The Crystal Structure of a hCA VII Variant Provides Insights into the Molecular Determinants Responsible for Its Catalytic Behavior
Abstract
1. Introduction
2. Results
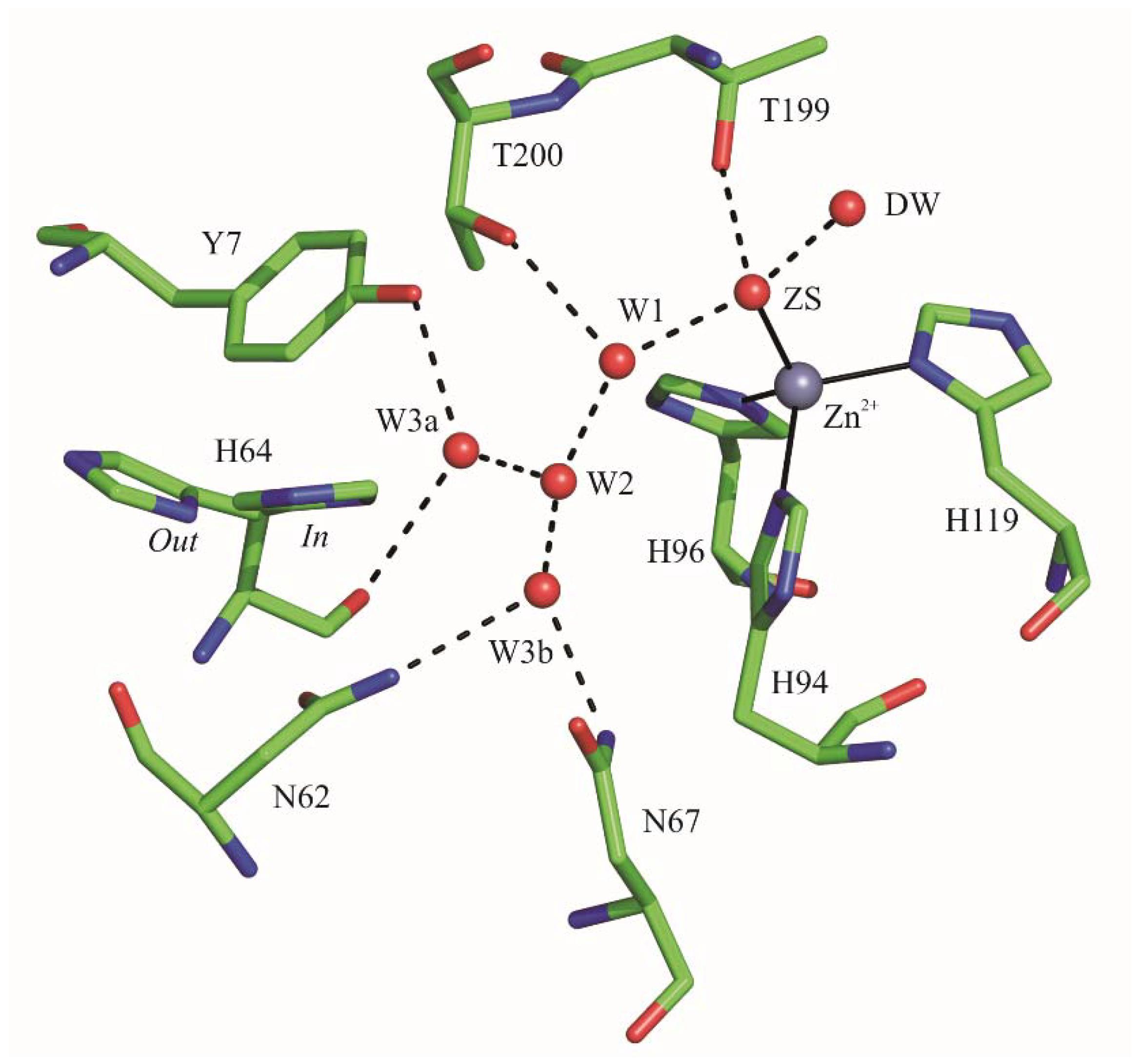
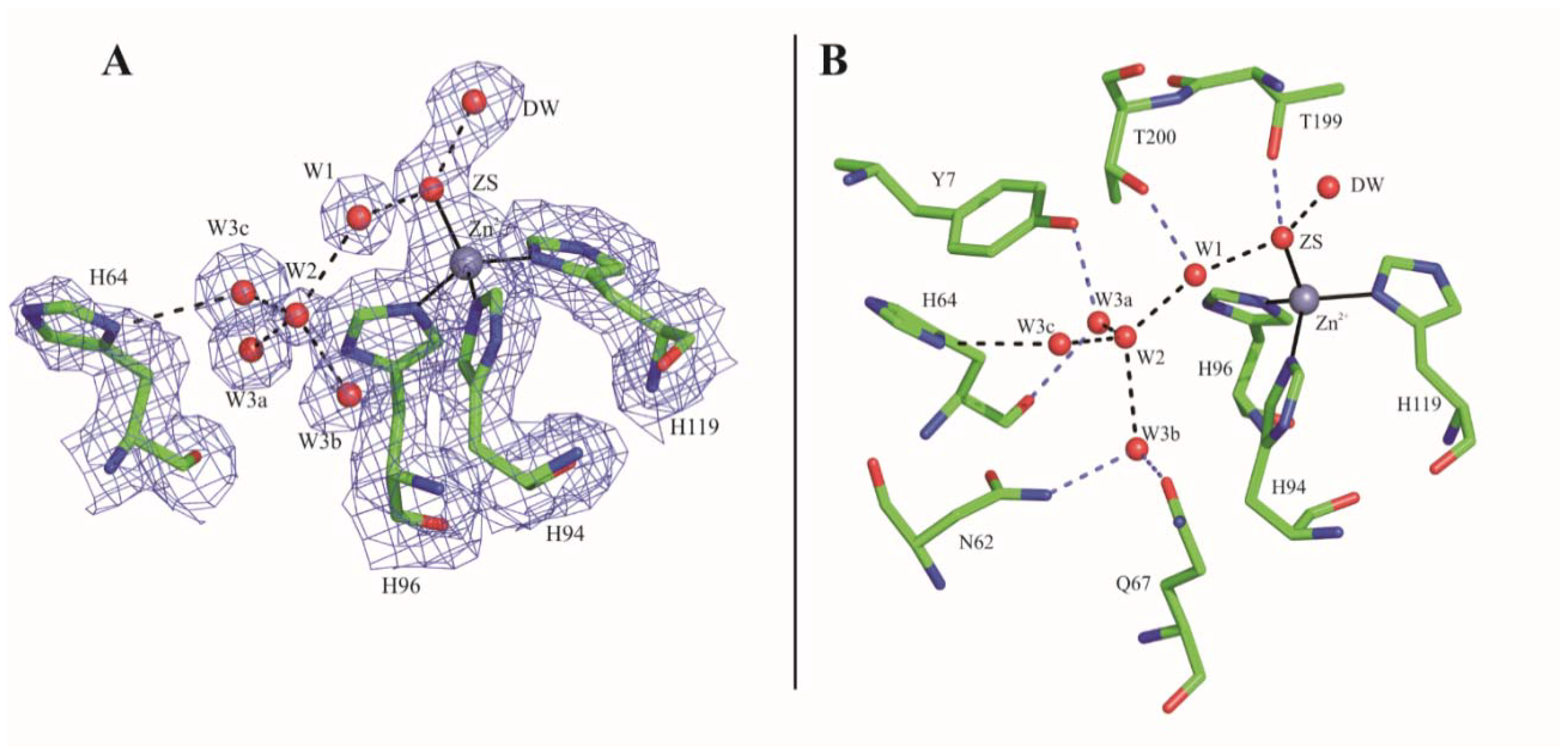
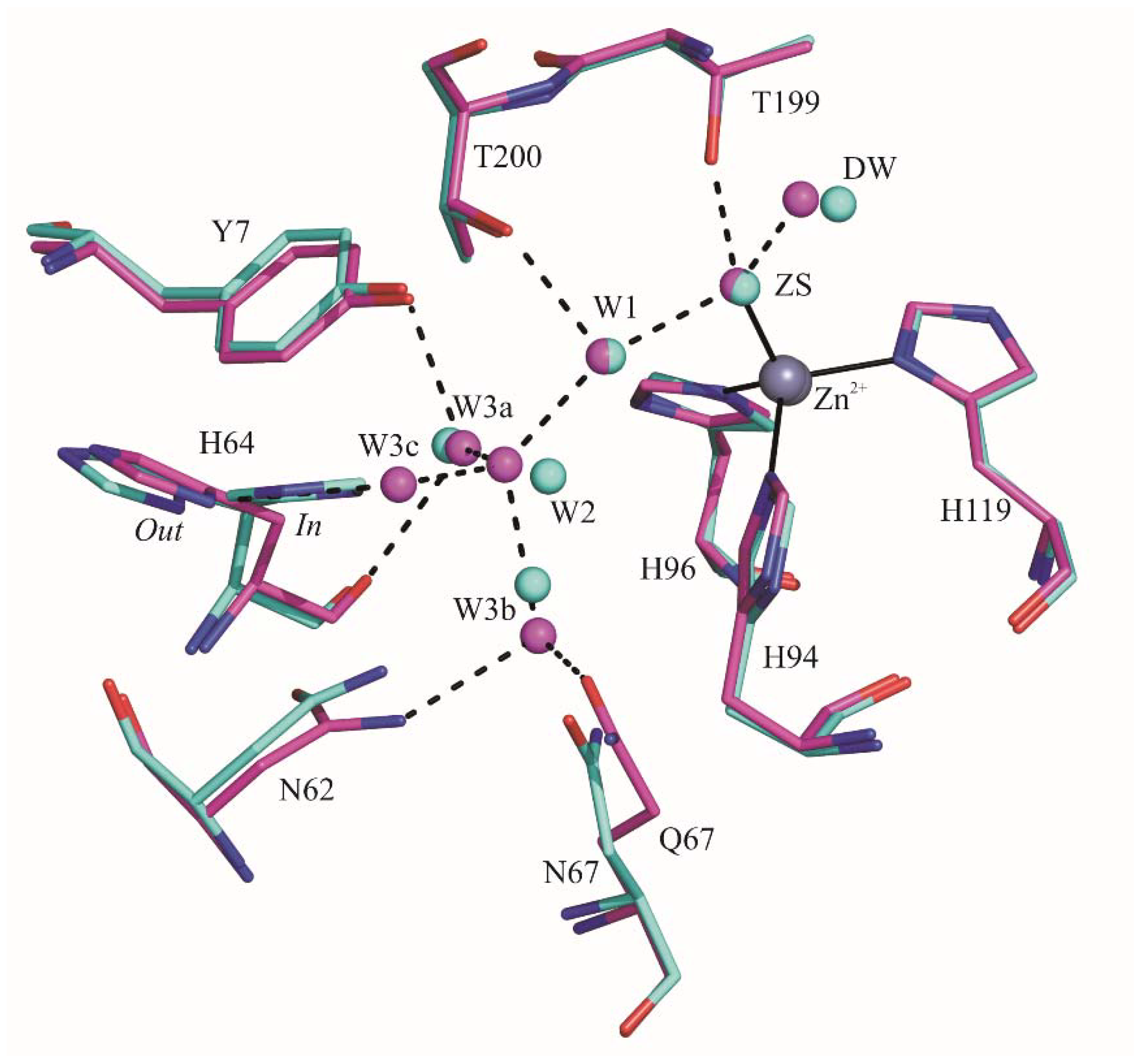
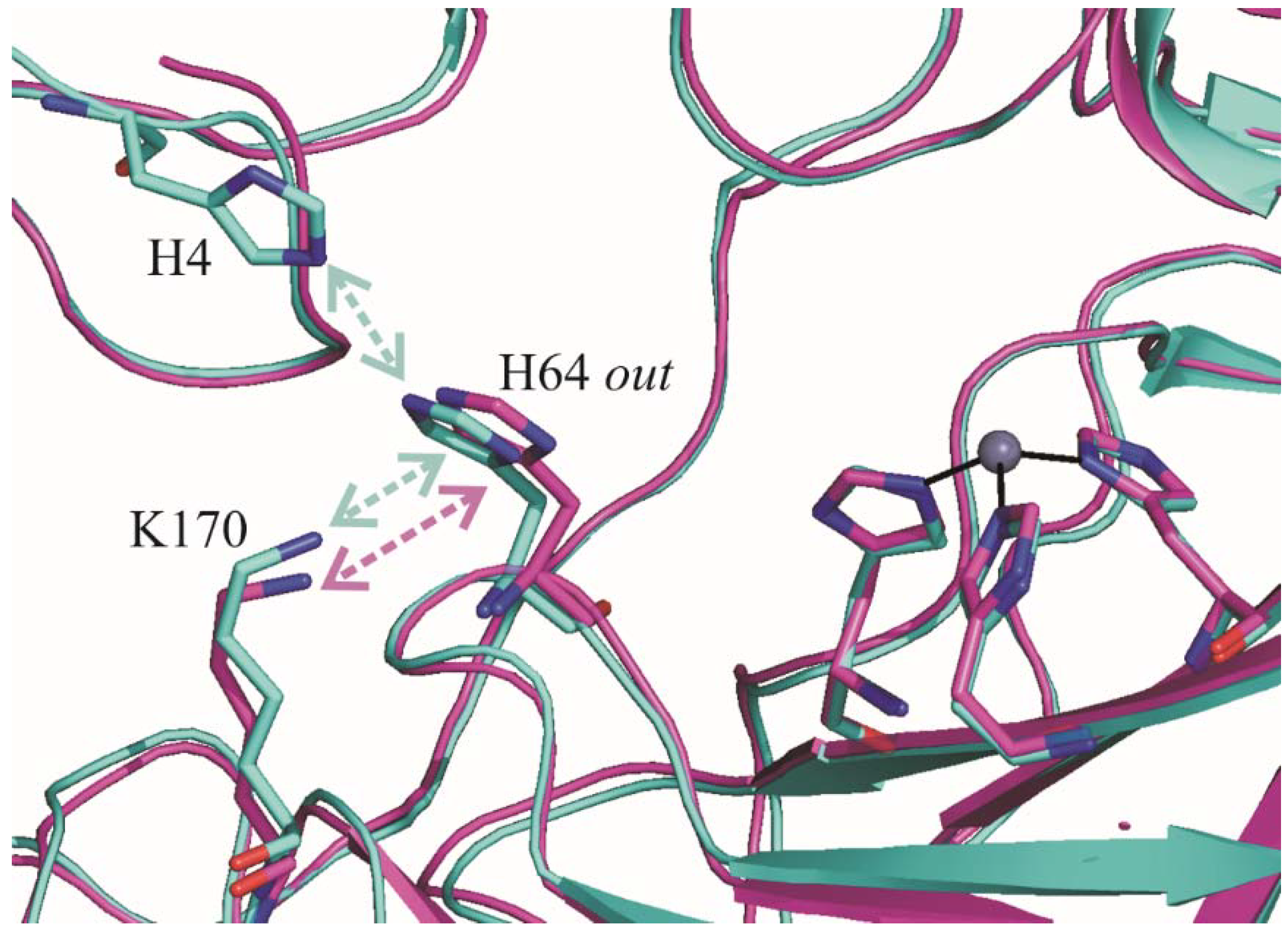
2.1. Crystallographic Studies
2.2. pKa Calculations
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
- F: 5′-CGCGCGCCATGGGCATGACCGGCCACCACG-3′
- R: 5′-CGCGCGCTCGAGGGCCCGGAAGGAGGC-3′
4.2. Crystallization and X-ray Data Collection
4.3. Structure Determination and Refinement
4.4. Theoretical pKa Calcuations
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Alterio, V.; Di Fiore, A.; D’Ambrosio, K.; Supuran, C.T.; De Simone, G. Multiple binding modes of inhibitors to carbonic anhydrases: How to design specific drugs targeting 15 different isoforms? Chem. Rev. 2012, 112, 4421–4468. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Fisher, G.M.; Andrews, K.T.; Poulsen, S.A.; Capasso, C.; Supuran, C.T. Discovery of a new family of carbonic anhydrases in the malaria pathogen plasmodium falciparum—The eta-carbonic anhydrases. Bioorg. Med. Chem. Lett. 2014, 24, 4389–4396. [Google Scholar] [CrossRef] [PubMed]
- Kikutani, S.; Nakajima, K.; Nagasato, C.; Tsuji, Y.; Miyatake, A.; Matsuda, Y. Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 2016, 113, 9828–9833. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Feng, L.; Jeffrey, P.D.; Shi, Y.; Morel, F.M. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 2008, 452, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Jakubzick, C.; Whittam, T.S.; Ferry, J.G. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc. Natl. Acad. Sci. USA 1999, 96, 15184–15189. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Monti, S.M.; De Simone, G. Thermal-stable carbonic anhydrases: A structural overview. In Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications; Frost, S.C., McKenna, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 75, pp. 387–404. [Google Scholar]
- Alterio, V.; Langella, E.; De Simone, G.; Monti, S.M. Cadmium-containing carbonic anhydrase CDCA1 in marine diatom Thalassiosira weissflogii. Mar. Drugs 2015, 13, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Di Fiore, A.; Capasso, C.; Supuran, C.T. The zinc coordination pattern in the eta-carbonic anhydrase from plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg. Med. Chem. Lett. 2015, 25, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; De Simone, G. (Eds.) Carbonic Anhydrases as Biocatalysts. From Theory to Medical and Industrial Applications; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Silverman, D.N.; McKenna, R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc. Chem. Res. 2007, 40, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Boone, C.D.; Kondeti, B.; McKenna, R. Structural annotation of human carbonic anhydrases. J. Enzym. Inhib. Med. Chem. 2013, 28, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.D.; Pinard, M.; McKenna, R.; Silverman, D. Catalytic mechanism of alpha-class carbonic anhydrases: CO2 hydration and proton transfer. Subcell. Biochem. 2014, 75, 31–52. [Google Scholar] [PubMed]
- Mikulski, R.L.; Silverman, D.N. Proton transfer in catalysis and the role of proton shuttles in carbonic anhydrase. Biochim. Biophys. Acta 2010, 1804, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, M.; Kondeti, B.; Tu, C.; Maupin, C.M.; Silverman, D.N.; McKenna, R. Structural insight into activity enhancement and inhibition of H64A carbonic anhydrase II by imidazoles. IUCrJ 2014, 1, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.K.; Silverman, D.N.; Forsman, C.; Jonsson, B.H.; Lindskog, S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry 1989, 28, 7913–7918. [Google Scholar] [CrossRef] [PubMed]
- Jewell, D.A.; Tu, C.K.; Paranawithana, S.R.; Tanhauser, S.M.; LoGrasso, P.V.; Laipis, P.J.; Silverman, D.N. Enhancement of the catalytic properties of human carbonic anhydrase III by site-directed mutagenesis. Biochemistry 1991, 30, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.; Tu, C.; Qian, M.; Laipis, P.; Agbandje-McKenna, M.; Silverman, D.N.; McKenna, R. Structural and kinetic analysis of the chemical rescue of the proton transfer function of carbonic anhydrase II. Biochemistry 2001, 40, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Christianson, D.W. Structural properties of human carbonic anhydrase II at pH 9.5. Biochem. Biophys. Res. Commun. 1991, 181, 579–584. [Google Scholar] [CrossRef]
- Fisher, Z.; Hernandez Prada, J.A.; Tu, C.; Duda, D.; Yoshioka, C.; An, H.; Govindasamy, L.; Silverman, D.N.; McKenna, R. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry 2005, 44, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.F.; Fierke, C.A.; Alexander, R.S.; Christianson, D.W. Conformational mobility of His-64 in the Thr-200→Ser mutant of human carbonic anhydrase II. Biochemistry 1991, 30, 9153–9160. [Google Scholar] [CrossRef] [PubMed]
- Taraphder, S.; Maupin, C.M.; Swanson, J.M.; Voth, G.A. Coupling protein dynamics with proton transport in human carbonic anhydrase II. J. Phys. Chem. B 2016, 120, 8389–8404. [Google Scholar] [CrossRef] [PubMed]
- Maupin, C.M.; Voth, G.A. Proton transport in carbonic anhydrase: Insights from molecular simulation. Biochim. Biophys. Acta 2010, 1804, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Maupin, C.M.; McKenna, R.; Silverman, D.N.; Voth, G.A. Elucidation of the proton transport mechanism in human carbonic anhydrase II. JACS 2009, 131, 7598–7608. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.U.; Song, H.; Avvaru, B.S.; Gruner, S.M.; Park, S.; McKenna, R. Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals. Proc. Natl. Acad. Sci. USA 2016, 113, 5257–5262. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.Z.; Tu, C.; Bhatt, D.; Govindasamy, L.; Agbandje-McKenna, M.; McKenna, R.; Silverman, D.N. Speeding up proton transfer in a fast enzyme: Kinetic and crystallographic studies on the effect of hydrophobic amino acid substitutions in the active site of human carbonic anhydrase II. Biochemistry 2007, 46, 3803–3813. [Google Scholar] [CrossRef] [PubMed]
- Domsic, J.F.; Williams, W.; Fisher, S.Z.; Tu, C.; Agbandje-McKenna, M.; Silverman, D.N.; McKenna, R. Structural and kinetic study of the extended active site for proton transfer in human carbonic anhydrase II. Biochemistry 2010, 49, 6394–6399. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Sjoblom, B.; Sauer-Eriksson, A.E.; Jonsson, B.H. Organization of an efficient carbonic anhydrase: Implications for the mechanism based on structure-function studies of a T199P/C206S mutant. Biochemistry 2002, 41, 7628–7635. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Avvaru, B.S.; Tu, C.; McKenna, R.; Silverman, D.N. Role of hydrophilic residues in proton transfer during catalysis by human carbonic anhydrase II. Biochemistry 2008, 47, 12028–12036. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, J.C.; Venta, P.J.; Eddy, R.L.; Fukushima, Y.S.; Shows, T.B.; Tashian, R.E. Characterization of the human gene for a newly discovered carbonic anhydrase, CA VII, and its localization to chromosome 16. Genomics 1991, 11, 835–848. [Google Scholar] [CrossRef]
- Earnhardt, J.N.; Qian, M.; Tu, C.; Lakkis, M.M.; Bergenhem, N.C.; Laipis, P.J.; Tashian, R.E.; Silverman, D.N. The catalytic properties of murine carbonic anhydrase VII. Biochemistry 1998, 37, 10837–10845. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Truppo, E.; Supuran, C.T.; Alterio, V.; Dathan, N.; Bootorabi, F.; Parkkila, S.; Monti, S.M.; De Simone, G. Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide. Bioorg. Med. Chem. Lett. 2010, 20, 5023–5026. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, R.; Monti, D.M.; Truppo, E.; Arciello, A.; Supuran, C.T.; De Simone, G.; Monti, S.M. Human carbonic anhydrase VII protects cells from oxidative damage. Biol. Chem. 2013, 394, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Monti, D.M.; De Simone, G.; Langella, E.; Supuran, C.T.; Di Fiore, A.; Monti, S.M. Insights into the role of reactive sulfhydryl groups of carbonic anhydrase III and VII during oxidative damage. J. Enzym. Inhib. Med. Chem. 2017, 32, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Truppo, E.; Supuran, C.T.; Sandomenico, A.; Vullo, D.; Innocenti, A.; Di Fiore, A.; Alterio, V.; De Simone, G.; Monti, S.M. Carbonic anhydrase VII is S-glutathionylated without loss of catalytic activity and affinity for sulfonamide inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 1560–1564. [Google Scholar] [CrossRef] [PubMed]
- Ruusuvuori, E.; Li, H.; Huttu, K.; Palva, J.M.; Smirnov, S.; Rivera, C.; Kaila, K.; Voipio, J. Carbonic anhydrase isoform VII acts as a molecular switch in the development of synchronous gamma-frequency firing of hippocampal CA1 pyramidal cells. J. Neurosci. 2004, 24, 2699–2707. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.; Voipio, J.; Kaila, K. Two developmental switches in GABAergic signalling: The K+–Cl− cotransporter KCC2 and carbonic anhydrase CA VII. J. Physiol. 2005, 562, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Bootorabi, F.; Janis, J.; Smith, E.; Waheed, A.; Kukkurainen, S.; Hytonen, V.; Valjakka, J.; Supuran, C.T.; Vullo, D.; Sly, W.S.; et al. Analysis of a shortened form of human carbonic anhydrase VII expressed in vitro compared to the full-length enzyme. Biochimie 2010, 92, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Asiedu, M.; Ossipov, M.H.; Kaila, K.; Price, T.J. Acetazolamide and midazolam act synergistically to inhibit neuropathic pain. Pain 2010, 148, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Avvaru, B.S.; Kim, C.U.; Sippel, K.H.; Gruner, S.M.; Agbandje-McKenna, M.; Silverman, D.N.; McKenna, R. A short, strong hydrogen bond in the active site of human carbonic anhydrase II. Biochemistry 2010, 49, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.Z.; Maupin, C.M.; Budayova-Spano, M.; Govindasamy, L.; Tu, C.; Agbandje-McKenna, M.; Silverman, D.N.; Voth, G.A.; McKenna, R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry 2007, 46, 2930–2937. [Google Scholar] [CrossRef] [PubMed]
- Hakansson, K.; Carlsson, M.; Svensson, L.A.; Liljas, A. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J. Mol. Biol. 1992, 227, 1192–1204. [Google Scholar] [CrossRef]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins 2005, 61, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H. Protein electrostatics and pKa blind predictions; contribution from empirical predictions of internal ionizable residues. Proteins 2011, 79, 3333–3345. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, R.; Domsic, J.F.; Ling, G.; Tu, C.; Robbins, A.H.; Silverman, D.N.; McKenna, R. Structure and catalysis by carbonic anhydrase II: Role of active-site tryptophan 5. Arch. Biochem. Biophys. 2011, 516, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, R.; West, D.; Sippel, K.H.; Avvaru, B.S.; Aggarwal, M.; Tu, C.; McKenna, R.; Silverman, D.N. Water networks in fast proton transfer during catalysis by human carbonic anhydrase II. Biochemistry 2013, 52, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, D.; Konig, P.; Guo, H.; Cui, Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry 2008, 47, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Cudney, R.; Patel, S.; Weisgraber, K.; Newhouse, Y.; McPherson, A. Screening and optimization strategies for macromolecular crystal growth. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Jancarik, J.; Kim, S.-H. Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Crystallogr. 1991, 24, 409–411. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Navaza, J. Amore: An automated package for molecular replacement. Acta Crystallogr. Sect. A Found. Crystallogr. 1994, 50, 157–163. [Google Scholar] [CrossRef]
- Brunger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar] [PubMed]
- Brunger, A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007, 2, 2728–2733. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.A.; Zou, J.Y.; Cowan, S.W.; Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991, 47 Pt 2, 110–119. [Google Scholar] [CrossRef]
- Hooft, R.W.; Vriend, G.; Sander, C.; Abola, E.E. Errors in protein structures. Nature 1996, 381, 272. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. Procheck: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]

| Cell Parameter | Value |
|---|---|
| Space group | P21212 |
| Unit cell parameters (Å) | a = 66.3 |
| b = 89.4 | |
| c = 44.4 | |
| Number of independent molecules | 1 |
| Data collection statistics | |
| Resolution limits (Å) | 31.5–1.91 |
| Wavelength (Å) | 1.54178 |
| Temperature (K) | 100 |
| Total reflections | 94217 |
| Unique reflections | 20846 |
| Redundancy | 4.5 |
| Completeness (%) | 98.8 (87.3) |
| R-merge (%) * | 0.079 (0.520) |
| <I>/<σ(I)> | 14.7 (2.0) |
| Refinement statistics | |
| Resolution limits (Å) | 31.5–1.91 |
| R-work ** (%) | 19.6 |
| R-free ** (%) | 24.4 |
| r.m.s.d. from ideal geometry: | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.4 |
| Number of protein atoms | 2063 |
| Number of water molecules | 162 |
| Average B factor (Å2) | |
| All atoms | 21.1 |
| Protein atoms | 20.7 |
| Water molecules | 25.6 |
| Enzyme | pKa | |
|---|---|---|
| His64 in | His64 out | |
| dmCA VII | 3.8 * | 4.6 |
| hCA II | 3.7 | 4.2 (4.3 #) |
| hCA II Structures | pKa |
|---|---|
| His64 out | |
| 1TE3 | 4.2 |
| 3KS3 (A) | 4.3 |
| 3KS3 (B) | 4.7 |
| 2CBA (A) | 3.9 |
| 2CBA (B) | 4.2 |
| 1TBT | 4.3 |
| 1TEQ | 4.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buonanno, M.; Di Fiore, A.; Langella, E.; D’Ambrosio, K.; Supuran, C.T.; Monti, S.M.; De Simone, G. The Crystal Structure of a hCA VII Variant Provides Insights into the Molecular Determinants Responsible for Its Catalytic Behavior. Int. J. Mol. Sci. 2018, 19, 1571. https://doi.org/10.3390/ijms19061571
Buonanno M, Di Fiore A, Langella E, D’Ambrosio K, Supuran CT, Monti SM, De Simone G. The Crystal Structure of a hCA VII Variant Provides Insights into the Molecular Determinants Responsible for Its Catalytic Behavior. International Journal of Molecular Sciences. 2018; 19(6):1571. https://doi.org/10.3390/ijms19061571
Chicago/Turabian StyleBuonanno, Martina, Anna Di Fiore, Emma Langella, Katia D’Ambrosio, Claudiu T. Supuran, Simona Maria Monti, and Giuseppina De Simone. 2018. "The Crystal Structure of a hCA VII Variant Provides Insights into the Molecular Determinants Responsible for Its Catalytic Behavior" International Journal of Molecular Sciences 19, no. 6: 1571. https://doi.org/10.3390/ijms19061571
APA StyleBuonanno, M., Di Fiore, A., Langella, E., D’Ambrosio, K., Supuran, C. T., Monti, S. M., & De Simone, G. (2018). The Crystal Structure of a hCA VII Variant Provides Insights into the Molecular Determinants Responsible for Its Catalytic Behavior. International Journal of Molecular Sciences, 19(6), 1571. https://doi.org/10.3390/ijms19061571









