The Expression Pattern of PLIN2 in Differentiated Adipocytes from Qinchuan Cattle Analysis of Its Protein Structure and Interaction with CGI-58
Abstract
:1. Introduction
2. Results
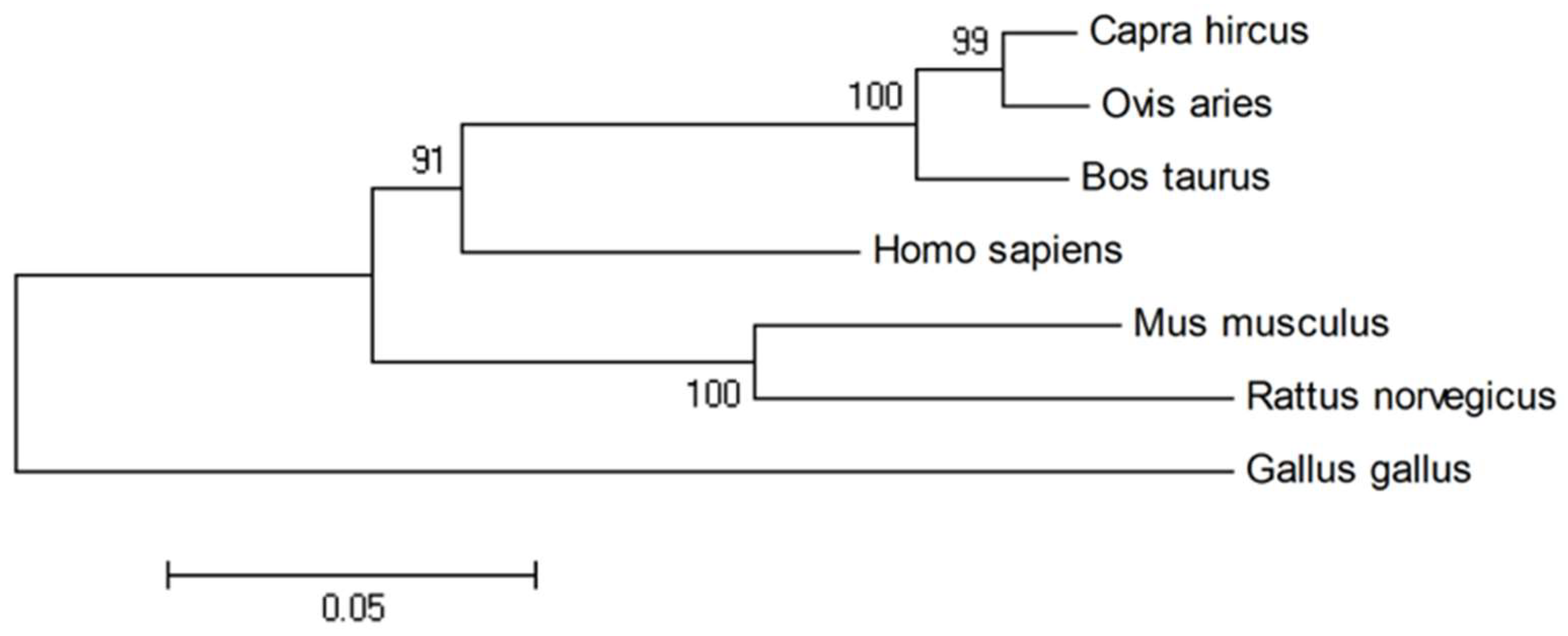
2.1. Sequence Homology, Inferred Phylogenetic Tree and Sequence Alignments
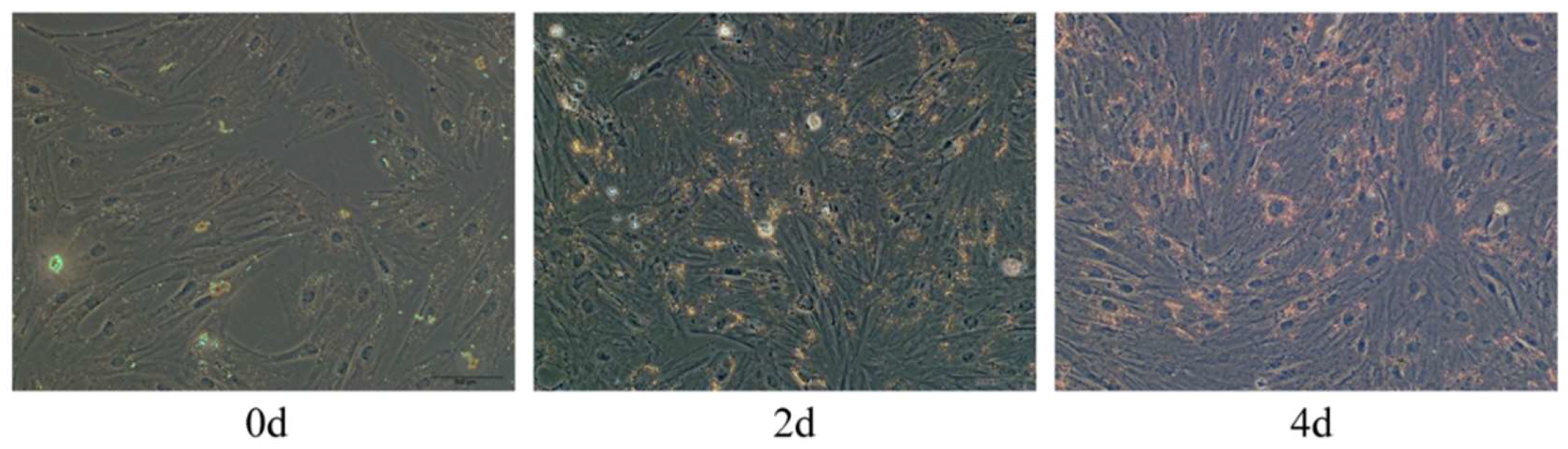
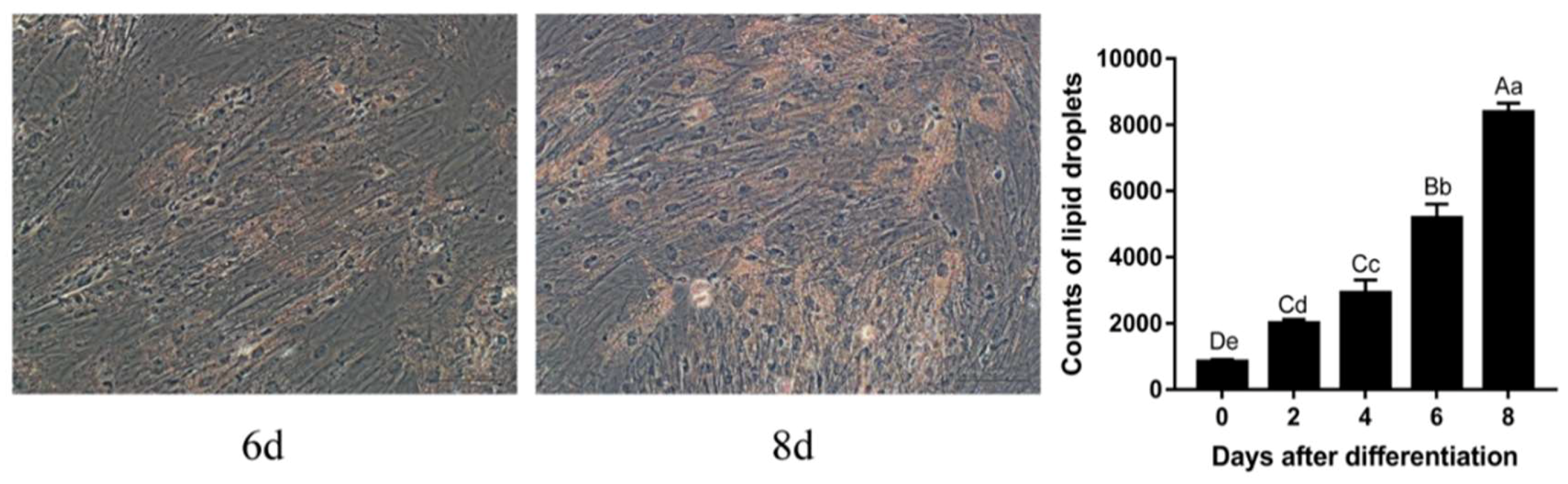
2.2. Oil Red O Staining
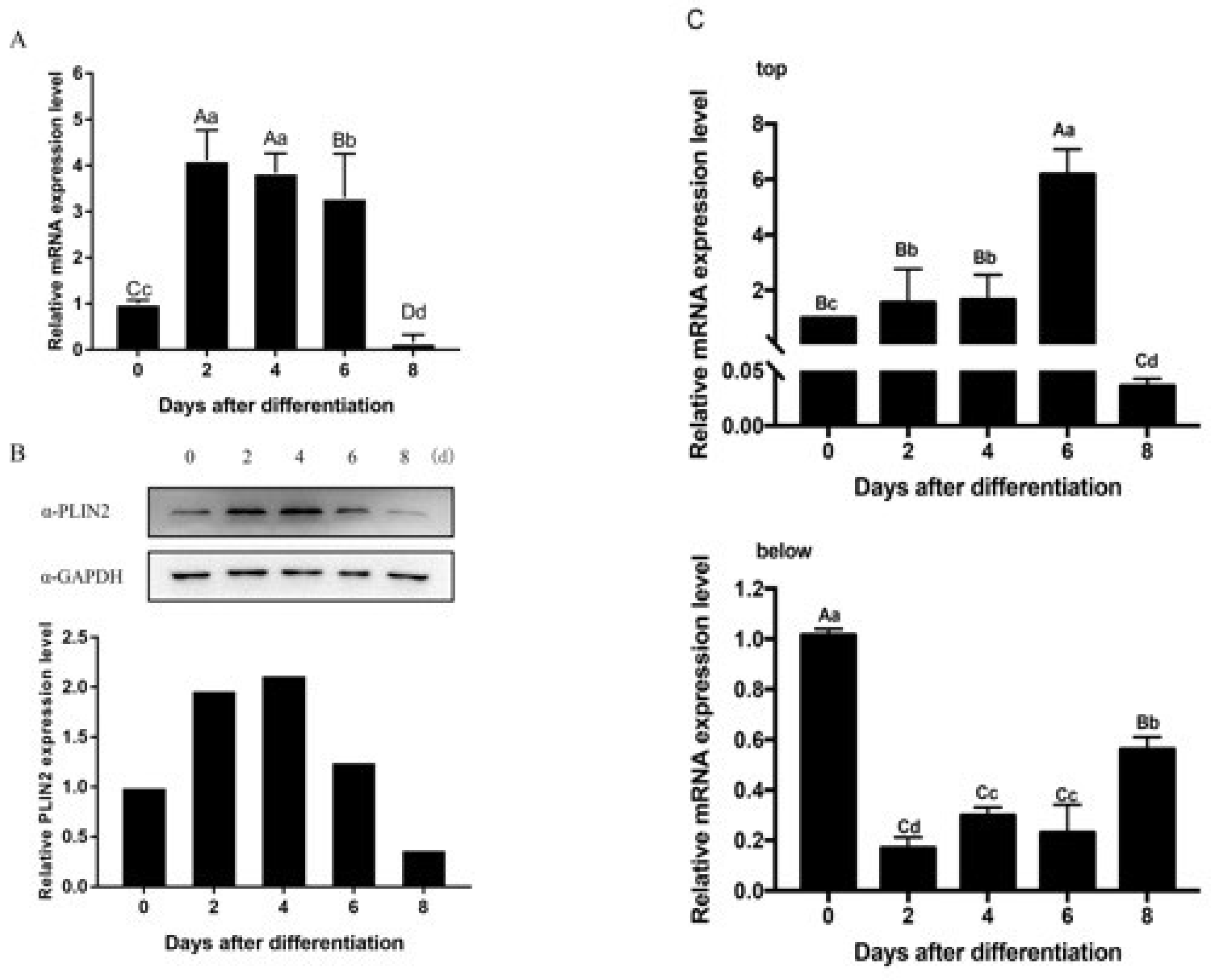
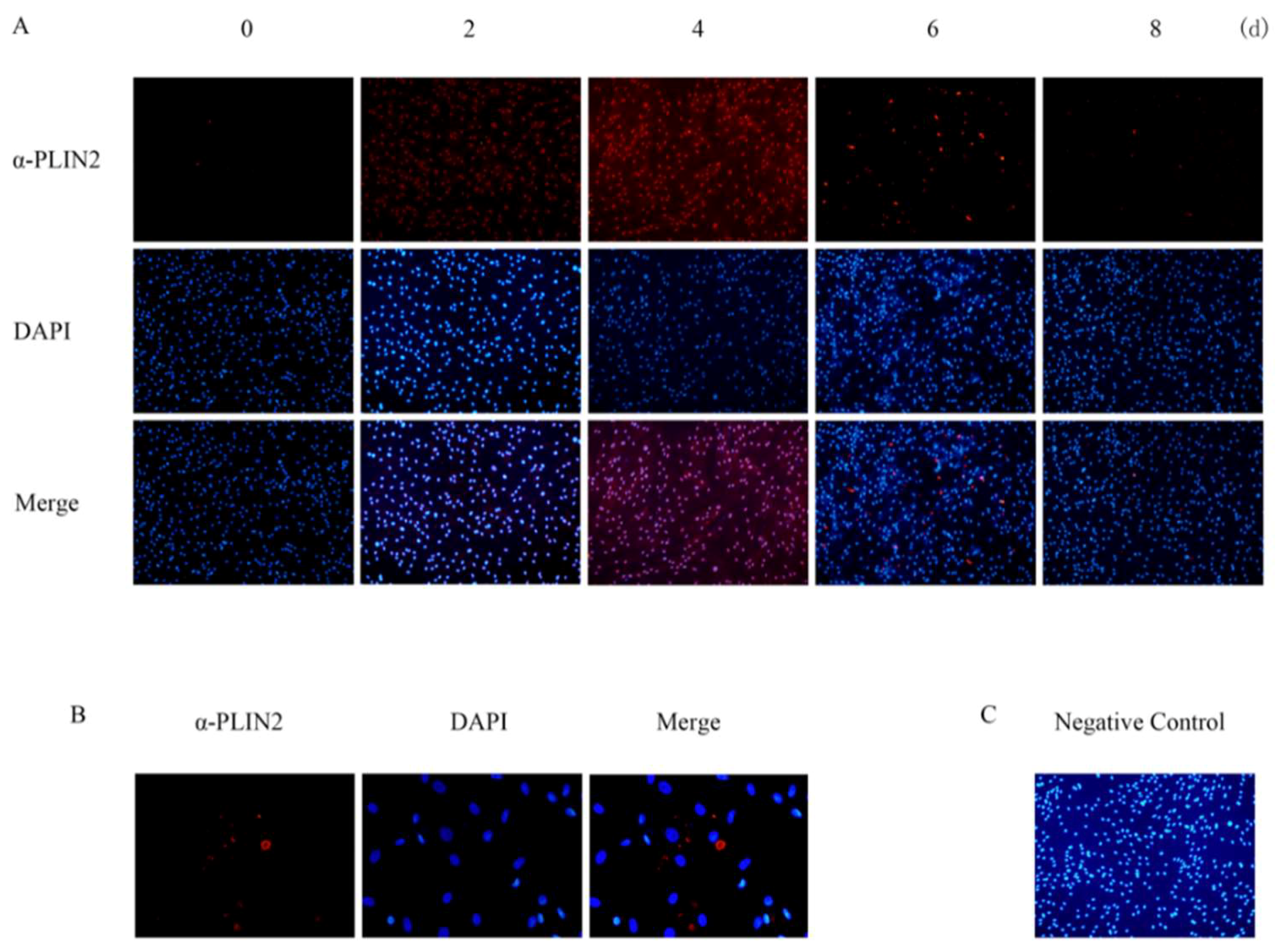
2.3. PLIN2 Expression and Location
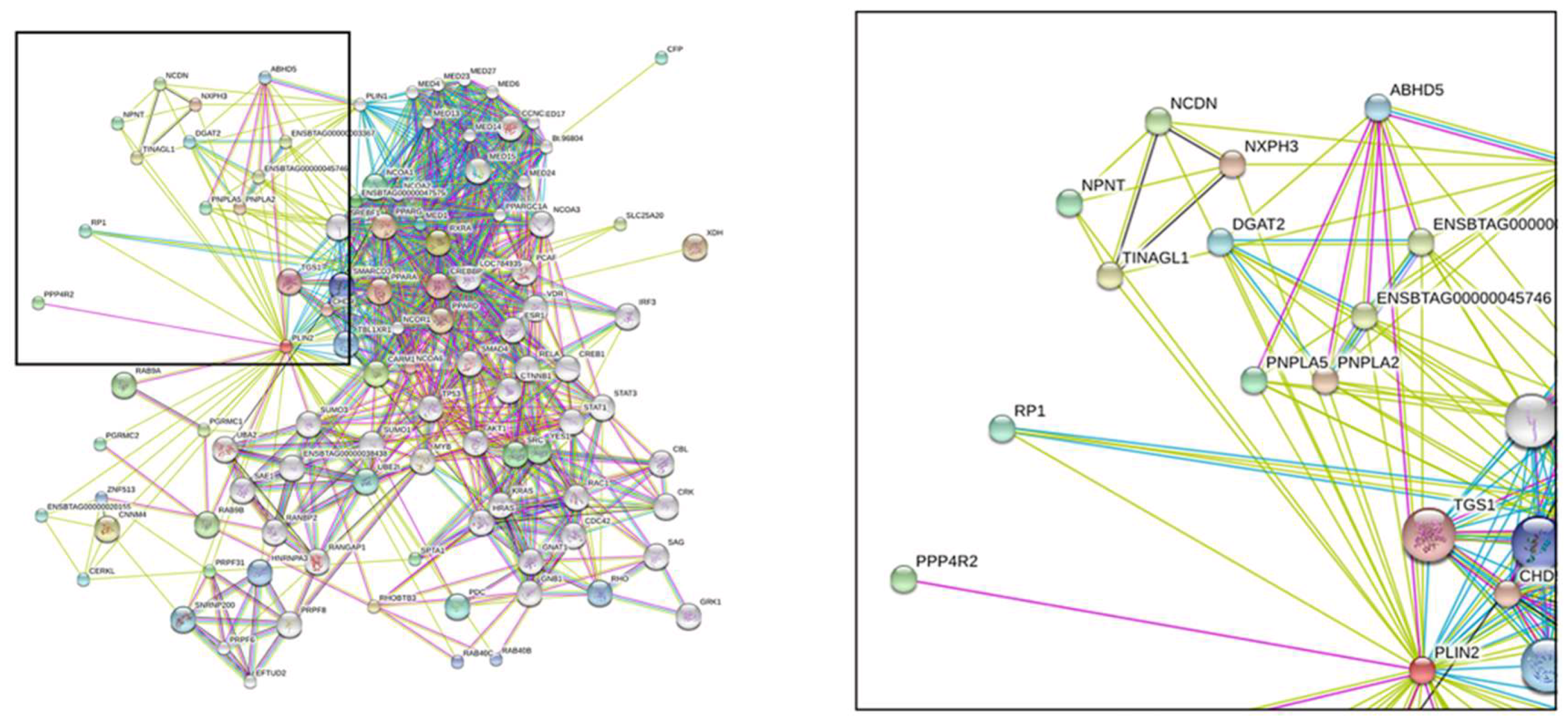
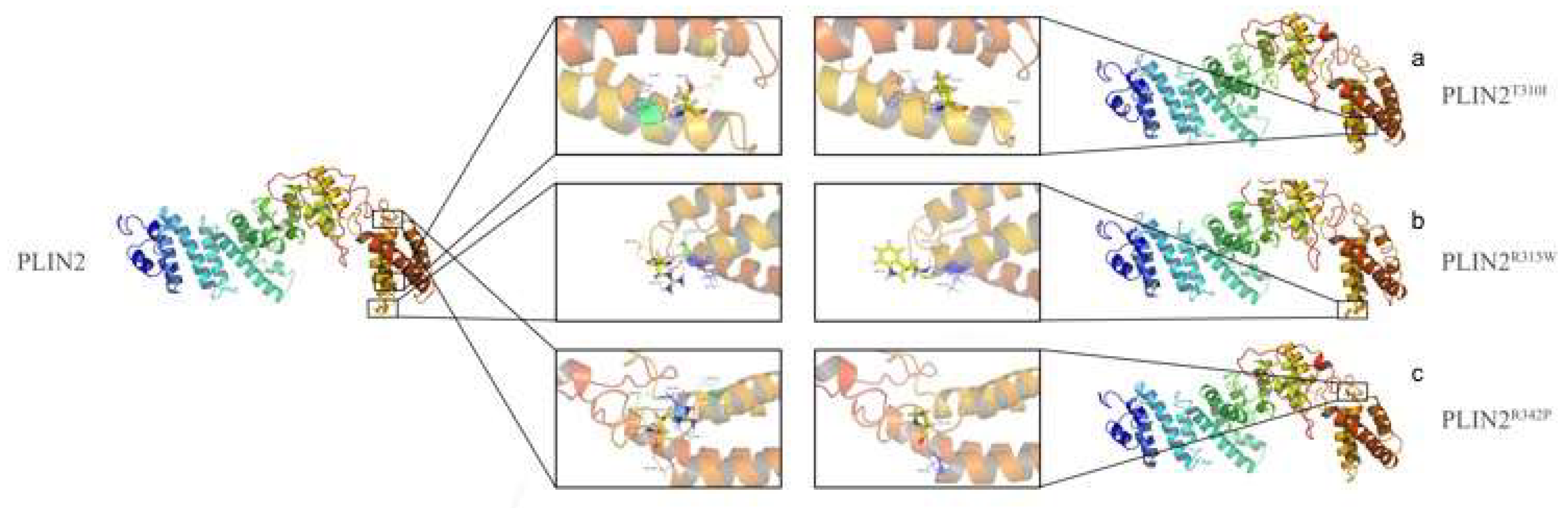
2.4. Protein Structure Analysis
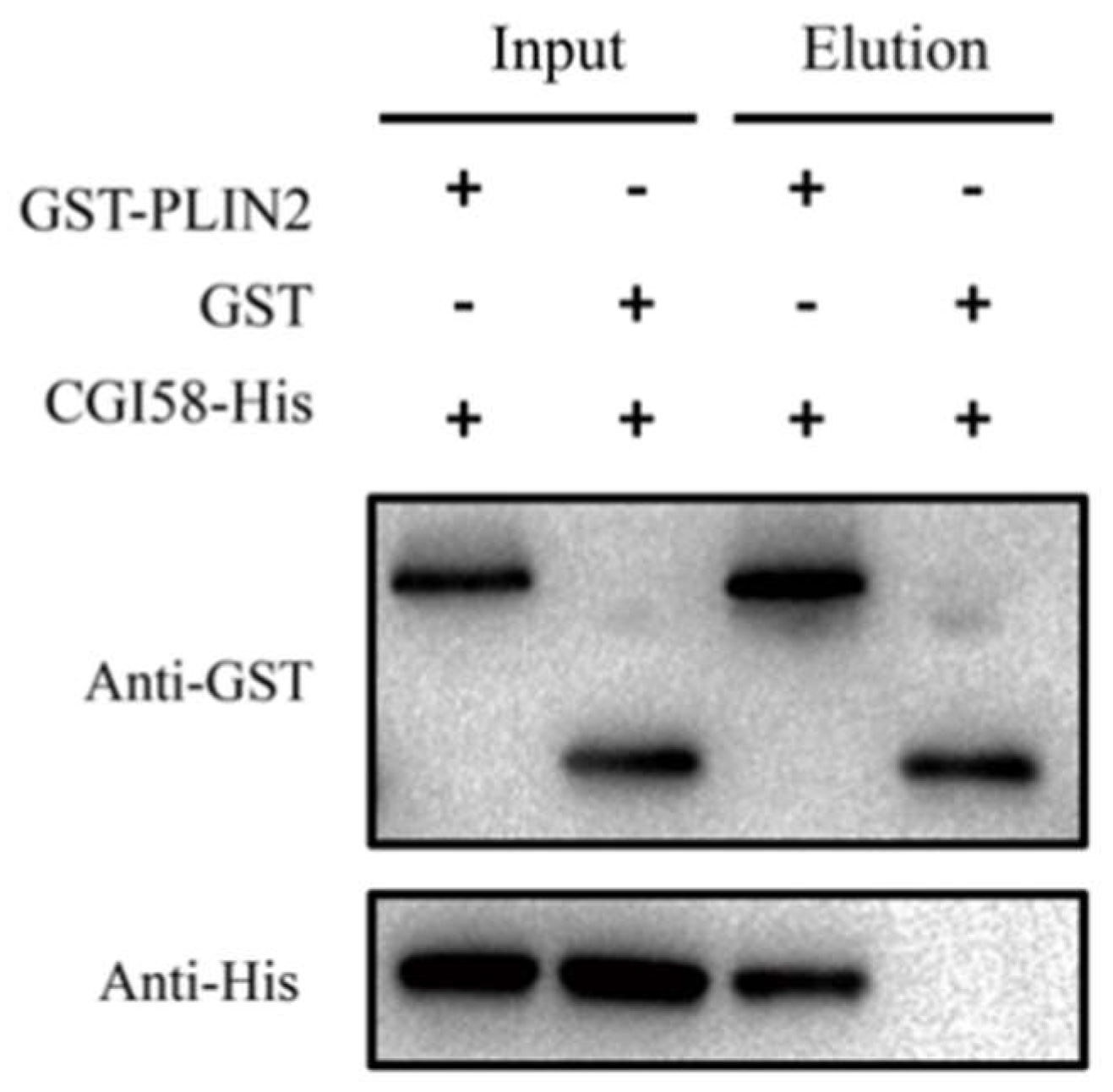
2.5. PLIN2 Interaction with CGI-58 Pull-Down Assay
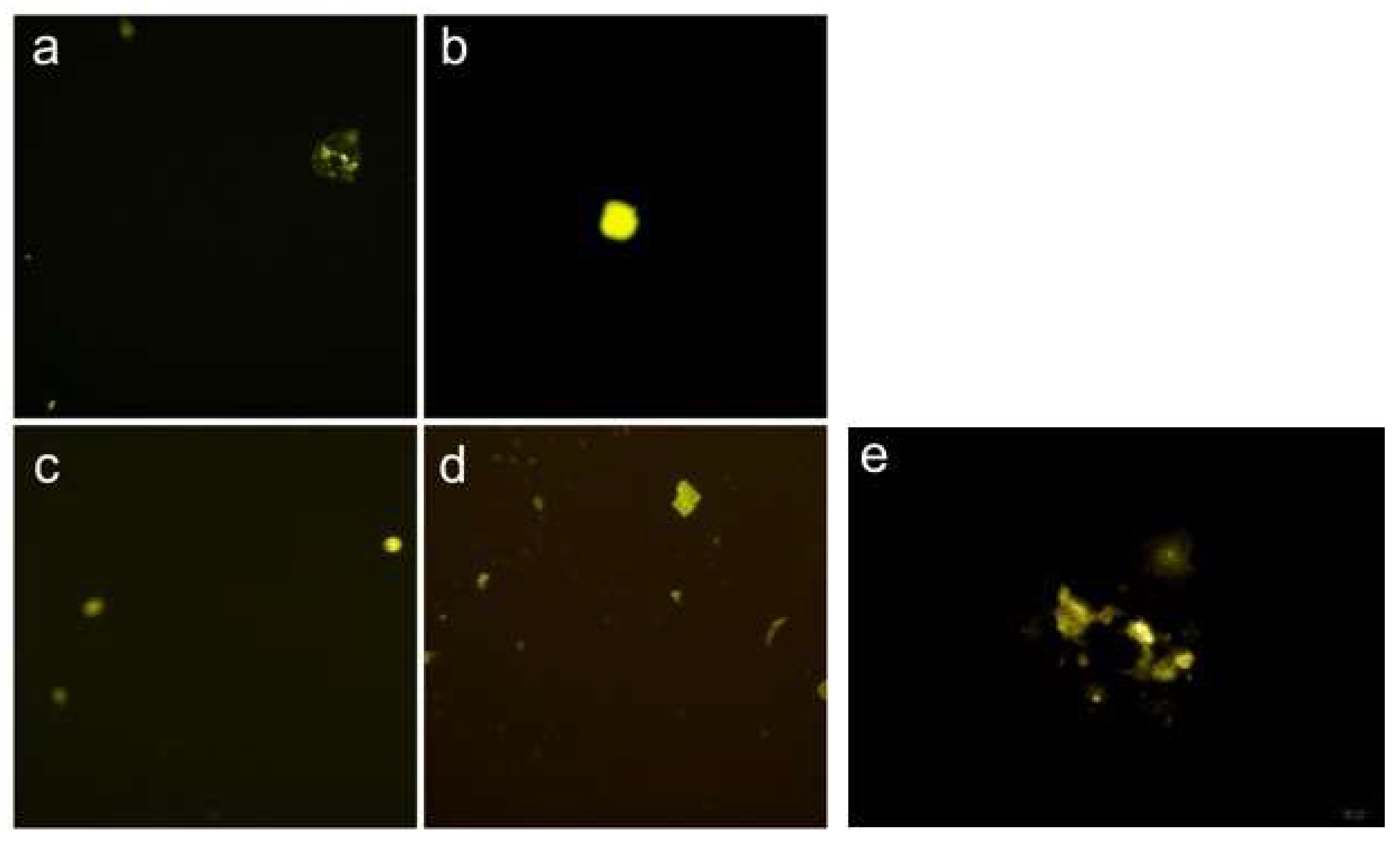
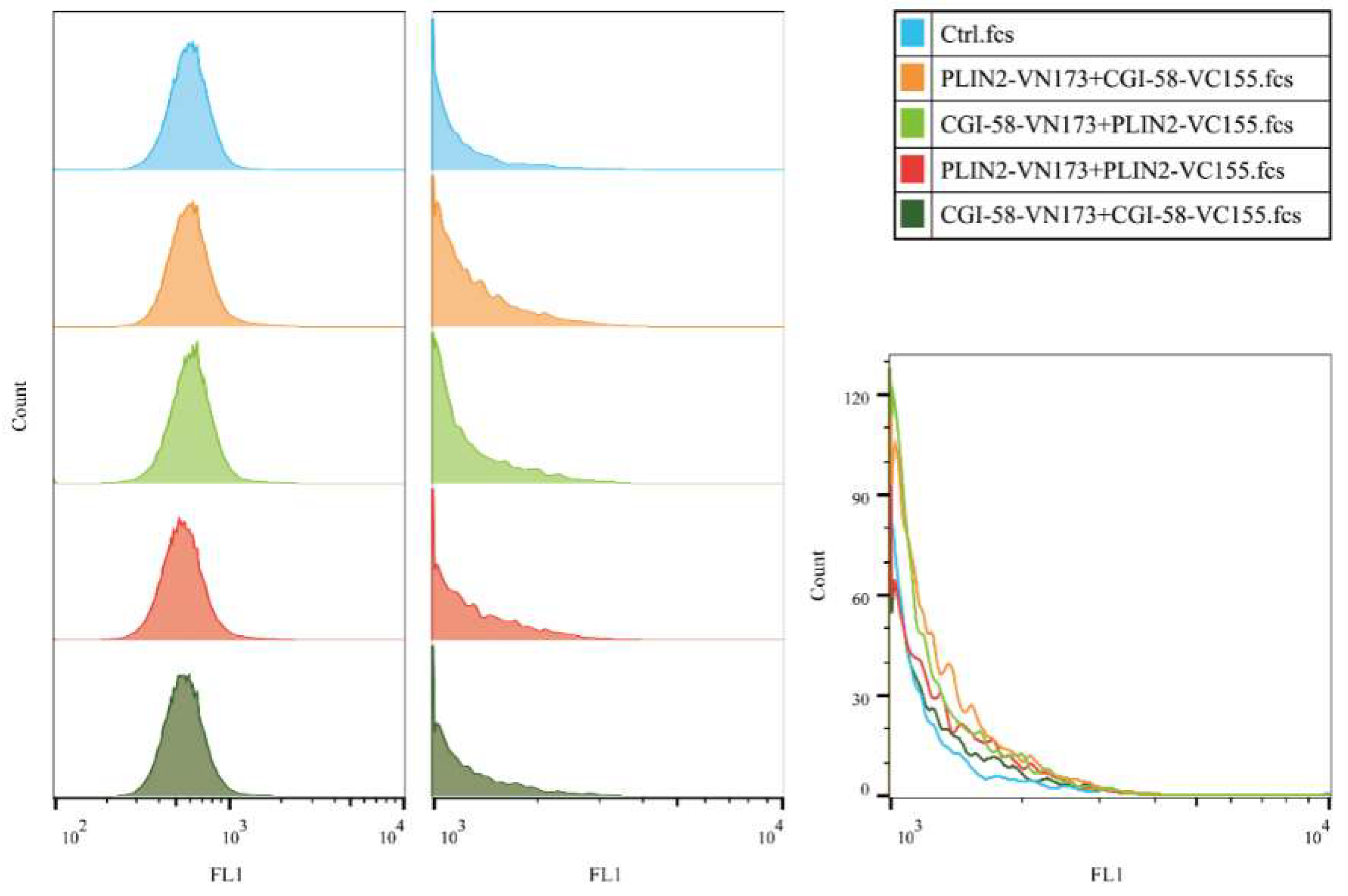
2.6. Bimolecular Fluorescence Complementation with Flow Cytometry (BiFC-FC)
2.7. Amino Acid Mutation Analysis
3. Discussion
4. Material and Methods
4.1. Isolation and Cell Culture of Bovine Adipocytes
4.2. Induction of Adipocytes Lipid Droplet Formation
4.3. Oil Red O Staining and Counting Lipid Droplets
4.4. Quantitative RT-PCR
4.5. Western Blot Analysis
4.6. Immunofluorescence
4.7. Bioinformatic Study
4.8. GST Pull-Down Assays
4.9. Bimolecular Fluorescence Complementation (BiFC)
4.10. Flow Cytometry
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Onal, G.; Kutlu, O.; Gozuacik, D.; Dokmeci Emre, S. Lipid Droplets in Health and Disease. Lipid Health Dis. 2017, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Torday, J.; Rehan, V. Neutral lipid trafficking regluates alveolar type 2 surfactant phospholipid and surfactant protein expression. Exp. Lung Res. 2011, 37, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Omatsu, N.; Omukae, A.; Osumi, T. Analysis of interaction partners for perilipin and ADRP on lipid droplets. Mol. Cell. Biochem. 2006, 284, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat Mobilization in Adipose Tissue is Promoted by Adipose Triglyceride Lipase. Science 2016, 306, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Bickel, P.E.; Tansey, J.T.; Welte, M.A. Pat proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 2009, 1791, 419–440. [Google Scholar] [CrossRef] [PubMed]
- Robenek, H.; Robenek, M.J.; Buers, I.; Lorkowski, S.; Hofnagel, O.; Troyer, D.; Severs, N.J. Lipid droplets gain pat family proteins by interaction with specialized plasma membrane domains. J. Biol. Chem. 2005, 280, 26330–26338. [Google Scholar] [CrossRef] [PubMed]
- Tauchi-Sato, K.; Ozeki, S.; Houjou, T.; Taguchi, R.; Fujimoto, T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 2002, 277, 44507–44512. [Google Scholar] [CrossRef] [PubMed]
- Kory, N.; Thiam, A.R.; Farese, R.V., Jr.; Walther, T.C. Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell 2015, 34, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, A.R.; Brasaemle, D.L.; McAndrews-Hill, M.; Sztalryd, C.; Londos, C. Adoption of perilipin as a unifying nomenclature for the mammalian pat-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010, 51, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Wolins, N.E.; D Brasaemle, D.L.; Bickel, P.E. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006, 580, 5484–5491. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.V.; Wei, S.; Duarte, M.; Du, M.; Jiang, Z.; Hausman, G.J.; Bergen, W.G. Cell supermarket: Adipose tissue as a source of stem cells. J. Genom. 2013, 1, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Sastre, D.; da Costa, N.N.; de Sa, A.L.; Conceicao, S.D.; Chiaratti, M.R.; Adona, P.R.; Guemra, S.; Meirelles, F.V.; Santos Sdo, S.; Sena, L.; et al. Expression of plin2 and plin3 during oocyte maturation and early embryo development in cattle. Theriogenology 2014, 81, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.M.; Hwang, R.D.; Greenberg, A.S.; Yeo, H.L. Temporal and spatial assembly of lipid droplet-associated proteins in 3t3-l1 preadipocytes. Histochem. Cell Biol. 2003, 120, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Betters, J.L.; Lord, C.; Ma, Y.; Han, X.; Yang, K.; Alger, H.M.; Melchior, J.; Sawyer, J.; Shah, R.; et al. Cgi-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J. Lipid Res. 2010, 51, 3306–3315. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L.; Subramanian, V.; Garcia, A.; Marcinkiewicz, A.; Rothenberg, A. Perilipin a and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 2009, 326, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by cgi-58 and defective in chanarin-dorfman syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Yang, H.; Wang, S.P.; Soni, K.G.; Brunel-Guitton, C.; Mitchell, G.A. Inborn errors of cytoplasmic triglyceride metabolism. J. Inherit. Metab. Dis. 2015, 38, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, M.; Schweiger, M.; Schreiber, R.; Haemmerle, G.; Lass, A.; Fledelius, C.; Jacobsen, P.; Tornqvist, H.; Zechner, R.; Zimmermann, R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 2006, 281, 40236–40241. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Itabe, H.; Odaki, M.; Hama, K.; Fujimoto, Y.; Mori, M.; Sasabe, N.; Aoki, J.; Arai, H.; Takano, T. Adrp/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J. Lipid Res. 2006, 47, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shinoda, A.; Kamada, H.; Shimizu, M.; Inoue, J.; Sato, R. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 2016, 6, 20975. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Shin, K.; Patterson, R.E.; Liu, X.Q.; Rainey, J.K. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem. Cell Biol. 2016, 94, 507–527. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, E.; Sansom, M.S. Membrane proteins: Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2008, 18, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.; Lloubes, R. Large complexes: Cloning strategy, production, and purification. Methods Mol. Biol. 2017, 1615, 299–309. [Google Scholar] [PubMed]
- Gui, L.; Jia, C.; Zhang, Y.; Zhao, C.; Zan, L. Association studies on the bovine lipoprotein lipase gene polymorphism with growth and carcass quality traits in qinchuan cattle. Mol. Cell. Probes 2016, 30, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T. Protein secondary structure prediction based on position- specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Alexei, V.F.; Oled, B.P. Protein Physics (Chinese version); Science Press: Beijing, China, 2012; pp. 104–112. [Google Scholar]
- Tuncbag, N.; Gursoy, A.; Nussinov, R.; Keskin, O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011, 9, 1341–1354. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Hu, C. Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 2012, 5, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 2006, 1, 61–66. [Google Scholar] [CrossRef]
- Dahlhoff, M.; Camera, E.; Picardo, M.; Zouboulis, C.C.; Chan, L.; Chang, B.H.; Schneider, M.R. Plin2, the major perilipin regulated during sebocyte differentiation, controls sebaceous lipid accumulation in vitro and sebaceous gland size in vivo. Biochim. Biophys. Acta 2013, 1830, 4642–4649. [Google Scholar] [CrossRef] [PubMed]
- Straub, B.K.; Gyoengyoesi, B.; Koenig, M.; Hashani, M.; Pawella, L.M.; Herpel, E.; Mueller, W.; Macher-Goeppinger, S.; Heid, H.; Schirmacher, P. Adipophilin/perilipin-2 as a lipid droplet-specific marker for metabolically active cells and diseases associated with metabolic dysregulation. Histopathology 2013, 62, 617–631. [Google Scholar] [CrossRef] [PubMed]
- McManaman, J.L.; Bales, E.S.; Orlicky, D.J.; Jackman, M.; MacLean, P.S.; Cain, S.; Crunk, A.E.; Mansur, A.; Graham, C.E.; Bowman, T.A.; et al. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J. Lipid Res. 2013, 54, 1346–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, L.; Dalen, K.; Dorward, H.; Marcinkiewicz, A.; Russell, D.; Gong, D.; Londos, C.; Yamaguchi, T.; Holm, C.; et al. Activation of hormone-sensitive lipase requires two steps, protein phosphorylation and binding to the pat-1 domain of lipid droplet coat proteins. J. Biol. Chem. 2009, 284, 32116–32125. [Google Scholar] [CrossRef] [PubMed]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell. Biol. 2008, 9, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Brites, D.; Fernandes, A. Neuroinflammation and depression: Microglia activation, extracellular microvesicles and microrna dysregulation. Front. Cell. Neurosci. 2015, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Sztalryd, C.; Lu, X.; Tansey, J.T.; Gan, J.; Dorward, H.; Kimmel, A.R.; Londos, C. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 2005, 280, 42841–42847. [Google Scholar] [CrossRef] [PubMed]
- Sriram, S.M.; Kim, B.Y.; Kwon, Y.T. The n-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011, 12, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Schmidt, R.; Bukau, B. The n-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef] [PubMed]
- AlSaleh, A.; Sanders, T.A.; O'Dell, S.D. Effect of interaction between pparg, ppara and adipoq gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc. Nutr. Soc. 2012, 71, 141–153. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, R.E.; Ramos, S.V.; Vandenboom, R.; Roy, B.D.; Peters, S.J. Skeletal muscle plin proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R644–R650. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L.; Barber, T.; Wolins, N.E.; Serrero, G.; Blanchette-Mackie, E.J.; Londos, C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet associated protein. J. Lipid Res. 1997, 38, 2249–2263. [Google Scholar] [PubMed]
- Mak, K.M.; Ren, C.; Ponomarenko, A.; Cao, Q.; Lieber, C.S. Adipose differentiation-related protein is a reliable lipid droplet marker in alcoholic fatty liver of rats. Alcohol. Clin. Exp. Res. 2008, 32, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Minnaard, R.; Sparks, L.M.; Schaart, G.; Losen, M.; de Baets, M.H.; Duimel, H.; Kersten, S.; Bickel, P.E.; Schrauwen, P.; et al. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 2012, 137, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Yuan, B.; Zhang, L.; Gao, Y.; Jiang, H.; Dai, L.; Zhang, J. miR-375 negatively regulates porcine preadipocyte differentiation by targeting BMPR2. FEBS Lett. 2016, 590, 1417–1427. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
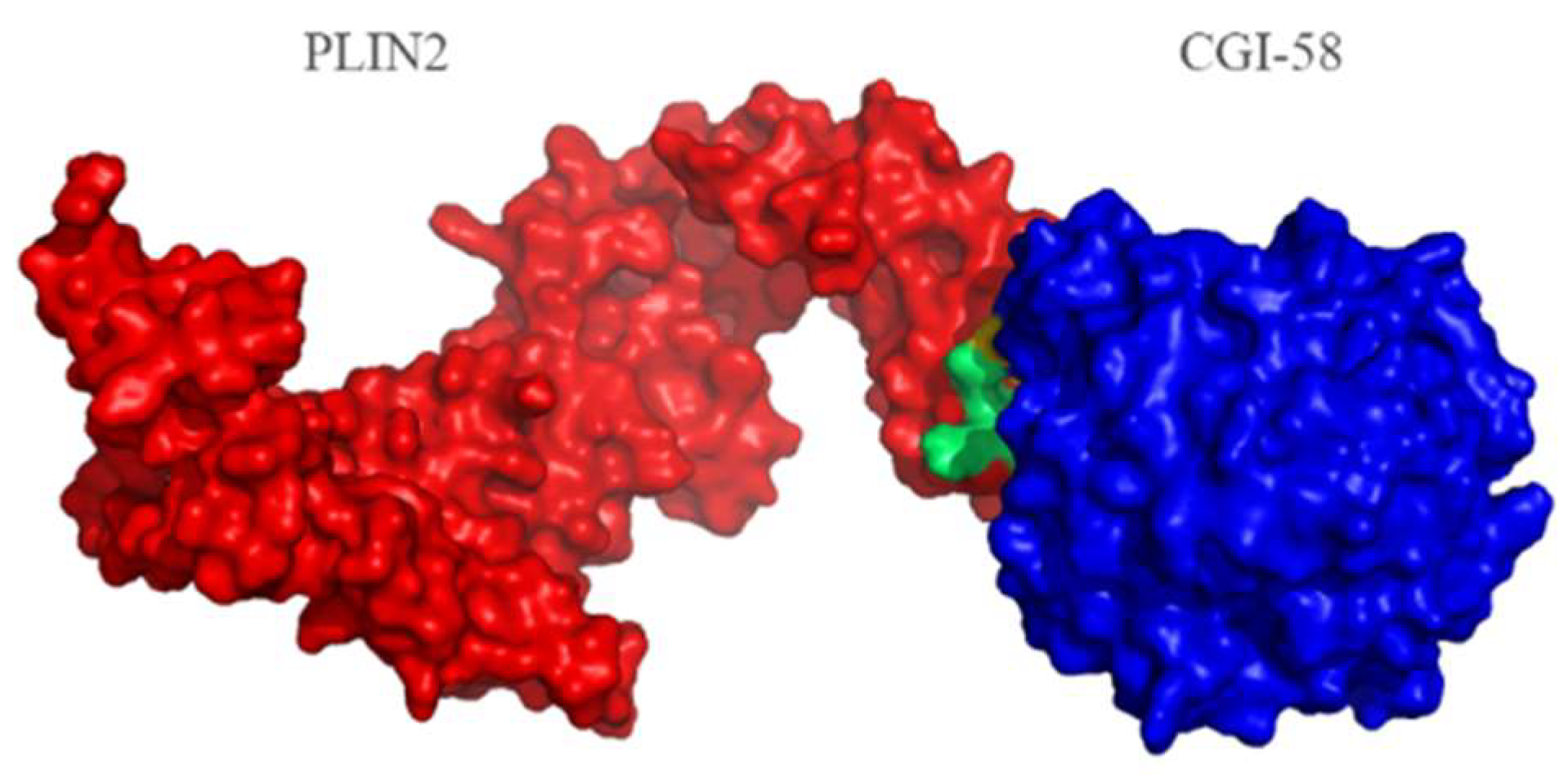
| CGI-58 | PLIN2 | |
|---|---|---|
| TRP_27 | <--> | LEU_371 |
| PHE_119 | <--> | SER_367 |
| TRP_27 | <--> | LYS_375 |
| PHE_119 | <--> | LYS_364 |
| PHE_119 | <--> | GLU_365 |
| CYS_28 | <--> | GLN_379 |
| PHE_119 | <--> | ASP_368 |
| GLU_259 | <--> | SER_367 |
| TRP_27 | <--> | ALA_314 |
| ARG_118 | <--> | ASP_368 |
| TRP_27 | <--> | ILE_313 |
| ARG_118 | <--> | SER_367 |
| PRO_25 | <--> | GLN_379 |
| ARG_118 | <--> | GLY_369 |
| THR_26 | <--> | GLY_376 |
| THR_26 | <--> | LYS_375 |
| THR_255 | <--> | ARG_315 * |
| THR_255 | <--> | ALA_314 |
| PRO_29 | <--> | LEU_311 |
| Vector Names | Efficiency |
|---|---|
| PLIN2-VN173+CGI-58-VC155 | ++ |
| PLIN2-VN173+PLIN2-VC155 | + |
| CGI-58-VN173+CGI-58-VC155 | + |
| CGI-58-VN173+PLIN2-VC155 | ++ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Wang, Y.; Zhang, L.; Ning, Y.; Zan, L. The Expression Pattern of PLIN2 in Differentiated Adipocytes from Qinchuan Cattle Analysis of Its Protein Structure and Interaction with CGI-58. Int. J. Mol. Sci. 2018, 19, 1336. https://doi.org/10.3390/ijms19051336
Li P, Wang Y, Zhang L, Ning Y, Zan L. The Expression Pattern of PLIN2 in Differentiated Adipocytes from Qinchuan Cattle Analysis of Its Protein Structure and Interaction with CGI-58. International Journal of Molecular Sciences. 2018; 19(5):1336. https://doi.org/10.3390/ijms19051336
Chicago/Turabian StyleLi, Peiwei, Yaning Wang, Le Zhang, Yue Ning, and Linsen Zan. 2018. "The Expression Pattern of PLIN2 in Differentiated Adipocytes from Qinchuan Cattle Analysis of Its Protein Structure and Interaction with CGI-58" International Journal of Molecular Sciences 19, no. 5: 1336. https://doi.org/10.3390/ijms19051336
APA StyleLi, P., Wang, Y., Zhang, L., Ning, Y., & Zan, L. (2018). The Expression Pattern of PLIN2 in Differentiated Adipocytes from Qinchuan Cattle Analysis of Its Protein Structure and Interaction with CGI-58. International Journal of Molecular Sciences, 19(5), 1336. https://doi.org/10.3390/ijms19051336




