Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency
Abstract
1. Introduction
2. Results and Discussion
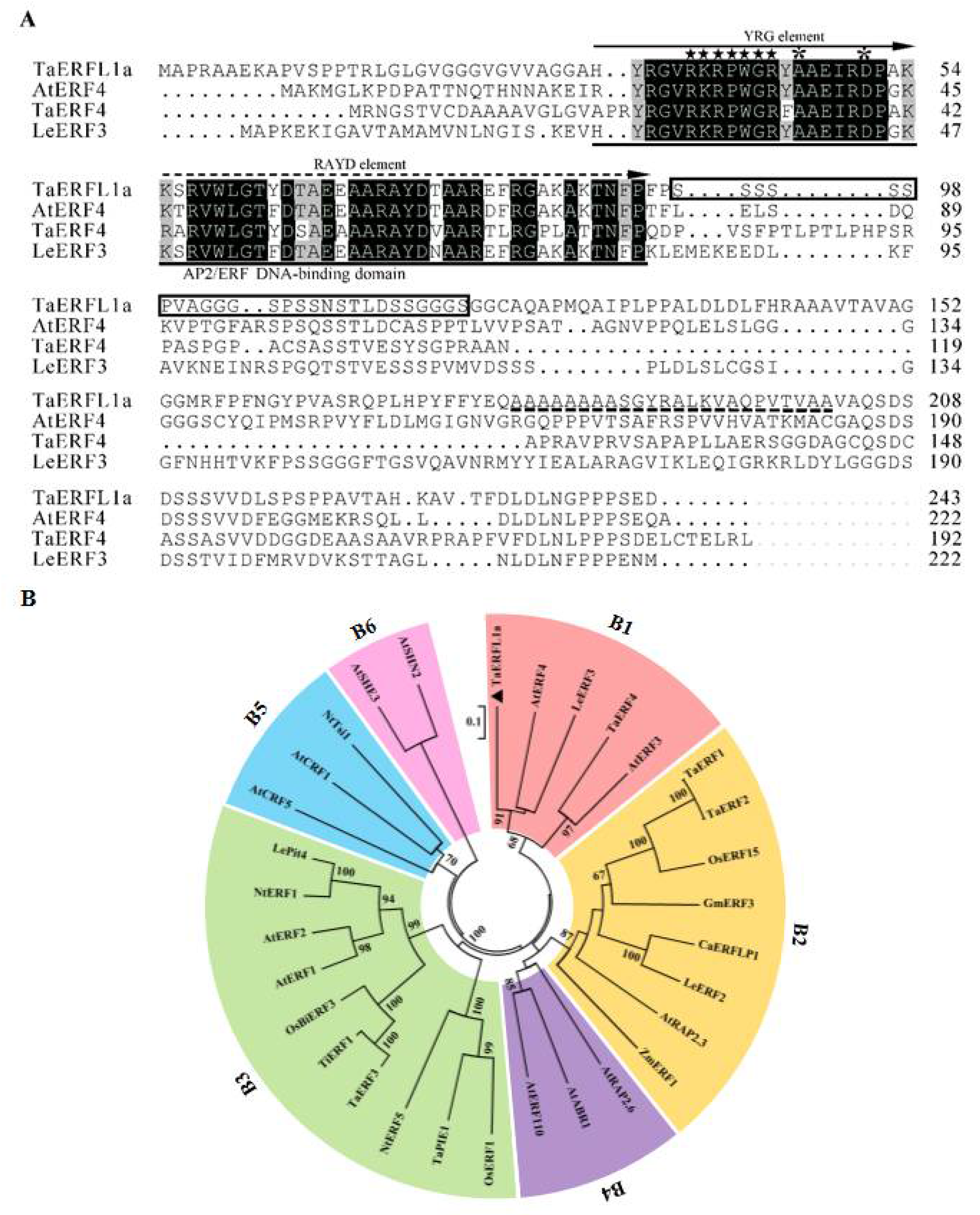
2.1. Identification of TaERFL1a
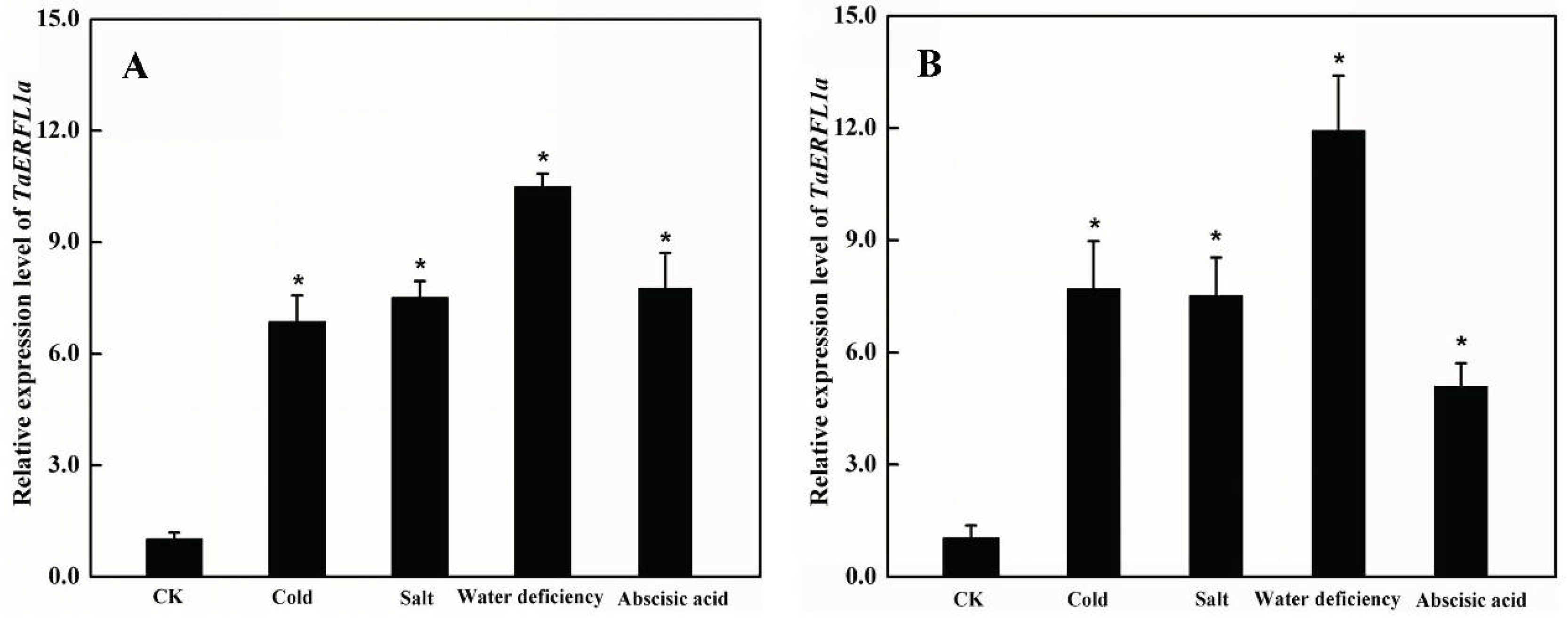
2.2. Expression Profiles of TaERFL1a during the Responses of Wheat to Abiotic Stresses and Abscisic Acid
2.3. Function of TaERFL1a in the Response of Wheat to Abiotic Stresses
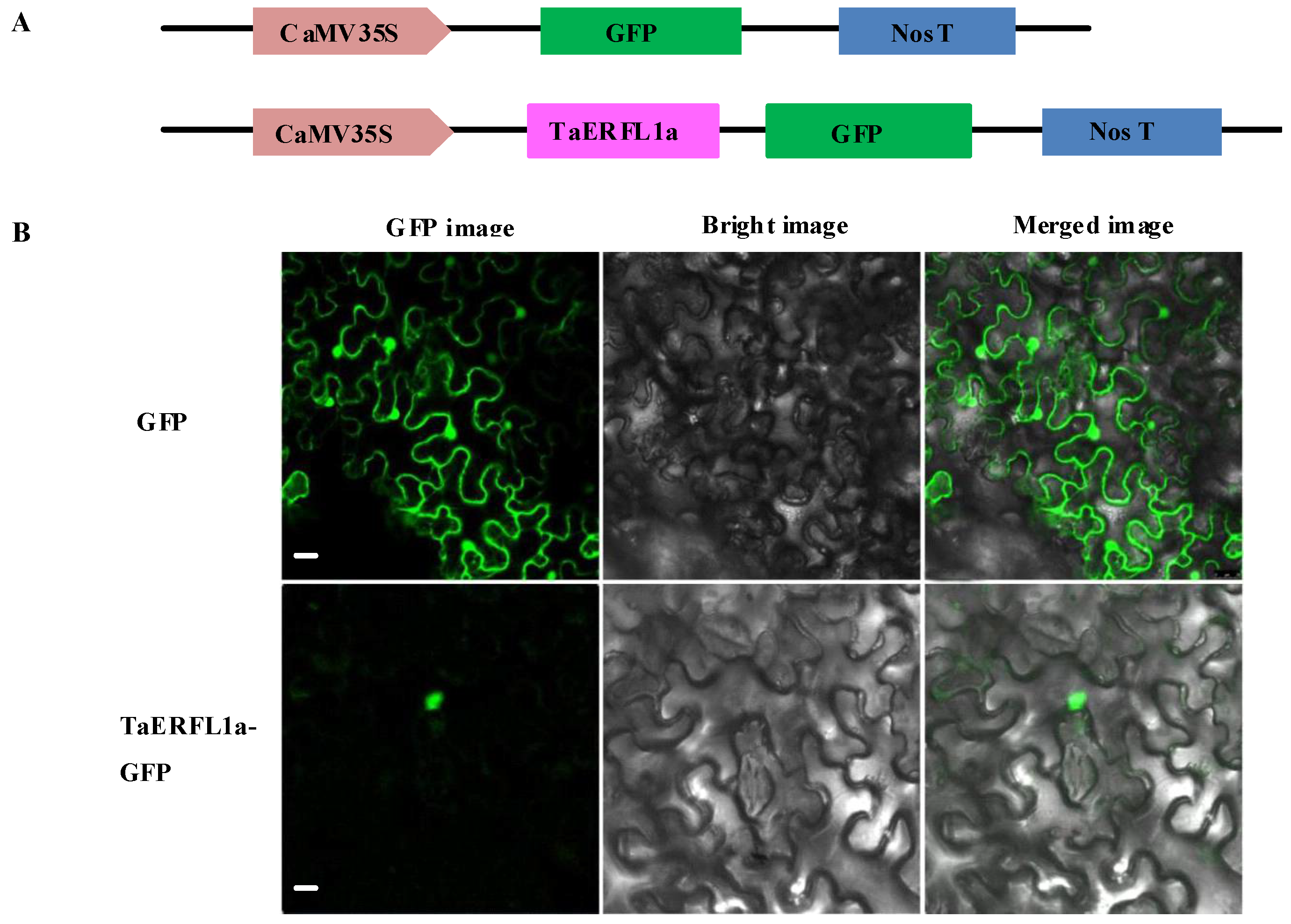
2.4. Subcellular Localization of TaERFL1a
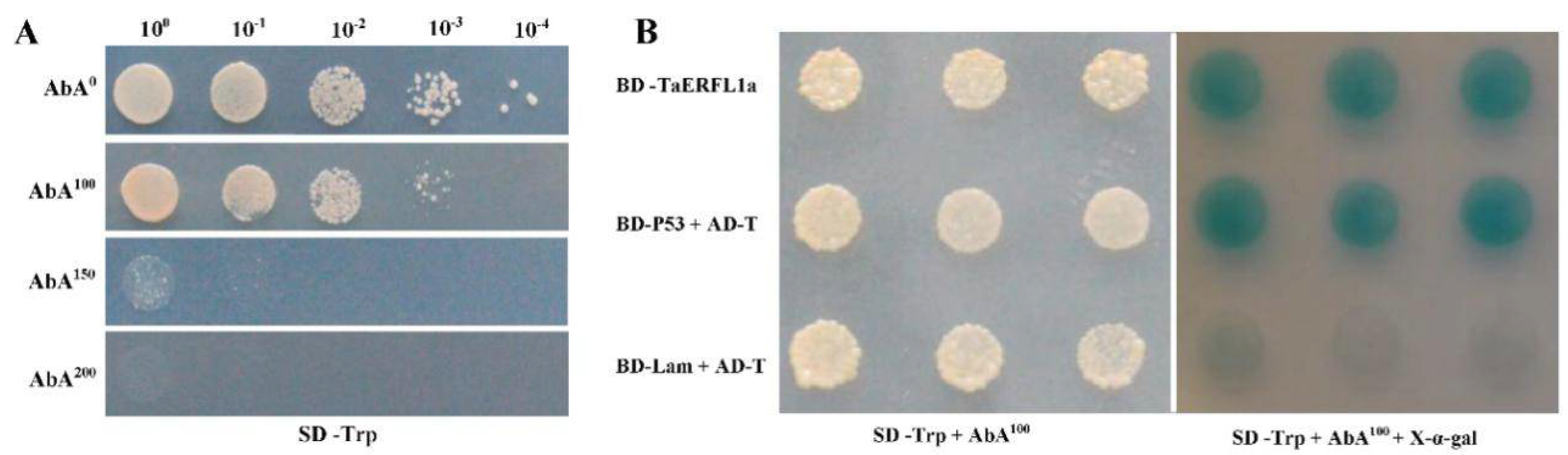
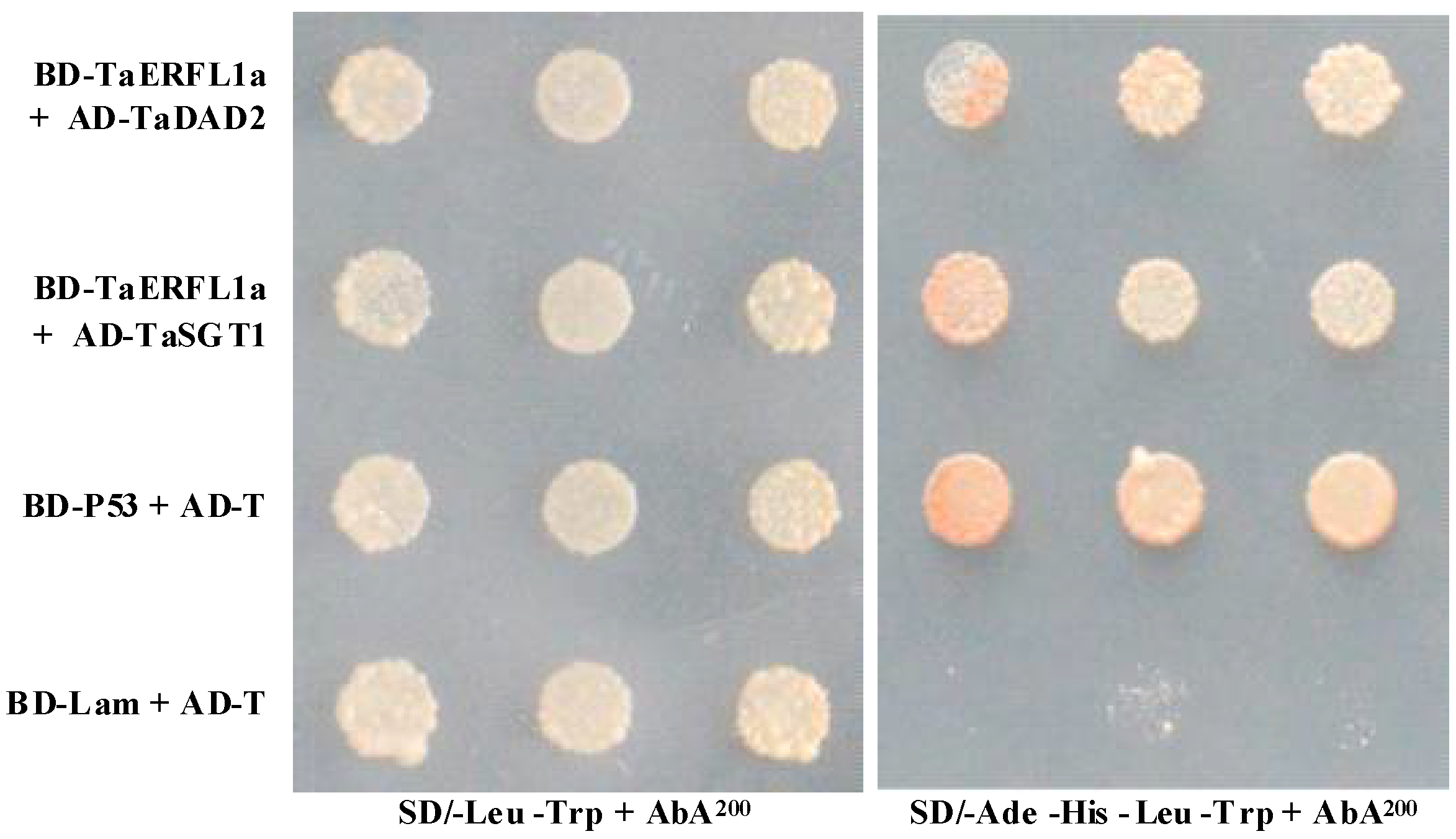
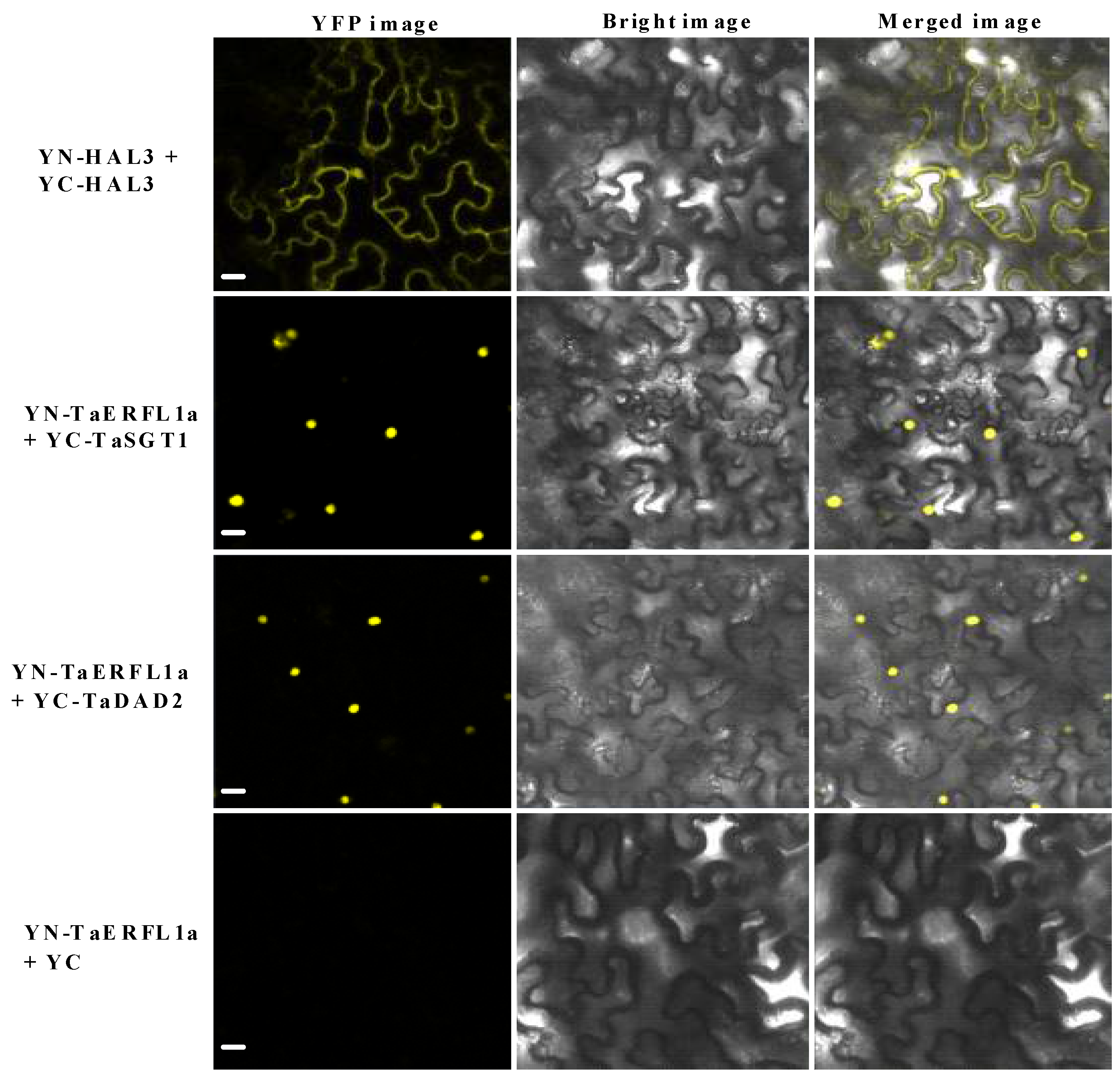
2.5. Interacting Proteins of TaERFL1a
3. Materials and Methods
3.1. Identification and Sequence Analysis of TaERFL1a
3.2. Plant Materials and Abiotic Stresses
3.3. Transcript Level of TaERFL1a
3.4. Transient Silencing of TaERFL1a
3.5. Subcellular Localization of TaERFL1a
3.6. Transcription Activation of, and Proteins Interacting with TaERFL1a
3.7. Transcript Levels of the Genes Encoding Six Identified Proteins
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kilian, J.; Peschke, F.; Berendzen, K.W.; Harter, K.; Wanke, D. Prerequisites, performance and profits of transcriptional profiling the abiotic stress response. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; Charlotte, O.; Krogh, J.M.; Skriver, S. Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Thirugnanasambantham, K.; Durairaj, S.; Saravanan, S.; Karikalan, K.; Muralidaran, S.; Ibrahim, V.; Islam, H. Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol. Biol. Rep. 2015, 33, 347. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.X.; Wang, Y.X.; Liu, X.F.; Li, J.Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Yang, N.; Pyvovarenko, T.; Pillman, K.; Bazanova, N.; Tikhomirov, N.; Eliby, S.; Shirley, N.; Sivasankar, S.; Tingey, S.; et al. Complex regulation by Apetala2 domain-containing transcription factors revealed through analysis of the stress-responsive TdCor410b promoter from durum wheat. PLoS ONE 2013, 8, e58713. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Shi, J.; Xu, W.; Li, H.; He, M.; Xu, Y.; Xu, T.; Yang, Y.; Cao, J.; Wang, Y. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J. Plant Physiol. 2013, 170, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Ai, X.; Xu, F.; Quan, T.; Liu, S.; Xia, G. Isolation and characterization of a bread wheat salinity responsive ERF transcription factor. Gene 2012, 511, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H.; Pan, X.; Chen, X.; Zhang, Z.; Lu, X.; Huang, R. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgen. Res. 2011, 20, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, W.; Chen, J.; Yang, Y.; Zhang, Z.; Wang, X.C.; Huang, R. Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 2007, 63, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.D.; Hu, S.J.; Zhang, Z.L.; Zhang, H.W.; Zhang, Z.J.; Huang, R.F. Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Xia, L.Q.; Chen, M.; Cheng, X.G.; Zhang, R.Y.; Li, L.C.; Zhao, Y.X.; Lu, Y.; Ni, Z.Y.; Liu, L.; et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 2007, 65, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Li, G.; Yang, W.; Han, Q.; Ma, H.; Wang, Y.; Ren, J.; Zhu, Y.; Guo, T. Transcriptional profile of the spring freeze response in the leaves of bread wheat. Acta Physiol. Plant. 2013, 35, 575–587. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat genome. Science 2014, 345, 1251788. [Google Scholar]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; International Wheat Genome Sequencing Consortium; Mayer, K.F.X.; Olsen, O.A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A.; Fahima, T.; Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 2012, 63, 5045–5059. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributiong to heat, and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.J.; Fitzgerald, T.L.; Stiller, J.; Berkman, P.J.; Gardiner, D.M.; Manner, J.M.; Henry, R.J.; Kazan, K. The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 2017, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Mahmood, T. Functional role of DREB and ERF transcription factors: Regulating stress-responsive network in plants. Acta Physiol. Plant. 2015, 37, 178. [Google Scholar] [CrossRef]
- Liu, W.; Karemera, N.J.U.; Wu, T.; Yang, Y.; Zhang, X.; Xu, X.; Wang, Y.; Han, Z. The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS ONE 2017, 12, e0186580. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, Z.; Grierson, D. Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J. Plant Physiol. 2008, 165, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yang, X.; Liu, Z.; Zhang, T.; Lu, S.; Liu, Y. Low yield gap of winter wheat in the North China plain. Eur. J. Agron. 2014, 59, 1–12. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Yang, Y.; Zhang, Y.; Zhang, Z.; Zhang, H.; Hu, X. Abscisic acid regulates TSRF1-mediated resistance to Ralstonia solanacearum by modifying the expression of GCC box-containing genes in tobacco. J. Exp. Bot. 2008, 59, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Tufan, H.A.; Stefanato, F.L.; McGrann, G.R.D.; MacCormack, R.; Boyd, L.A. The barley stripe mosaic virus system used for virus-induced gene silencing in cereals differentially affects susceptibility to fungal pathogens in wheat. J. Plant Physiol. 2011, 168, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates Brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Pan, L.; Miao, J.; Liu, T. Identification of interacting proteins with aryl hydrocarbon receptor in scallop Chlamys farreri by yeast two hybrid screening. Ecotoxicol. Environ. Safe 2016, 133, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shen, Z.; Meng, G.; Lu, Y.; Wang, Y. Genome-wide analysis of the Brachypodium distachyon (L.) P. Beauv. Hsp90 gene family reveals molecular evolution and expression profiling under drought and salt stresses. PLoS ONE 2017, 12, e0189187. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; Fan, R.; Wang, X.; Wang, D.; Zhang, X. TaRAR1 and TaSGT1 associate with TaHSP90 to function in bread wheat (Triticum aestivum L.) seedling growth and stripe rust resistance. Plant Mol. Biol. 2015, 87, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, C.; Zhang, H.; Xu, J.R.; Liu, B.; Lv, J.; Han, D.; Huang, L.; Kang, Z. TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus. Mol. Plant Microbe Interact. 2011, 24, 79–90. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shanmugam, A.; Thamilarasan, S.G.; Park, J.I.; Jung, M.Y.; Nou, I.S. Characterization and abiotic stress-responsive expression analysis of SGT1 genes in Brassica oleracea. Genome 2016, 59, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, Y. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta.-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Huseynova, I.M.; Aliyeva, D.R.; Aliyev, J.A. Subcellular localization and responses of superoxide dismutase isoforms in local wheat varieties subjected to continuous soil drought. Plant Physiol. Biochem. 2014, 81, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Calif. Agric. Exp. Stat. Circ. 1950, 347, 357–359. [Google Scholar]
- Wei, L.; Wang, L.; Yang, Y.; Liu, G.; Wu, Y.; Guo, T.; Kang, G. Abscisic acid increases leaf starch content of polyethylene glycol-treated wheat seedlings by temporally increasing transcripts of genes encoding starch synthesis enzymes. Acta Physiol. Plant. 2015, 37, 206. [Google Scholar] [CrossRef]
- Li, G.; Wu, Y.; Liu, G.; Xiao, X.; Wang, P.; Gao, T.; Xu, M.; Han, Q.; Wang, Y.; Guo, T.; et al. Large-scale proteomics combined with transgenic experiments demonstrates an important role of jasmonic acid in potassium deficiency response in wheat and rice. Mol. Cell. Proteom. 2017, 16, 1889–1905. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.Z.; Liu, G.Q.; Li, C.W.; Kang, G.Z.; Guo, T.C. Identification of the TaBTF3 gene in wheat (Triticum aestivum L.) and the effect of its silencing on wheat chloroplast, mitochondria and mesophyll cell development. Biochem. Biophys. Res. Commun. 2012, 426, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wu, Y.; Xu, M.; Gao, T.; Wang, P.; Wang, L.; Guo, T.; Kang, G. Virus-induced gene silencing identifies an important role of the TaRSR1 transcription factor in starch synthesis in bread wheat. Int. J. Mol. Sci. 2016, 17, 1557. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Rudd, J.J.; Kanyuka, K. Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance. Fungal Genet. Biol. 2015, 79, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.; Li, G.; Xu, W.; Peng, X.; Han, Q.; Zhu, Y.; Guo, T. Proteomics reveals the effects of salicylic acid on growth and tolerance to subsequent drought stress in wheat. J. Proteome Res. 2012, 11, 6066–6079. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Cao, M.J.; Liu, Y.; Yang, J.L.; Zheng, S.J. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 2015, 208, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene expression and yeast two-hybrid studies of 1R-MYB transcription factor mediating drought stress response in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2015, 6, 1117. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Shan, J.X.; Gao, J.P.; Lin, H.X. OsHAL3, a blue light-responsive protein, interacts with the floral regulator Hd1 to activate flowering in rice. Mol. Plant 2016, 6, 233–244. [Google Scholar] [CrossRef] [PubMed]
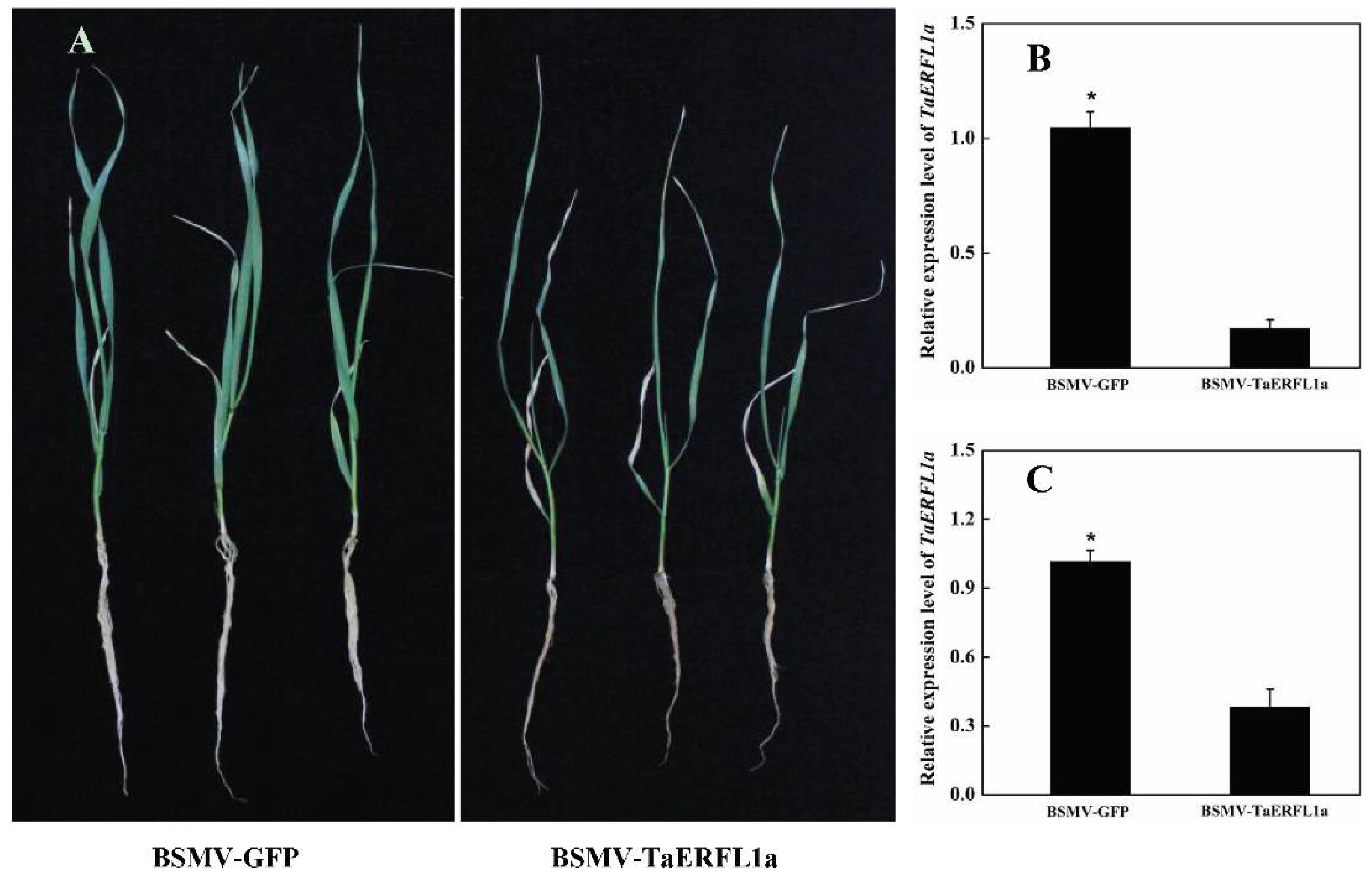
| Treatments | Fresh Weights (g/Plant) | Dry Weights (g/Plant) | Absolute Water Contents (%) | MDA Concentrations (nmol·L−1 FW) |
|---|---|---|---|---|
| BSMV-GFP | 0.49 ± 0.024 a | 0.08 ± 0.007 a | 84.42 ± 0.650 a | 18.98 ± 1.144 a |
| BSMV-TaERFL1a | 0.22 ± 0.006 b | 0.06 ± 0.020 b | 74.56 ± 0.912 b | 31.80 ± 0.639 b |
| No. | Homology Accession No. | Homologous Protein | Plant Species | Clone Numbers | Homology |
|---|---|---|---|---|---|
| 1 | JQ269674 | Cu/Zn Superoxide dismutase | Triticum aestivum | 1 | 97.8% |
| 2 | KJ907387.1 | SGT1 | Triticum aestivum | 1 | 99.3% |
| 3 | GU564291 | Defender against cell death 2 | Triticum aestivum | 1 | 100.0% |
| 4 | XM_020309713 | Pyrrolidone-carboxylate peptidase | Aegilops tauschii | 1 | 85.6% |
| 5, 6, 7 | AM087943 | Avenin-like a precursor | Aegilops tauschii | 3 | 93.2% |
| 8, 9, 10 | XM_020326125 | MOB kinase activator-like | Aegilops tauschii | 3 | 95.3% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, T.; Li, G.-Z.; Wang, C.-R.; Dong, J.; Yuan, S.-S.; Wang, Y.-H.; Kang, G.-Z. Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency. Int. J. Mol. Sci. 2018, 19, 1465. https://doi.org/10.3390/ijms19051465
Gao T, Li G-Z, Wang C-R, Dong J, Yuan S-S, Wang Y-H, Kang G-Z. Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency. International Journal of Molecular Sciences. 2018; 19(5):1465. https://doi.org/10.3390/ijms19051465
Chicago/Turabian StyleGao, Tian, Ge-Zi Li, Chuan-Ren Wang, Jie Dong, Sha-Sha Yuan, Yong-Hua Wang, and Guo-Zhang Kang. 2018. "Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency" International Journal of Molecular Sciences 19, no. 5: 1465. https://doi.org/10.3390/ijms19051465
APA StyleGao, T., Li, G.-Z., Wang, C.-R., Dong, J., Yuan, S.-S., Wang, Y.-H., & Kang, G.-Z. (2018). Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency. International Journal of Molecular Sciences, 19(5), 1465. https://doi.org/10.3390/ijms19051465




