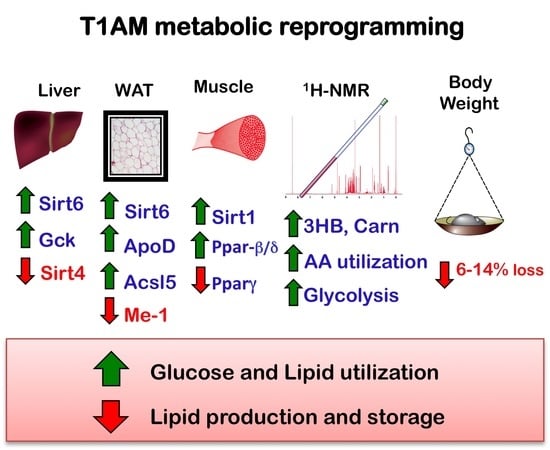
Metabolic Reprogramming by 3-Iodothyronamine (T1AM): A New Perspective to Reverse Obesity through Co-Regulation of Sirtuin 4 and 6 Expression
Abstract
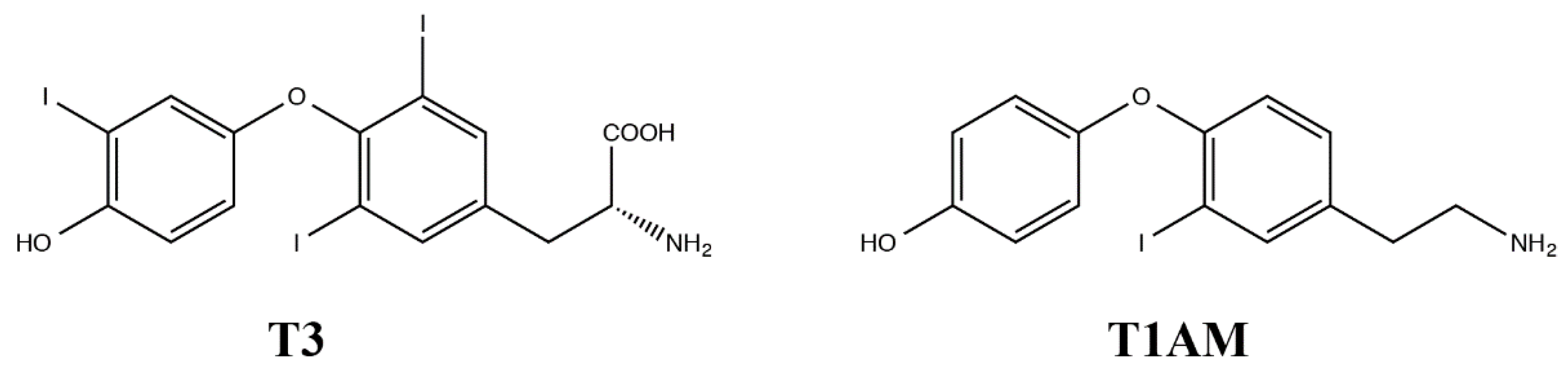
1. Introduction
2. Results
2.1. Food Consumption, Activity Levels, Weight Loss and Lipid Profiles
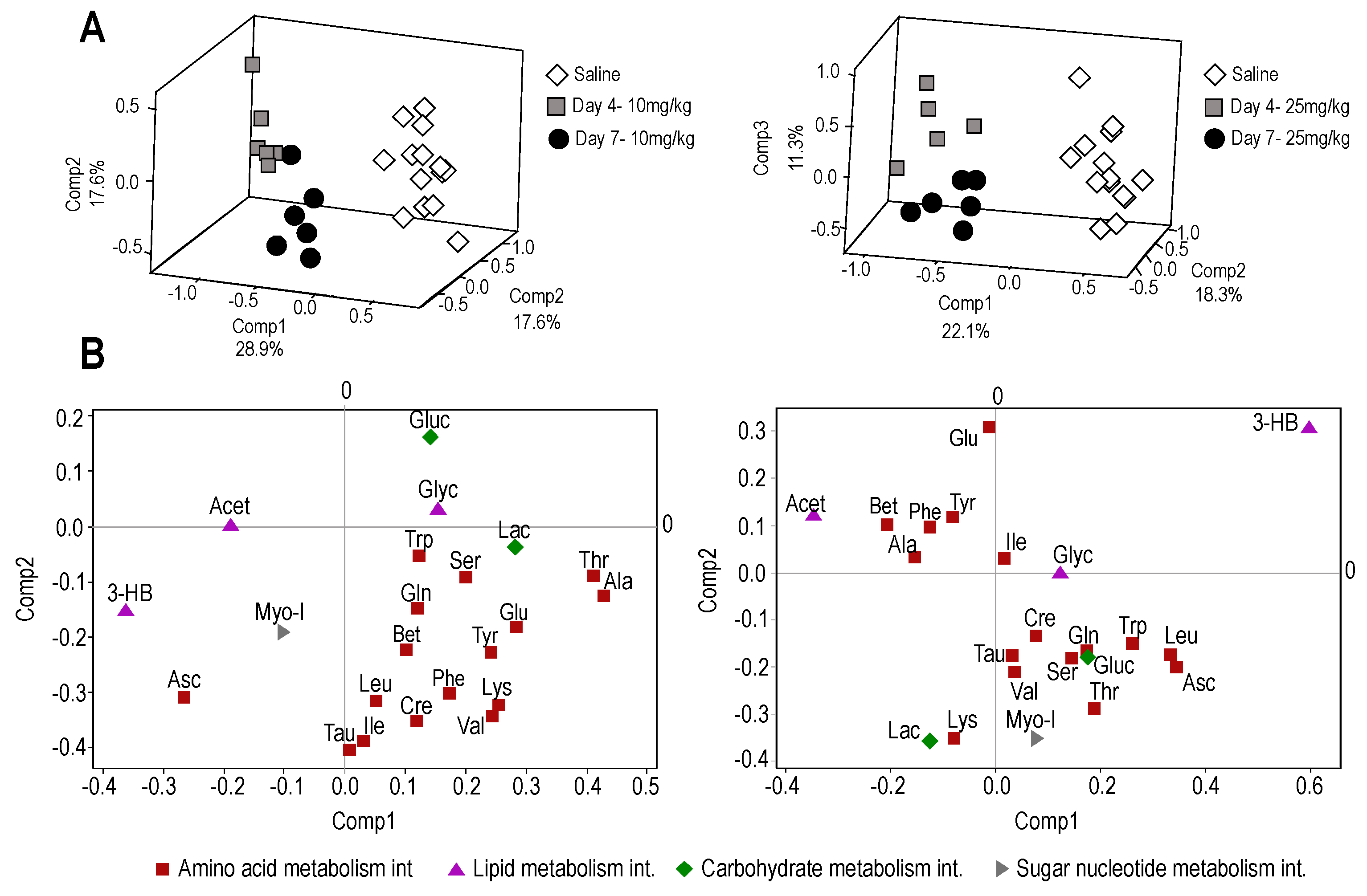
2.2. Nuclear Magnetic Resonance (NMR) Metabolome Analysis
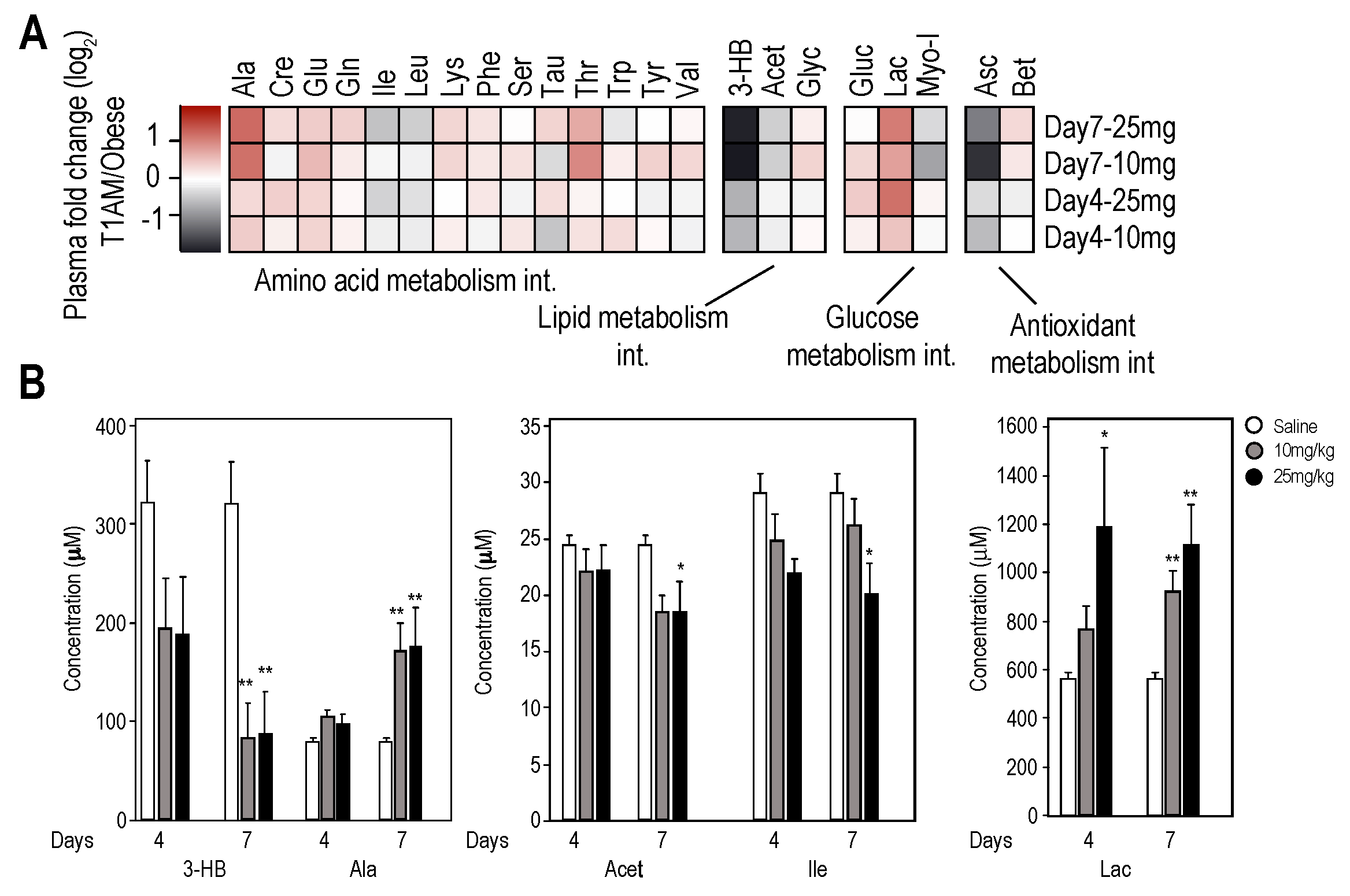
2.2.1. Plasma Metabolome
2.2.2. Liver and Muscle Metabolome Profiles
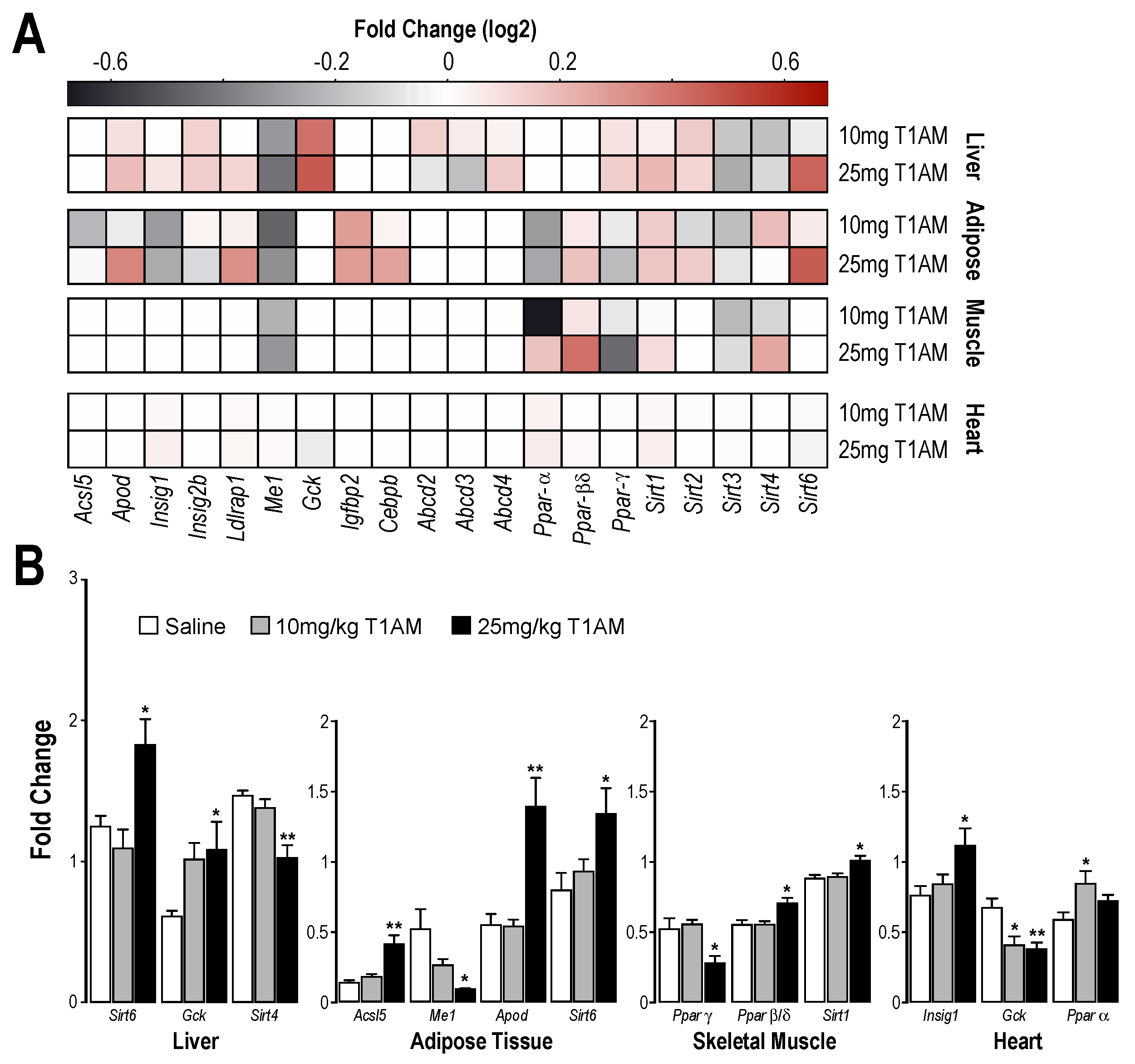
2.3. RT-qPCR Results
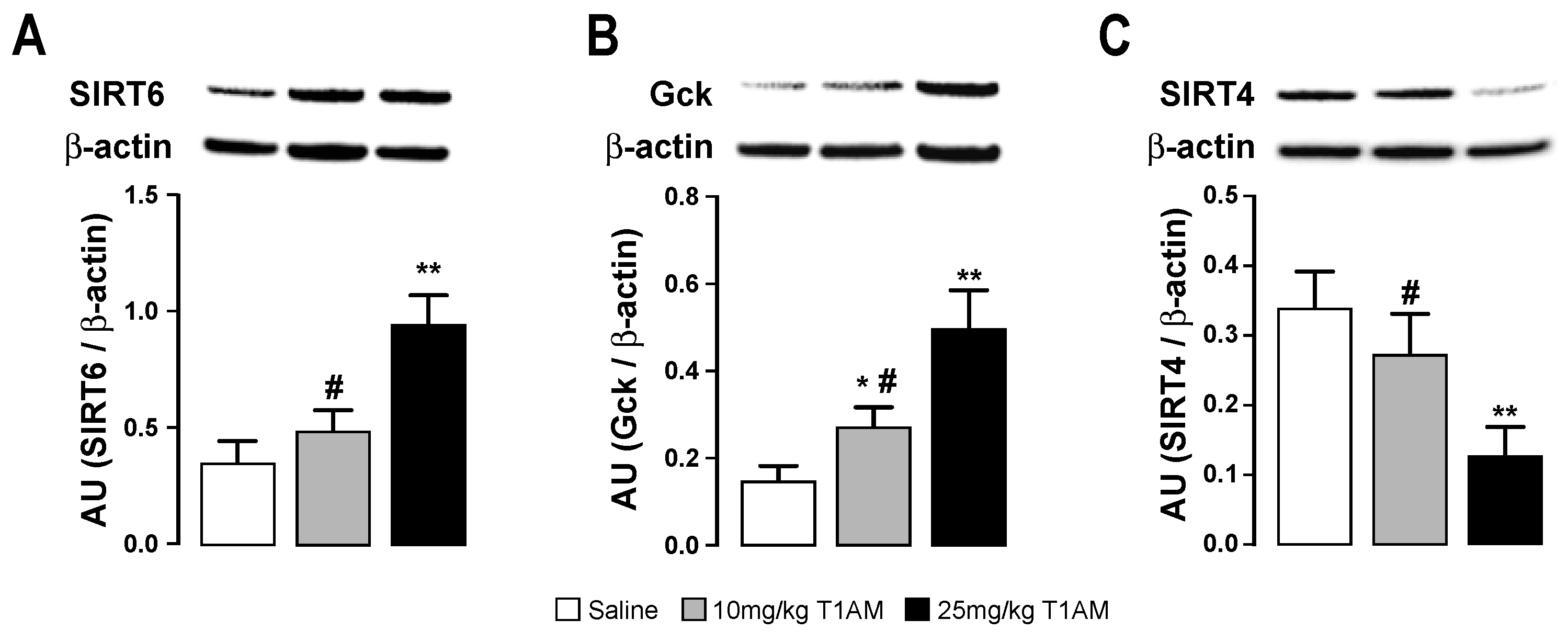
2.4. Western Blot of Tissues
2.5. Tissue T1AM Concentration
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animal Study
4.3. Plasma NMR Samples Preparation
4.4. Tissue NMR Samples Preparation
4.5. NMR Data Collection and Analysis
4.6. Tissue Preparation for Gene and Protein Expression Analyses
4.7. Gene Expression Analysis
4.8. Western Blotting Analysis
4.9. LC/MS-MS Assay
4.10. Biochemical Assays
4.11. Statistical Analysis
Metabolomics
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Scanlan, T.S.; Suchland, K.L.; Hart, M.E.; Chiellini, G.; Huang, Y.; Kruzich, P.J.; Frascarelli, S.; Crossley, D.A.; Bunzow, J.R.; Ronca-Testoni, S.; et al. 3-Iodothyronamine is an endogenous and rapid-acting derivative of thyroid hormone. Nat. Med. 2004, 10, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Bunzow, J.R.; Sonders, M.S.; Arttamangkul, S.; Harrison, L.M.; Zhang, G.; Quigley, D.I.; Darland, T.; Suchland, K.L.; Pasumamula, S.; Kennedy, J.L.; et al. Amphetamine, 3,4-methylenedioxymethampheta-mine, lysergic acid diethylamide and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol. Pharmacol. 2001, 60, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Borowsky, B.; Adham, N.; Jones, K.A.; Raddaz, R.; Artymyshyn, R.; Ogozalek, K.L.; Durkin, M.M.; Lakhlani, P.P.; Bonini, J.A.; Pathirana, S.; et al. Trace amines: Identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 8966–8971. [Google Scholar] [CrossRef] [PubMed]
- Saba, A.; Chiellini, G.; Frascarelli, S.; Marchini, M.; Ghelardoni, S.; Raffaelli, A.; Tonacchera, M.; Vitti, P.; Scanlan, T.S.; Zucchi, R. Tissue distribution and cardiac metabolism of 3-iodothyronamine. Endocrinology 2010, 151, 5063–5073. [Google Scholar] [CrossRef] [PubMed]
- Hoefig, C.S.; Köhrle, J.; Brabant, G.; Dixit, K.; Yap, B.; Strasburger, C.J.; Wu, Z. Evidence for extrathyroidal formation of 3-iodothyronamine in humans as provided by a novel monoclonal antibody-based chemiluminescent serum immunoassay. J. Clin. Endocrinol. Metab. 2011, 96, 1864–1872. [Google Scholar] [CrossRef] [PubMed]
- Hackenmueller, S.A.; Marchini, M.; Saba, A.; Zucchi, R.; Scanlan, T.S. Biosynthesis of 3-iodothyronamine (T1AM) is dependent on the sodium-iodide symporter and thyroperoxidase but does not involve extrathyroidal metabolism of T4. Endocrinology. 2012, 153, 5659–5667. [Google Scholar] [CrossRef] [PubMed]
- Dinter, J.; Muhlhaus, J.; Wienchol, C.L.; Cöster, M.; Hermsdorf, T.; Stäubert, C.; Köhrle, J.; Schöneberg, T.; Kleinau, G.; Mergler, S.; et al. The Multitarget Ligand 3-Iodothyronamine Modulates β-Adrenergic Receptor 2 Signaling. Eur. Thyroid J. 2015, 4, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Dinter, J.; Muhlhaus, J.; Jacobi, S.F.; Wienchol, C.L.; Cöster, M.; Meister, J.; Hoefig, C.S.; Müller, A.; Köhrle, J.; Grüters, A.; et al. 3-Iodothyronamine differentially modulates alpha-2A-adrenergic receptor-mediated signaling. J. Mol. Endocrinol. 2015, 54, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Snead, A.N.; Santos, M.S.; Seal, R.P.; Miyakawa, M.; Edwards, R.H.; Scanlan, T.S. Thyronamines inhibit plasma membrane and vesicular monoamine transport. ACS Chem. Biol. 2007, 2, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Chiellini, G.; Frascarelli, S.; Ghelardoni, S.; Carnicelli, V.; Tobias, S.C.; de Barber, A.; Brogioni, S.; Ronca-Testoni, S.; Cerbai, E.; Grandy, D.K.; et al. Cardiac effects of 3-iodothyronamine: A new aminergic system modulating cardiac function. FASEB J. 2007, 21, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Ghelardoni, S.; Suffredini, S.; Frascarelli, S.; Brogioni, S.; Chiellini, G.; Ronca-Testoni, S.; Grandy, D.K.; Scanlan, T.S.; Cerbai, E.; Zucchi, R. Modulation of cardiac ionic homeostasis by 3-iodothyronamine. J. Cell. Mol. Med. 2009, 13, 3082–3090. [Google Scholar] [CrossRef] [PubMed]
- Zucchi, R.; Accorroni, A.; Chiellini, G. Update on 3-iodothyronamine and its neurological and metabolic actions. Front. Physiol. 2014, 5, 402. [Google Scholar] [CrossRef] [PubMed]
- Accorroni, A.; Chiellini, G.; Origlia, N. Effects of Thyroid Hormones and their Metabolites on Learning and Memory in Normal and Pathological Conditions. Curr. Drug Metab. 2017, 18, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Braulke, L.J.; Klingenspor, M.; de Barber, A.; Tobias, S.C.; Grandy, D.K.; Scanlan, T.S.; Heldmaier, G. 3-Iodothyronamine: A novel hormone controlling the balance between glucose and lipid utilisation. J. Comp. Physiol. B. 2008, 178, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Haviland, J.A.; Reiland, H.; Butz, D.E.; Tonelli, M.; Porter, W.P.; Zucchi, R.; Scanlan, T.S.; Chiellini, G.; Assadi-Porter, F.M. NMR-based metabolomics and breath studies show lipid and protein catabolism during low dose chronic T1AM treatment. Obesity 2013, 21, 2538–2544. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.L.; Kiersgaard, M.K.; Sørensen, D.B.; Mikkelsen, L.F. Fasting of mice: A review. Lab. Anim. 2013, 47, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell. Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.; German, N.J.; Saha, A.K.; de Boer, V.C.; Davies, M.; Koves, T.R.; Dephoure, N.; Fischer, F.; Boanca, G.; Vaitheesvaran, B.; et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 2013, 50, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Soupene, E.; Kuypers, F.A. Mammalian Long-Chain Acyl-CoA Synthetases. Exp. Biol. Med. 2008, 233, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, G.; Henry Dong, H. Apolipoprotein D in lipid metabolism and its functional implication in atherosclerosis and aging. Aging 2009, 1, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kanfi, Y.; Peshti, V.; Gil, R.; Naiman, S.; Nahum, L.; Levin, E.; Kronfeld-Schor, N.; Cohen, H.Y. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010, 9, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Cui, X.; Chen, Q.; Yang, X.; Fang, F.; Zhang, J.; Liu, G.; Jin, W.; Chang, Y. Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep. 2017, 20, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, E.; Guzmán-Jofre, L.; Moore-Carrasco, R.; Palomo, I. Role of PPARs in inflammatory processes associated with metabolic syndrome. Mol. Med. Rep. 2013, 8, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Ghanian, Z.; Maleki, S.; Reiland, H.; Bütz, D.E.; Chiellini, G.; Assadi-Porter, F.M.; Ranji, M. Optical imaging of mitochondrial redox state in rodent models with 3-iodothyronamine. Exp. Biol. Med. 2014, 239, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, V.; Melissari, E.; Iofrida, C.; Righi, M.; di Russo, M.; Donzelli, R.; Saba, A.; Frascarelli, S.; Chiellini, G.; Zucchi, R.; et al. Modulation of gene expression by 3-iodothyronamine: Genetic evidence for a lipolytic pattern. PLoS ONE 2014, 9, e106923. [Google Scholar] [CrossRef] [PubMed]
- Haigis, M.C.; Guarente, L.P. Mammalian sirtuins–emerging roles in physiology, aging and calorie restriction. Genes Dev. 2006, 20, 2913–2922. [Google Scholar] [CrossRef] [PubMed]
- Guarente, L. Sirtuins as potential targets for metabolic syndrome. Nature 2006, 444, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Huynh, F.K.; Hershberger, K.A.; Hirschey, M.D. Targeting sirtuins for the treatment of diabetes. Diabetes Manag. 2013, 3, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The histone deacetylase Sirt6 regulates glucose homeostasis via hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Tasselli, L.; Zheng, W.; Chua, K.F. SIRT6: Novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 2017, 28, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Nasrin, N.; Wu, X.; Fortier, E.; Feng, Y.; Barè, O.C.; Chen, S.; Ren, X.; Wu, Z.; Streeper, R.S.; Bordone, L. SIRT4 Regulates Fatty Acid Oxidation and Mitochondrial Gene Expression in Liver and Muscle Cells. J. Biol. Chem. 2010, 285, 31995–32002. [Google Scholar] [CrossRef] [PubMed]
- Batatinha, H.A.; Lima, E.A.; Teixeira, A.A.; Souza, C.O.; Biondo, L.A.; Silveira, L.S.; Lira, F.S.; Rosa Neto, J.C. Association Between Aerobic Exercise and Rosiglitazone Avoided the NAFLD and Liver Inflammation Exacerbated in PPAR-α Knockout Mice. J. Cell. Physiol. 2017, 232, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Geisler, C.E.; Hepler, C.; Higgins, M.R.; Renquist, B.J. Hepatic adaptations to maintain metabolic homeostasis in response to fasting and refeeding in mice. Nutr. Metab. 2016, 13, 62. [Google Scholar] [CrossRef] [PubMed]
- De Lange, P.; Farina, P.; Moreno, M.; Ragni, M.; Lombardi, A.; Silvestri, E.; Burrone, L.; Lanni, A.; Goglia, F. Sequential changes in the signal transduction responses of skeletal muscle following food deprivation. FASEB J. 2006, 20, 2579–2581. [Google Scholar] [CrossRef] [PubMed]
- Chiellini, G.; Erba, P.; Carnicelli, V.; Manfredi, C.; Frascarelli, S.; Ghelardoni, S.; Mariani, G.; Zucchi, R. Distribution of exogenous [125I]-3-iodothyronamine in mouse in vivo: Relationship with trace amine-associated receptors. J. Endocrinol. 2012, 213, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Selen, E.S.; Bolandnazar, Z.; Tonelli, M.; Bütz, D.E.; Haviland, J.A.; Porter, W.P.; Assadi-Porter, F.M. NMR Metabolomics Show Evidence for Mitochondrial Oxidative Stress in a Mouse Model of Polycystic Ovary Syndrome. J. Proteome Res. 2015, 14, 3284–3291. [Google Scholar] [CrossRef] [PubMed]
- Haviland, J.A.; Tonelli, M.; Haughey, D.T.; Porter, W.P.; Assadi-Porter, F.M. Novel diagnostics of metabolic dysfunction detected in breath and plasma by selective isotope-assisted labeling. Metabolism 2012, 61, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, F.; Hansen, B.; Karcher, W.; Sjostrom, M.; Eriksson, L. Model validation by permutation tests: Applications to variable selection. J. Chemometr. 1996, 10, 521–532. [Google Scholar] [CrossRef]
- Teng, Q. NMR-Based Metabolomics. In Structural Biology-Practical NMR Applications, 2nd ed.; Springer Science + Business Media Inc.: New York, NY, USA, 2013; pp. 311–392. ISBN 978-1-4614-3964-6. [Google Scholar]
- Pattini, L.; Mazzara, S.; Conti, A.; Iannaccone, S.; Cerutti, S.; Alessio, M. An integrated strategy in two-dimensional electrophoresis analysis able to identify discriminants between different clinical conditions. Exp. Biol. Med. 2008, 233, 483–491. [Google Scholar] [CrossRef] [PubMed]

| Assay | Saline | 10 mg/kg/day | 25 mg/kg/day |
|---|---|---|---|
| Weight loss (g) | −1.6 ± 2.2 | −3.8 ± 0.7 b | −8.1 ± 3.2 a |
| Serum Cholesterol (mg/dL) | 90.8 ± 8.57 | 86.7 ± 7.84 | 77.7 ± 3.61 a |
| Serum TG (mg/dL) | 49.0 ± 3.46 | 52.11 ± 10.61 | 67.3 ± 4.23 a |
| Serum glucose (mg/dL) | 151 ± 21.60 | 165 ± 35.74 | 161.52 ± 24.02 |
| Tissue | Saline | Low T1AM (10 mg/kg/day) | High T1AM (25 mg/kg/day) |
|---|---|---|---|
| Liver + | 7.68 ± 0.85 | 318.3 ± 35.09 a | 767.0 ± 165.40 b |
| White Adipose + | 0.493 ± 0.17 | 1.71 ± 0.30 b | 16.96 ± 3.96 b |
| Muscle + | 19.84 ± 3.57 | 56.52 ± 12.64 | 412.86 ± 109.14 b |
| Heart + | 18.15 ± 4.38 | 37.62 ± 0.76 | 68.19 ± 14.10 b |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Assadi-Porter, F.M.; Reiland, H.; Sabatini, M.; Lorenzini, L.; Carnicelli, V.; Rogowski, M.; Selen Alpergin, E.S.; Tonelli, M.; Ghelardoni, S.; Saba, A.; et al. Metabolic Reprogramming by 3-Iodothyronamine (T1AM): A New Perspective to Reverse Obesity through Co-Regulation of Sirtuin 4 and 6 Expression. Int. J. Mol. Sci. 2018, 19, 1535. https://doi.org/10.3390/ijms19051535
Assadi-Porter FM, Reiland H, Sabatini M, Lorenzini L, Carnicelli V, Rogowski M, Selen Alpergin ES, Tonelli M, Ghelardoni S, Saba A, et al. Metabolic Reprogramming by 3-Iodothyronamine (T1AM): A New Perspective to Reverse Obesity through Co-Regulation of Sirtuin 4 and 6 Expression. International Journal of Molecular Sciences. 2018; 19(5):1535. https://doi.org/10.3390/ijms19051535
Chicago/Turabian StyleAssadi-Porter, Fariba M., Hannah Reiland, Martina Sabatini, Leonardo Lorenzini, Vittoria Carnicelli, Micheal Rogowski, Ebru S. Selen Alpergin, Marco Tonelli, Sandra Ghelardoni, Alessandro Saba, and et al. 2018. "Metabolic Reprogramming by 3-Iodothyronamine (T1AM): A New Perspective to Reverse Obesity through Co-Regulation of Sirtuin 4 and 6 Expression" International Journal of Molecular Sciences 19, no. 5: 1535. https://doi.org/10.3390/ijms19051535
APA StyleAssadi-Porter, F. M., Reiland, H., Sabatini, M., Lorenzini, L., Carnicelli, V., Rogowski, M., Selen Alpergin, E. S., Tonelli, M., Ghelardoni, S., Saba, A., Zucchi, R., & Chiellini, G. (2018). Metabolic Reprogramming by 3-Iodothyronamine (T1AM): A New Perspective to Reverse Obesity through Co-Regulation of Sirtuin 4 and 6 Expression. International Journal of Molecular Sciences, 19(5), 1535. https://doi.org/10.3390/ijms19051535