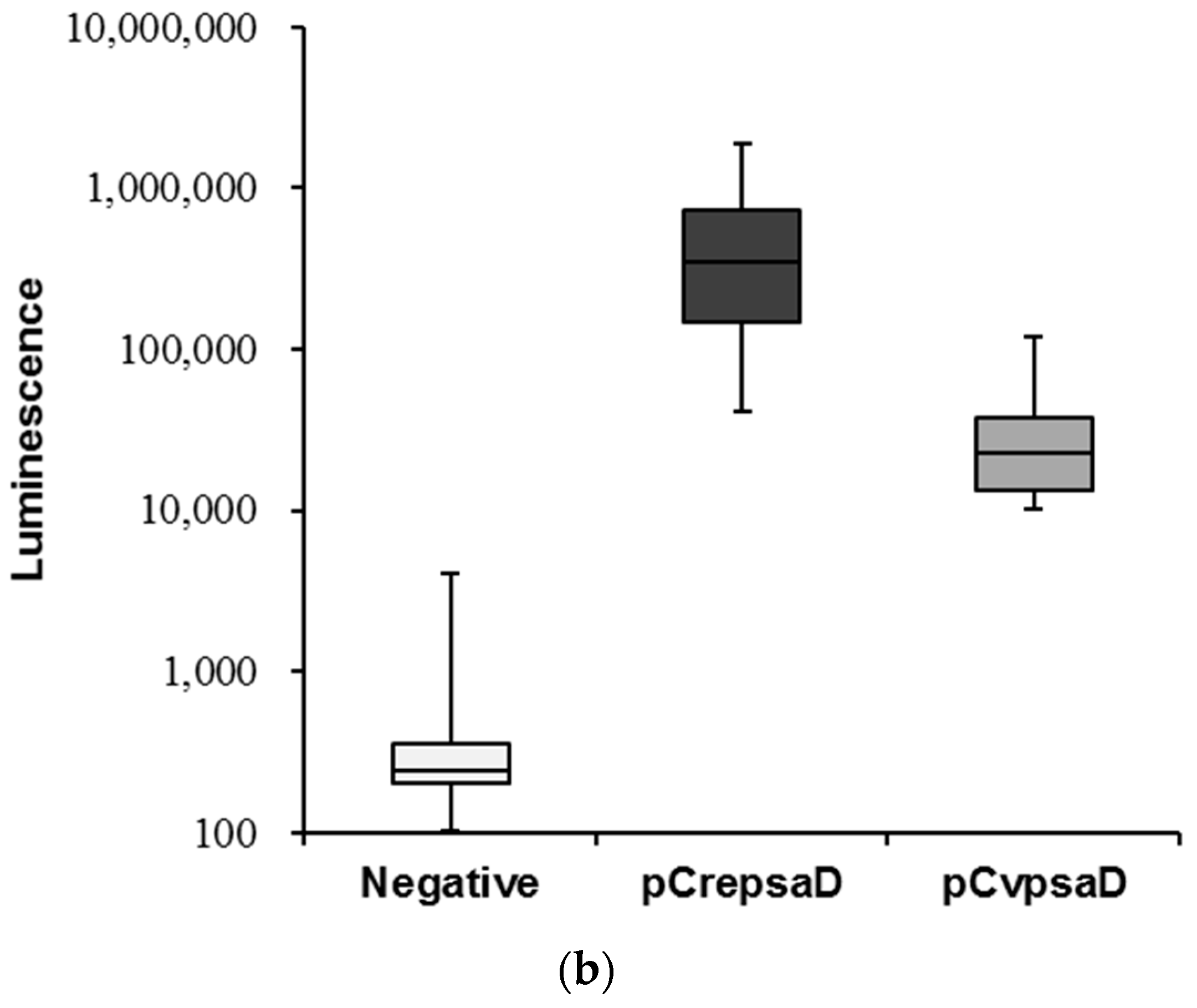
2.3. Validation of the CvpsaD Promoter Efficiency in C. reinhardtii
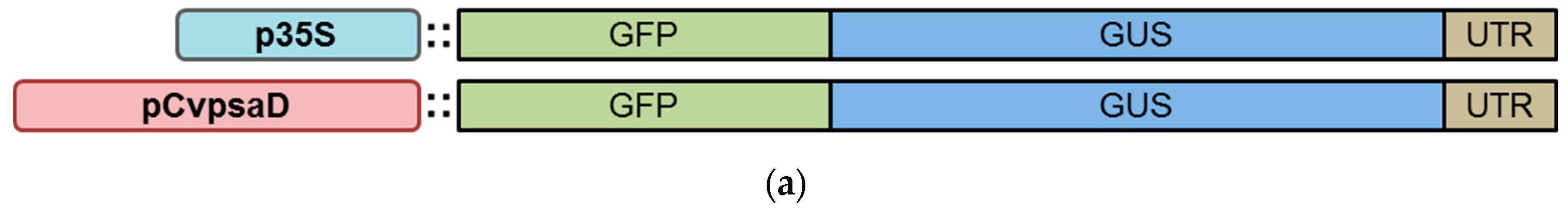
2.4. Confirmation of the CvpsaD Promoter Functionality by Expression of GFP-GUS Activity in Planta
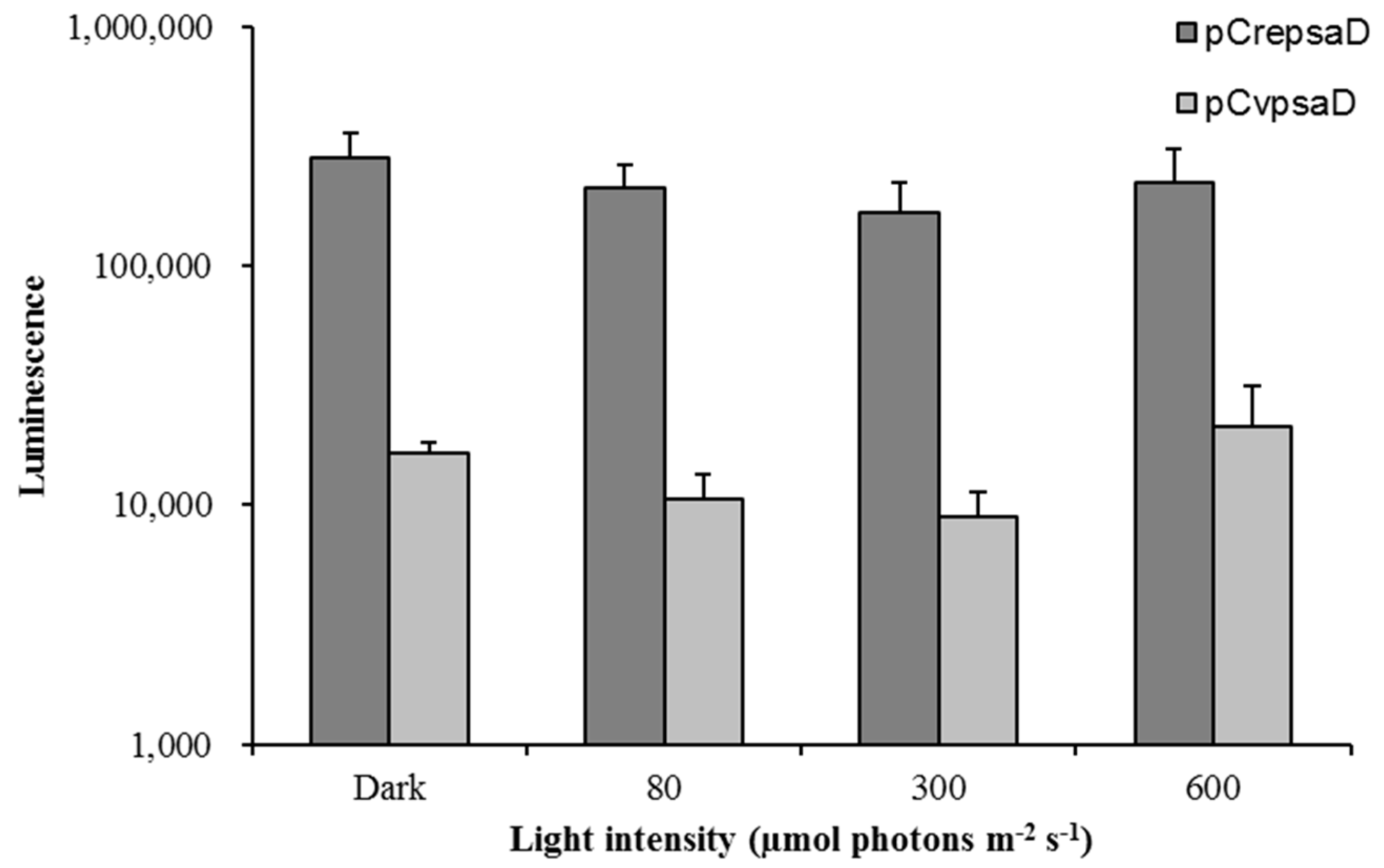
2.5. Light Inducibility of the CvpsaD Promoter under Various Light Conditions
3. Materials and Methods
3.1. Microalgae Cell Culture
3.2. Isolation and Sequence Analysis of Putative CvpsaD Gene
3.3. Expression Vector Construction
3.4. Generation of Transgenic Microalgae
3.5. Functional Analysis of the CvpsaD Promoter in Microalgae
3.5.1. Quantitative Real-Time PCR for Analysis of Transgene RNA Expression
3.5.2. Western Blot Analysis
3.5.3. Luciferase Assay
3.6. Analysis of the Function of the CvpsaD Promoter in Plants
3.7. Analysis of Light Inducibility of the CvpsaD Promoter
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Pulz, O.; Gross, W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 2004, 65, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Leu, S.; Boussiba, S. Advances in the production of high-value products by microalgae. Ind. Biotechnol. 2014, 10, 169–183. [Google Scholar] [CrossRef]
- Yu, X.; Chen, L.; Zhang, W. Chemicals to enhance microalgal growth and accumulation of high-value bioproducts. Front. Microbiol. 2015, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Gimpel, J.A.; Henríquez, V.; Mayfield, S.P. In metabolic engineering of eukaryotic microalgae: Potential and challenges come with great diversity. Front. Microbiol. 2015, 6, 1376. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sarkany, N.; Cui, Y. Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol. Lett. 2009, 31, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, O.; Escalante, F.M.; de-Bashan, L.E.; Bashan, Y. Heterotrophic cultures of microalgae: Metabolism and potential products. Water Res. 2011, 45, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, J.; Jiang, Y.; Chen, F. Chlorella species as hosts for genetic engineering and expression of heterologous proteins: Progress, challenge and perspective. Biotechnol. J. 2016, 11, 1244–1261. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.-C.; Tung, W. Electrotransformation of Chlorella vulgaris. Plant Cell Rep. 1999, 18, 778–780. [Google Scholar] [CrossRef]
- Kim, D.-H.; Kim, Y.T.; Cho, J.J.; Bae, J.-H.; Hur, S.-B.; Hwang, I.; Choi, T.-J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Zhang, Y.; Chen, X.; Zhang, P.; Ma, S. Development of a new method for genetic transformation of the green alga chlorella ellipsoidea. Mol. Biotechnol. 2013, 54, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.L.; Harikrishna, J.A.; Bakar, F.A.; Yeo, C.C.; San Cha, T. Heterologous expression of the streptococcus pneumoniae yoeb and pezt toxin genes is lethal in Chlorella vulgaris. Algal Res. 2016, 19, 21–29. [Google Scholar] [CrossRef]
- San Cha, T.; Yee, W.; Aziz, A. Assessment of factors affecting agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 2012, 28, 1771–1779. [Google Scholar]
- Liu, J.; Sun, Z.; Gerken, H.; Huang, J.; Jiang, Y.; Chen, F. Genetic engineering of the green alga chlorella zofingiensis: A modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol. 2014, 98, 5069–5079. [Google Scholar] [CrossRef] [PubMed]
- Run, C.; Fang, L.; Fan, J.; Fan, C.; Luo, Y.; Hu, Z.; Li, Y. Stable nuclear transformation of the industrial alga chlorella pyrenoidosa. Algal Res. 2016, 17, 196–201. [Google Scholar] [CrossRef]
- Talebi, A.F.; Tohidfar, M.; Tabatabaei, M.; Bagheri, A.; Mohsenpor, M.; Mohtashami, S.K. Genetic manipulation, a feasible tool to enhance unique characteristic of Chlorella vulgaris as a feedstock for biodiesel production. Mol. Biol. Rep. 2013, 40, 4421–4428. [Google Scholar] [CrossRef] [PubMed]
- Thanh, T.; Chi, V.T.Q.; Omar, H.; Abdullah, M.P.; Napis, S. Sequence analysis and potentials of the native rbcs promoter in the development of an alternative eukaryotic expression system using green microalga ankistrodesmus convolutus. Int. J. Mol. Sci. 2012, 13, 2676–2691. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Jeon, H.; Hwang, S.; Jin, E.; Chang, K.S. Development of a new constitutive expression system for the transformation of the diatom phaeodactylum tricornutum. Algal Res. 2015, 11, 50–54. [Google Scholar] [CrossRef]
- Hallmann, A. Algal transgenics and biotechnology. Transgen. Plant J. 2007, 1, 81–98. [Google Scholar]
- Franklin, S.; Ngo, B.; Efuet, E.; Mayfield, S.P. Development of a gfp reporter gene for chlamydomonas reinhardtii chloroplast. Plant J. 2002, 30, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Radakovits, R.; Jinkerson, R.E.; Darzins, A.; Posewitz, M.C. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 2010, 9, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Kucho, K.-I.; Kakoi, K.; Yamaura, M.; Iwashita, M.; Abe, M.; Uchiumi, T. Codon-optimized antibiotic resistance gene improves efficiency of transient transformation in frankia. J. Biosci. 2013, 38, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Bock, R. A codon-optimized luciferase from gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga chlamydomonas reinhardtii. Curr. Genet. 2008, 53, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Lumbreras, V.; Stevens, D.R.; Purton, S. Efficient foreign gene expression in chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998, 14, 441–447. [Google Scholar] [CrossRef]
- Schroda, M.; Blöcker, D.; Beck, C.F. The hsp70a promoter as a tool for the improved expression of transgenes in chlamydomonas. Plant J. 2000, 21, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Falcao, V.R.; Sayre, R.T. Evaluating nuclear transgene expression systems in chlamydomonas reinhardtii. Algal Res. 2013, 2, 321–332. [Google Scholar] [CrossRef]
- Wu, J.; Hu, Z.; Wang, C.; Li, S.; Lei, A. Efficient expression of green fluorescent protein (gfp) mediated by a chimeric promoter in chlamydomonas reinhardtii. Chin. J. Oceanol. Limnol. 2008, 26, 242–247. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Georgianna, D.R.; Lofgren, S.M.; Li, D.; Ellis, R.C.; Carruthers, D.N.; Dräger, A.; Masica, D.L.; Mayfield, S.P. Synthetic promoters capable of driving robust nuclear gene expression in the green alga chlamydomonas reinhardtii. Algal Res. 2016, 15, 135–142. [Google Scholar] [CrossRef]
- Baek, K.; Lee, Y.; Nam, O.; Park, S.; Sim, S.J.; Jin, E. Introducing dunaliella lip promoter containing light-inducible motifs improves transgenic expression in chlamydomonas reinhardtii. Biotechnol. J. 2016, 11, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Flieger, K.; Wicke, A.; Herrmann, R.; Oelmüller, R. Promoter and leader sequences of the spinach psad and psaf genes direct an opposite light response in tobacco cotyledons: Psad sequences downstream of the atg codon are required for a positive light response. Plant J. 1994, 6, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, V.P.; Ke, A.; Chitnis, P.R. The psad subunit of photosystem i (mutations in the basic domain reduce the level of psad in the membranes). Plant Physiol. 1997, 115, 1699–1705. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Rochaix, J.-D. The flanking regions of psad drive efficient gene expression in the nucleus of the green alga chlamydomonas reinhardtii. Mol. Genet. Genom. 2001, 265, 888–894. [Google Scholar] [CrossRef]
- Nakamura, M.; Tsunoda, T.; Obokata, J. Photosynthesis nuclear genes generally lack tata-boxes: A tobacco photosystem i gene responds to light through an initiator. Plant J. 2002, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Yu, Y.; Fu, Y.; Foley, J.; Halees, A.; Weng, Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of yy1. Genome Res. 2007, 17, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; Kadonaga, J.T. The rna polymerase ii core promoter: A key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Wang, Y.; Hou, H.; Qian, Y. Characterization of sequence elements from malvastrum yellow vein betasatellite regulating promoter activity and dna replication. Virol. J. 2012, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, S.; Das, S.G.; Chakraborty, H.J.; Gupta, S.; Datta, A. Identification of regulatory sequence signatures in microrna precursors implicated in neurological disorders. Adv. Biosci. Biotechnol. 2013, 4, 26. [Google Scholar] [CrossRef]
- Li, T.; Gong, C.; Wang, T. The rice light-regulated gene ra68 encodes a novel protein interacting with oxygen-evolving complex psbo mature protein. Plant Mol. Biol. Rep. 2010, 28, 136. [Google Scholar] [CrossRef]
- Teakle, G.R.; Manfield, I.W.; Graham, J.F.; Gilmartin, P.M. Arabidopsis thaliana gata factors: Organisation, expression and dna-binding characteristics. Plant Mol. Biol. 2002, 50, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.E.; Quail, P.H. Identification of promoter motifs involved in the network of phytochrome a-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003, 133, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, Y.; Lee, J.H.; Jin, E. Expression of the high light-inducible dunaliella lip promoter in chlamydomonas reinhardtii. Planta 2013, 238, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; López, R.C.; Gómez-Gómez, L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron’s apocarotenoid precursors. J. Exp. Bot. 2009, 61, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cui, Y.; Gan, Z.; Shi, C.; Shi, X. Isolation and analysis of the cppsy gene and promoter from chlorella protothecoides cs-41. Mar. Drugs 2015, 13, 6620–6635. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H. Pathogen-and nacl-induced expression of the scam-4 promoter is mediated in part by a gt-1 box that interacts with a gt-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. Wrky transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Yu, G.; Zhang, X.; Jia, C.; Qin, J.; Pan, H. Functional analysis of the maize c-repeat/dre motif-binding transcription factor cbf3 promoter in response to abiotic stress. Int. J. Mol. Sci. 2015, 16, 12131–12146. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.-L.; Gou, W.; Han, X.-L.; Qin, C.; Zhang, L.-X.; Abomohra, A.E.-F.; Ashraf, M. Cloning and functional analysis of phosphoethanolamine methyltransferase promoter from maize (Zea mays L.). Int. J. Mol. Sci. 2018, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Berthold, P.; Schmitt, R.; Mages, W. An engineered streptomyces hygroscopicus aph 7 gene mediates dominant resistance against hygromycin b in chlamydomonas reinhardtii. Protist 2002, 153, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Ma, L.; Strickland, E.; Deng, X.W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and arabidopsis. Plant Cell 2005, 17, 3239–3256. [Google Scholar] [CrossRef] [PubMed]
- Kindle, K.L. High-frequency nuclear transformation of chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [PubMed]
- Shtaida, N.; Khozin-Goldberg, I.; Solovchenko, A.; Chekanov, K.; Didi-Cohen, S.; Leu, S.; Cohen, Z.; Boussiba, S. Downregulation of a putative plastid pdc e1α subunit impairs photosynthetic activity and triacylglycerol accumulation in nitrogen-starved photoautotrophic chlamydomonas reinhardtii. J. Exp. Bot. 2014, 65, 6563–6576. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A. Assaying chimeric genes in plants: The gus gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]


| No | Name | Sequence | Function |
|---|---|---|---|
| 1 | BRE | GGGCGCC | TFIIB recognition element |
| 2 | CAAT box | GGCAAT CAAT CAAAT | Common cis-acting element in promoter and enhancer regions |
| 3 | CCAAT box | CAACGG | MYBHv1 binding site |
| 4 | CATT box | GCATTC | Part of light-responsive element |
| 5 | CCCAT motif | CCCAT | Putative core element, sap |
| 6 | CGTCA-motif | CGTCA | MeJA-responsiveness |
| 7 | DPE | GGCACC | Downstream promoter element, requires an Inr |
| 8 | G-box | CACGTG | cis-acting regulatory element involved in light responsiveness |
| 9 | GATA box | GATA | Binding with ASF-2, required for high level, light-regulated, and tissue-specific expression |
| 10 | GT-1 motif | GATAAT GAAAA | Pathogen- and salt-induced element |
| 11 | Inr | CTCATTCC | Initiator element, core promoter element |
| 12 | MNF1 | GTGCCCTAAAGCTGCA | Light-responsive element |
| 13 | SORLIP1AT/2AT | GCCAC GGGCC | Light-responsive elements |
| 14 | Sp1 | CCACCC | Light-responsive element |
| 15 | TATA box | TATA | Core promoter element around −30 of transcription start |
| 16 | TGACG-motif | TGACG | MeJA-responsiveness |
| 17 | W box | TGACT TTGAC | cis-regulatory element, recognized by WRKY transcription factors, stress responsive |
| 18 | 5’ untranslated region (UTR) Py-rich stretch | TTTCTTCTGT | Confers high transcription levels without the need for other upstream cis-acting elements |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Liu, L.; Hu, Z.; Jin, E. Identification and Functional Analysis of the psaD Promoter of Chlorella vulgaris Using Heterologous Model Strains. Int. J. Mol. Sci. 2018, 19, 1969. https://doi.org/10.3390/ijms19071969
Kim J, Liu L, Hu Z, Jin E. Identification and Functional Analysis of the psaD Promoter of Chlorella vulgaris Using Heterologous Model Strains. International Journal of Molecular Sciences. 2018; 19(7):1969. https://doi.org/10.3390/ijms19071969
Chicago/Turabian StyleKim, Jongrae, Linpo Liu, Zanmin Hu, and EonSeon Jin. 2018. "Identification and Functional Analysis of the psaD Promoter of Chlorella vulgaris Using Heterologous Model Strains" International Journal of Molecular Sciences 19, no. 7: 1969. https://doi.org/10.3390/ijms19071969
APA StyleKim, J., Liu, L., Hu, Z., & Jin, E. (2018). Identification and Functional Analysis of the psaD Promoter of Chlorella vulgaris Using Heterologous Model Strains. International Journal of Molecular Sciences, 19(7), 1969. https://doi.org/10.3390/ijms19071969





