Lack of the α1,3-Fucosyltransferase Gene (Osfuct) Affects Anther Development and Pollen Viability in Rice
Abstract
1. Introduction
2. Results
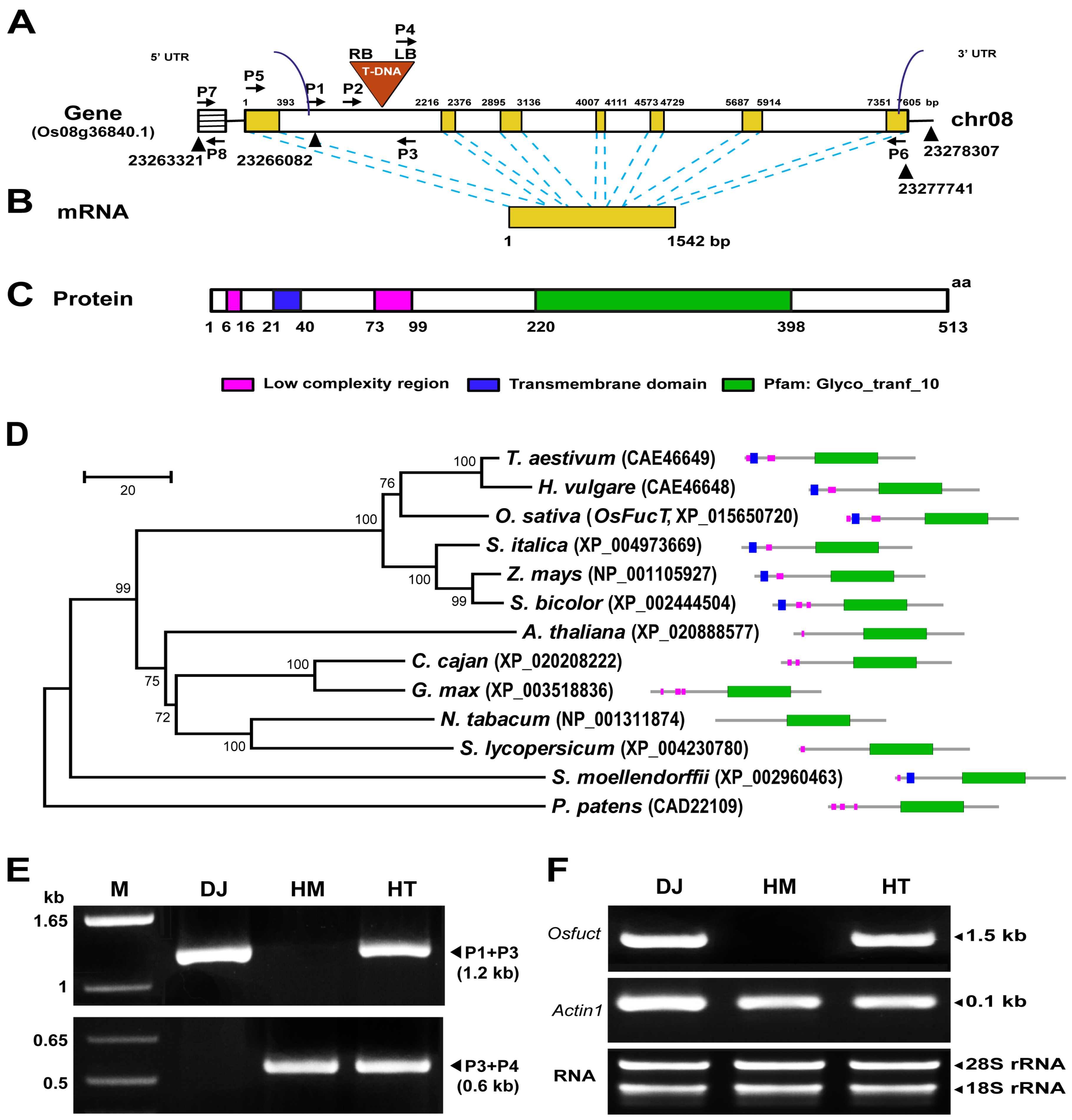
2.1. Identification and Isolation of a T-DNA Inserted Allele in the Rice Osfuct Gene
2.2. Osfuct Mutant Significantly Affects Anther Development
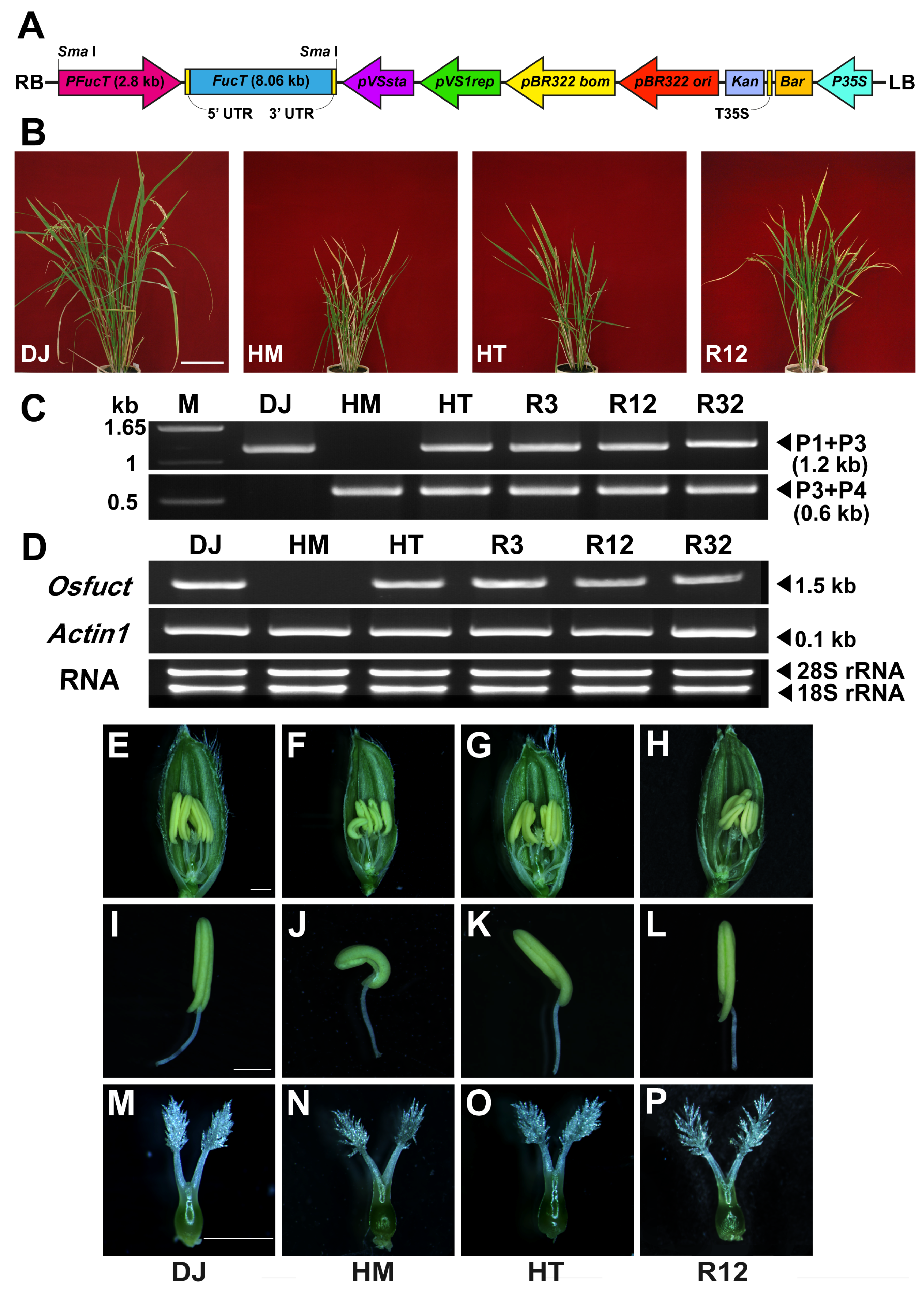
2.3. Rescue of Mutant Phenotype by Wild-Type Osfuct Transgene
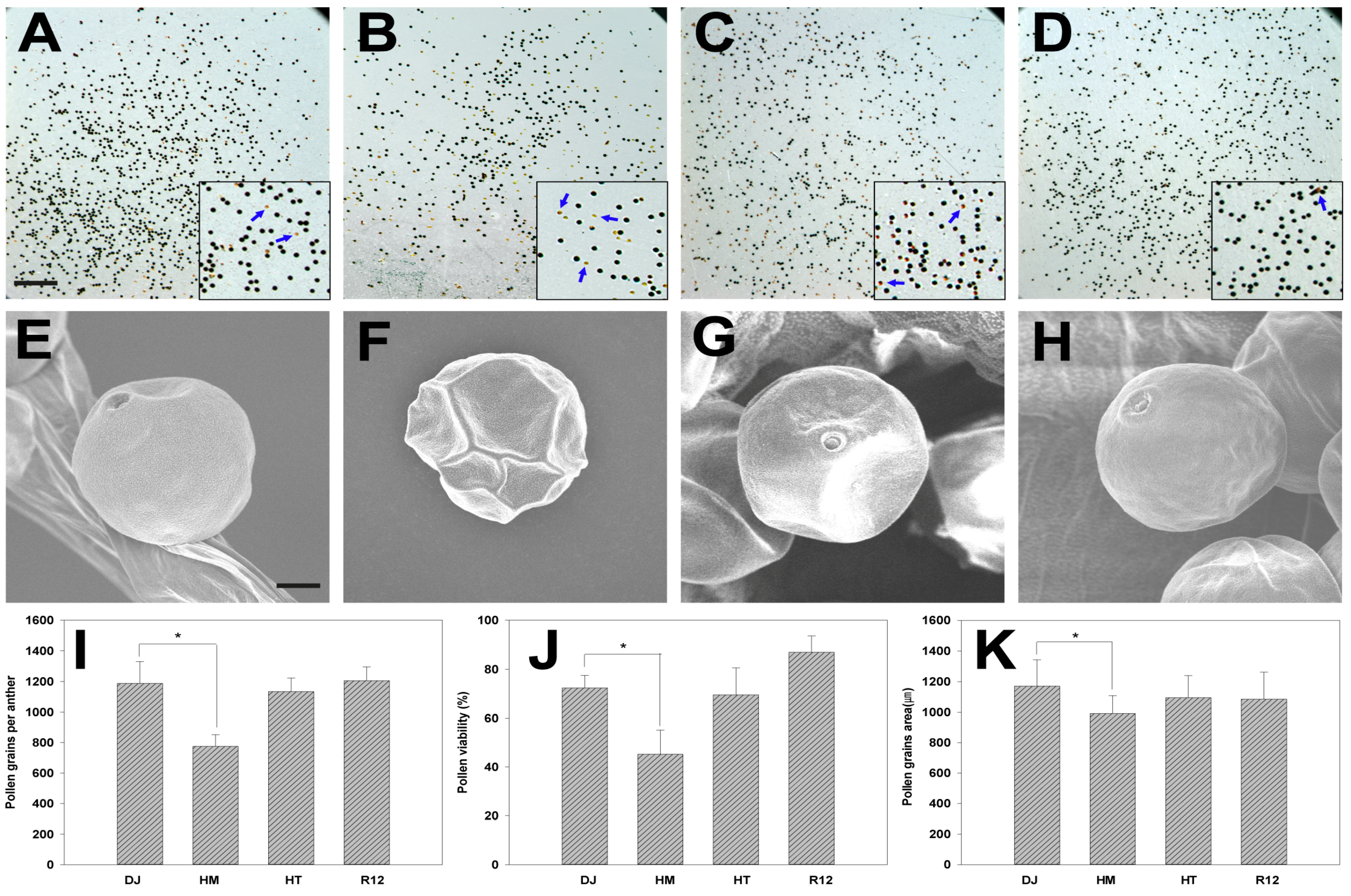
2.4. Effect of Osfuct Mutation on Pollen Development
2.5. The Glycosylation Pattern Is Altered in the Osfuct Mutant
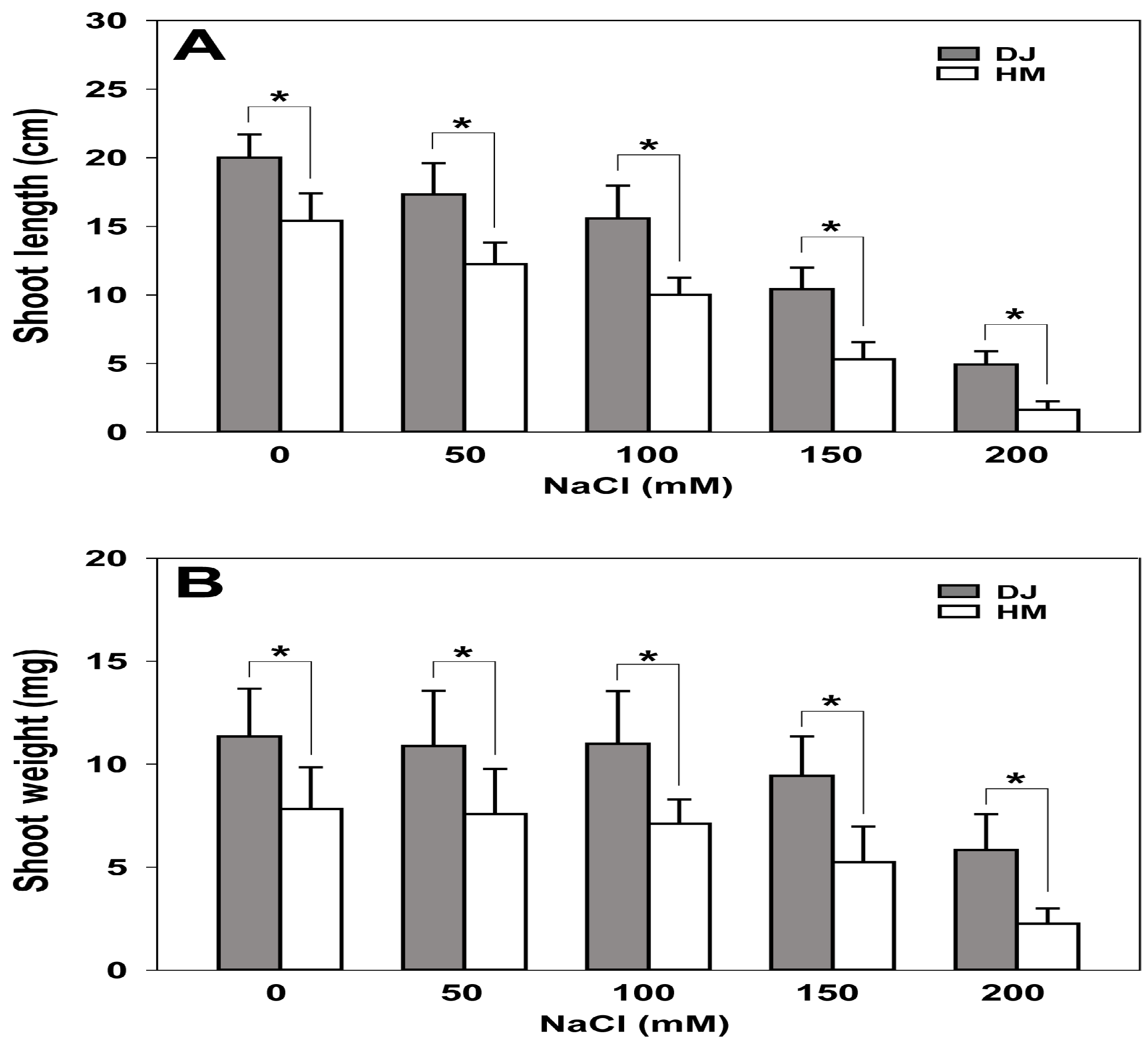
2.6. Osfuct Mutant Exhibits Sensitivity to Salt Stress
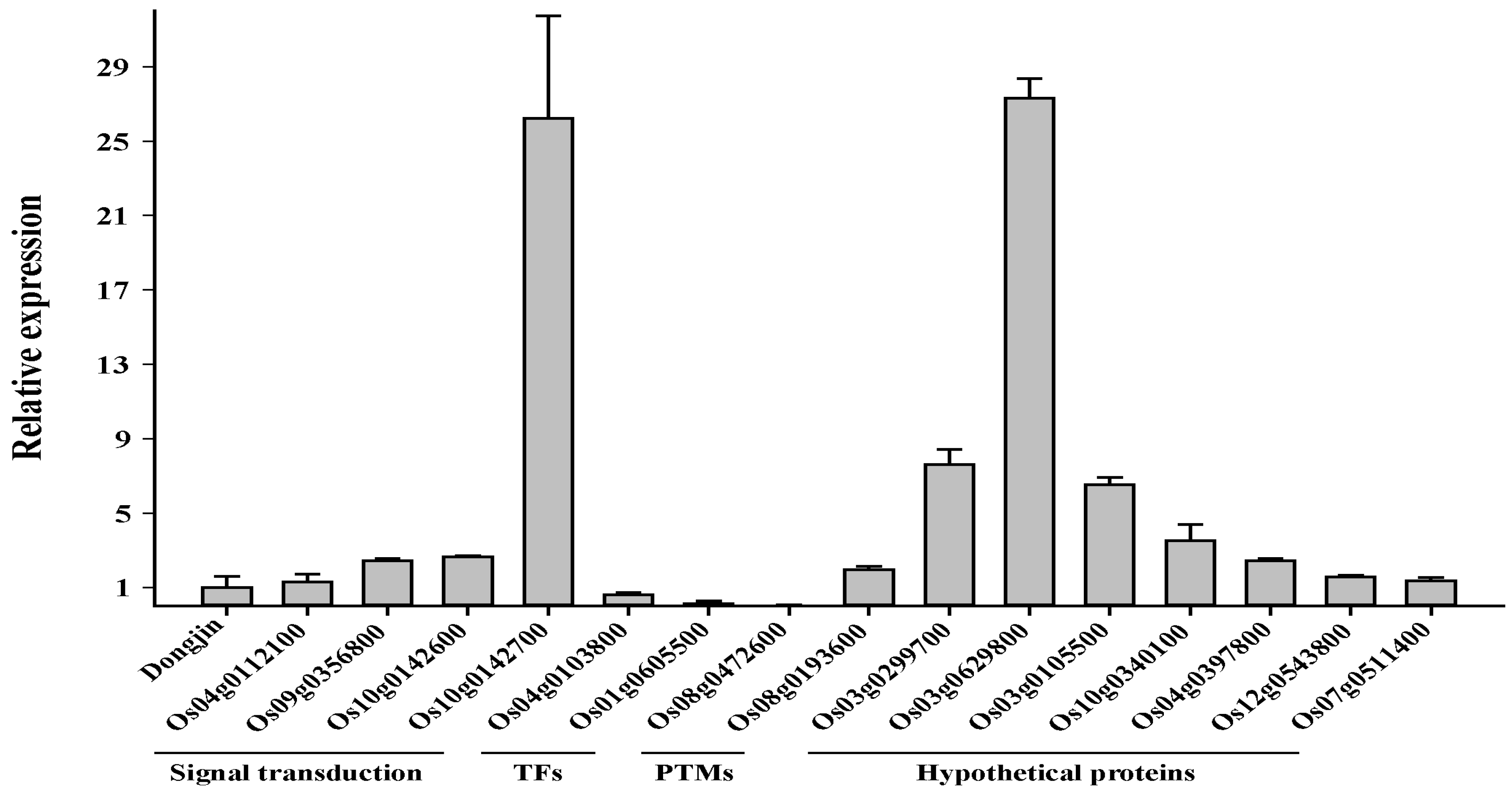
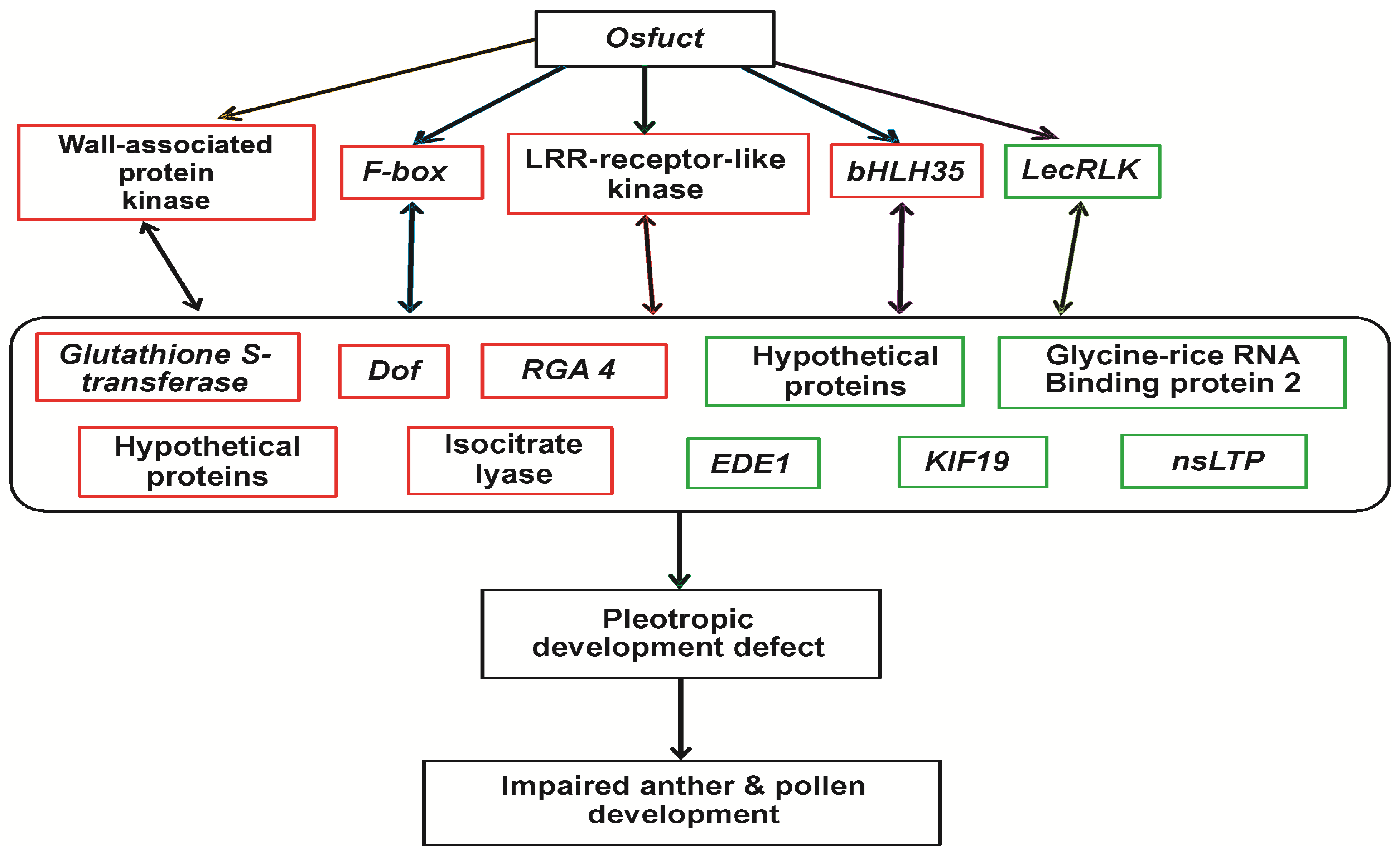
2.7. Transcriptomic Profiling of the Osfuct Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions for Mutant Screening
4.2. Genotyping and Phylogenic Analysis of the Osfuct Mutant
4.3. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Quantitative Real-Time PCR (qPCR) Analysis
4.4. Phenotypic Analyses of the Osfuct Mutant
4.5. Assessment of Reproductive Development of the Osfuct Mutant
4.6. Vector Construction for Complementation Analysis of Osfuct Disrupted Mutant
4.7. N-Glycan Profiling of the Osfuct Mutant
4.8. Salt Tolerance Assay of the Osfuct Mutant
4.9. Transcriptome Analyses of the Osfuct Mutant
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| OsFucT | α1,3-fucosyltransferase |
| MALDI-TOF | Matrix-assisted laser desorption/ionization time-of-flight |
| ORF | Open reading frame |
| HM | Homozygote |
| HT | Heterozygote |
| SEM | Scanning electron microscopy |
| GEO | Gene Expression Omnibus |
References
- Nagae, M.; Yamaguchi, Y. Function and 3D structure of the N-glycans on glycoproteins. Int. J. Mol. Sci. 2012, 13, 8398–8429. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Plant protein glycosylation. Glycobiology 2016, 26, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Goettig, P. Effects of glycosylation on the enzymatic activity and mechanisms of proteases. Int. J. Mol. Sci. 2018, 17, 1969. [Google Scholar] [CrossRef] [PubMed]
- Lerouxel, O.; Mouille, G.; Andeme-Onzighi, C.; Bruyant, M.P.; Seveno, M.; Loutelier-Bourhis, C.; Driouich, A.; Hofte, H.; Lerouge, P. Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein under glycosylation and defects in cell differentiation and growth. Plant J. 2005, 42, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Bakker, H.; Rouwendal, G.J.; Karnoup, A.S.; Florack, D.E.; Stoopen, G.M.; Helsper, J.P.; van Ree, R.; van Die, I.; Bosch, D. An antibody produced in tobacco expressing a hybrid β-1,4-galactosyltransferase is essentially devoid of plant carbohydrate epitopes. Proc. Natl. Acad. Sci. USA 2006, 103, 7577–7582. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Ko, K.S.; Seo, H.K.; Park, S.; Fanata, W.I.; Harmoko, R.; Ramasamy, N.K.; Thulasinathan, T.; Menqiste, T.; Lim, J.M.; et al. Limited addition of the 6-Arm β1,2-linked N-acetylglucosamine (GlcNAc) residue facilitates the formation of the largest N-Glycan in plants. J. Biol. Chem. 2015, 290, 16560–16572. [Google Scholar] [CrossRef] [PubMed]
- Foetisch, K.; Westphal, S.; Lauer, I.; Retzek, M.; Altmann, F.; Kolarich, D.; Scheurer, S.; Vieths, S. Biological activity of IgE specific for cross-reactive carbohydrate determinants. J. Allergy Clin. Immunol. 2003, 111, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, Y.; Gan, J.; Wang, W.; Zhang, H.; Liu, Y.; Wu, P. OsDGL1, a homolog of an oligosaccharyltransferase complex subunit, is involved in N-glycosylation and root development in rice. Plant Cell Physiol. 2013, 54, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Lalanne, E.; Honys, D.; Johnson, A.; Borner, G.H.; Lilley, K.S.; Dupree, P.; Grossniklaus, U.; Twell, D. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 2004, 16, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Farid, A.; Pabst, M.; Schoberer, J.; Altmann, F.; Glössl, J.; Strasser, R. Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J. 2011, 68, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Frank, J.; Kang, C.H.; Kajiura, H.; Vikram, M.; Ueda, A.; Kim, S.; Bahk, J.D.; Triplett, B.; Fujiyama, K.; et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 2008, 105, 5933–5938. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Altmann, F.; Mach, L.; Glössl, J.; Steinkellner, H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core α1,3-linked fucose. FEBS Lett. 2004, 561, 132–136. [Google Scholar] [CrossRef]
- Koprivova, A.; Stemmer, C.; Altmann, F.; Hoffmann, A.; Kopriva, S.; Gorr, G.; Reski, R.; Decker, E.L. Targeted knockouts of Physcomitrella lacking plant-specific immunogenic N-glycans. Plant Biotechnol. J. 2004, 2, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Mercx, S.; Smargiasso, N.; Chaumont, F.; De Pauw, E.; Boutry, M.; Navarre, C. Inactivation of the β(1,2)-xylosyltransferase and the α(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 2017, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Harmoko, R.; Yoo, J.Y.; Ko, K.S.; Ramasamy, N.K.; Hwang, B.Y.; Lee, E.J.; Kim, H.S.; Lee, K.J.; Oh, D.B.; Kim, D.Y.; et al. N-Glycan containing a core α1,3-fucose residue is required for basipetal auxin transport and gravitropic response in rice (Oryza sativa). New Phytol. 2016, 212, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.S.; Lee, S.; Jung, K.H.; Jun, S.H.; Jeong, D.H.; Lee, J.; Kim, C.; Jang, S.; Yang, K.; Nam, J.; et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000, 22, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR—Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Dievart, A.; Clark, S.E. LRR-containing receptors regulating plant development and defense. Development 2004, 131, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Abdin, M.Z. Exploring the role of wall associated receptor like kinases (Waks) during plant-fungal mutual interaction. Curr. Trends Biomed. Eng. Biosci. 2017, 4, 1–3. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Laudencla-Chlngcuanco, D.L.; Comal, L.; LI, M.; Harada, J.J. Isocitrate lyase and malate synthase genes from Brassica napus are active in pollen. Plant Physiol. 1994, 104, 857–864. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, D.; Hallett, M.A.; Zhu, W.; Rubart, M.; Liu, Y.; Yang, Z.; Chen, H.; Haneline, L.S.; Chan, R.J.; Schwartz, R.J.; et al. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development 2011, 138, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Roxas, V.P.; Smith, R.K., Jr.; Allen, E.R.; Allen, R.D. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997, 15, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.M.; Trevizan, C.B.; dos Santos, T.B.; de Souza, S.G.H. Genome-wide identification and characterization of the Dof transcription factor gene gamily in Phaseolus vulgaris L. Am. J. Plant Sci. 2017, 8, 3233–3257. [Google Scholar] [CrossRef]
- Okamoto, T.; Yamamoto, S.; Watanabe, Y.; Ohta, T.; Hanaoka, F.; Roeder, R.G.; Ohkuma, Y. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEβ subunit. J. Biol. Chem. 1998, 273, 19866–19876. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N.; Vashisht, A.A.; Tuteja, R. Translation initiation factor 4A: A prototype member of dead-box protein family. Physiol. Mol. Biol. Plants 2008, 14, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Veltman, D.M.; Insall, R.H. WASP family proteins: Their evolution and its physiological implications. Mol. Biol. Cell 2010, 21, 2880–2893. [Google Scholar] [CrossRef] [PubMed]
- Niwa, S.; Nakajuma, K.; Miki, H.; Minato, Y.; Wang, D.; Hirokawa, N. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev. Cell 2012, 23, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Pignocchi, C.; Minns, G.E.; Nesi, N.; Koumproglou, R.; Kitsios, G.; Benning, C.; Lloyd, C.W.; Doonan, J.H.; Hills, M.J. ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 2009, 21, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.H.; Ahn, S.J.; Goh, C.H.; Cho, K.; Han, O.; Kang, H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 2007, 50, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bouwmeester, K. l-type lectin receptor kinases: New forces in plant immunity. PLoS Pathog. 2017, 13, e1006433. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007, 50, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Qian, X.; Chen, M.; Fei, Q.; Meyers, B.C.; Liang, W.; Zhang, D. Regulatory role of a receptor-like kinase in specifying anther cell identity. Plant Physiol. 2016, 171, 2085–2100. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhang, S.; Wang, G.; Fan, S.; Li, M.; Chen, W.; Tu, B.; Tan, J.; Wang, Y.; Ma, B. Down-regulation of OsEMF2b caused semi-sterility due to anther and pollen development defects in rice. Front. Plant Sci. 2017, 8, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Farquharson, K.L. A domain in the bHLH transcription factor DYT1 is critical for anther development. Plant Cell 2016, 28, 997–998. [Google Scholar] [CrossRef] [PubMed]
- Brock, M.T.; Kover, P.X.; Weinig, C. Natural variation in GA1 associates with floral morphology in Arabidopsis thaliana. New Phytol. 2012, 195, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, C.; Lin, J.; Liu, J.; Liu, B.; Wang, J.; Huang, A.; Li, H.; Zhao, T. OsMPH1 regulates plant height and improves grain yield in rice. PLoS ONE 2017, 12, e0180825. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, G.; Zhao, J.; Chu, H.; Lin, W.; Zhang, D.; Wang, Z.; Liang, W. Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 2014, 10, e1004154. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Sun, F.; Wang, Q.; Chen, M.; Huang, Y.; Feng, Y.Q.; Luo, X.; Yang, J. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, D.; Li, D.; Liu, X.; Zhao, X.; Li, X.; Li, S.; Zhu, L. Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.). Front. Plant Sci. 2015, 6, 833. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Joshi, D.; Yadav, P.K.; Gupta, A.K.; Bhatt, T.K. Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 2016, 7, 806. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.E.; Olszewski, N.E. Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiol. 1991, 97, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xu, X.H.; Lu, X.; Xie, L.; Bai, B.; Zheng, C.; Sun, H.; He, Y.; Xie, X.Z. The rice phytochrome genes, PHYA and PHYB, have synergistic effects on anther development and pollen viability. Sci. Rep. 2017, 7, 6439. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Ishikawa, M.; Kitamura, S.; Takahashi, Y.; Soyano, T.; Machida, C.; Machida, Y. The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 2004, 9, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, Y.; Li, W.; Zhao, Z.; Ren, Y.; Wang, Y.; Gu, S.; Lin, Q.; Wang, D.; Jiang, L.; et al. Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 2011, 23, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; An, S.; Kang, H.G.; Moon, S.; Han, J.J.; Park, S.; Lee, H.S.; An, K.; An, G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002, 130, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.J.; Amtmann, A. N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 2009, 14, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.S.; Xing, X.J.; Peng, R.H.; Zhu, B.; Gao, J.J.; Yao, Q.H. Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant. 2016, 156, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Ronald, P.C. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 1999, 17, 53–57. [Google Scholar] [CrossRef]
- Davis, K.R.; Ausubel, F.M. Characterization of elicitor-induced defense responses in suspension-cultured cells of Arabidopsis. Mol. Plant-Microbe Interact. 1989, 2, 363–368. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Karg, S.R.; Frey, A.D.; Ferrara, C.; Streich, D.K.; Umana, P.; Kallio, P.T. A small-scale method for the preparation of plant N-linked glycans from soluble proteins for analysis by MALDI-TOF mass spectrometry. Plant Physiol. Biochem. 2009, 47, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.D.; Karg, S.R.; Kallio, P.T. Expression of rat β(1,4)-N-acetylglucosaminyltransferase III in Nicotiana tabacum remodels the plant-specific N-glycosylation. Plant Biotechnol. J. 2009, 7, 33–48. [Google Scholar] [CrossRef] [PubMed]
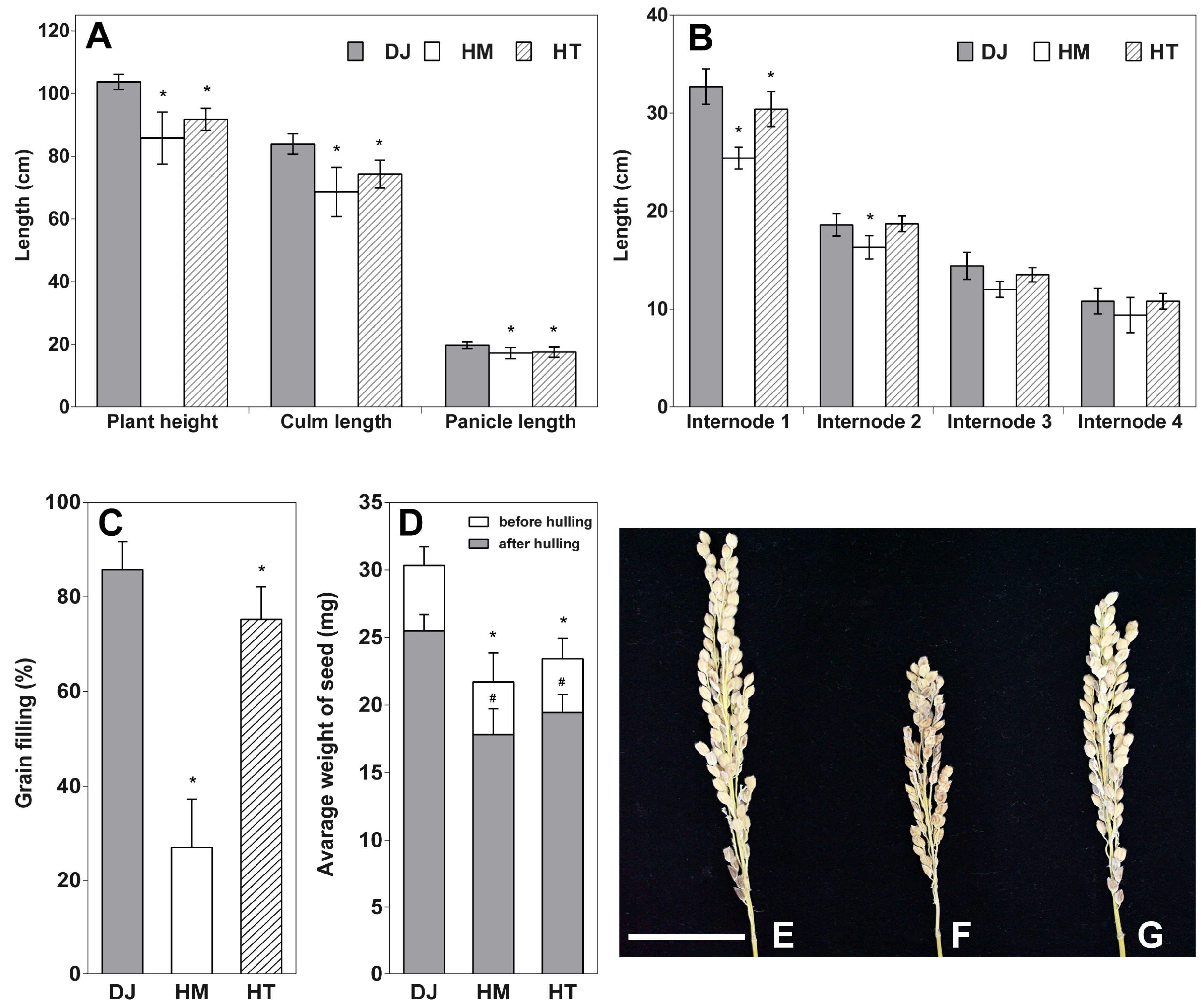
| Floral Parts | Dimensions (mm) | DJ | HM | HT | R12 |
|---|---|---|---|---|---|
| Mature flower | Length (mm) | 6.78 ± 0.28 | 6.12 ± 0.20 * | 6.29 ± 0.32 * | 6.45 ± 0.29 |
| Width (mm) | 2.65 ± 0.22 | 2.61 ± 0.25 | 2.66 ± 0.34 | 2.62 ± 0.22 | |
| Anther | Length (mm) | 2.03 ± 0.14 | 1.35 ± 0.27 * | 1.85 ± 0.11 * | 1.81 ± 0.07 * |
| Width (mm) | 0.38 ± 0.03 | 0.35 ± 0.03 | 0.39 ± 0.04 | 0.38 ± 0.01 | |
| Pistil | Length of pistil (mm) | 1.74 ± 0.15 | 1.47 ± 0.22 * | 1.79 ± 0.20 | 1.77 ± 0.15 |
| Width of the ovule (mm) | 0.38 ± 0.02 | 0.39 ± 0.02 | 0.40 ± 0.03 | 0.38 ± 0.01 |
| SN | Gene ID | Gene Location | DJ vs. HM (log2 Fold Change) | Gene Descriptions |
|---|---|---|---|---|
| 1 | Os03g0299700 | 10495846..10498081 | 46.75 | Hypothetical protein |
| 2 | - | - | 32.96 | Hygromycin * |
| 3 | Os03g0629800 | 24806433..24807496 | 24.06 | Hypothetical protein |
| 4 | Os01g0965300 | 42554248..42554866 | 14.77 | Hypothetical protein |
| 5 | Os03g0105500 | 338364..338744 | 8.03 | Hypothetical protein |
| 6 | Os02g0269650 | 9689236..9689758 | 5.41 | Hypothetical protein |
| 7 | Os10g0340100 | 9458993..9461016 | 5.14 | Hypothetical protein |
| 8 | Os04g0112100 | 692745..695801 | 4.55 | Putative disease resistance protein RGA4 |
| 9 | Os09g0356800 | 11483610..11506480 | 4.38 | LRR receptor-like serine/threonine-protein kinase |
| 10 | Os04g0397800 | 19645507..19646962 | 4.32 | Hypothetical protein |
| 11 | Os12g0543800 | 21881810..21883709 | 4.29 | Hypothetical protein |
| 12 | Os07g0511400 | 20194023..20194735 | 3.48 | Hypothetical protein |
| 13 | Os10g0142600 | 2591399..2596634 | 3.20 | Putative wall-associated protein kinase |
| 14 | Os10g0142700 | 2573722..2575920 | 3.05 | Putative wall-associated protein kinase (non-protein coding RNA) |
| 15 | Os08g0193600 | 5457436..5458183 | 2.72 | F-box protein-like |
| 16 | Os01g0204900 | 5756471..5757587 | 2.44 | Hypothetical protein |
| 17 | Os01g0605500 | 23850865..23854747 | −5.39 | Kinesin-like protein KIF19 |
| 18 | Os04g0116800 | 984402..985981 | −5.90 | Hypothetical protein |
| 19 | Os04g0103800 | 250445..251491 | −8.59 | WASH complex subunit CCDC53 homolog |
| 20 | Os08g0472600 | 23273330..23281518 | −42.45 | Glycoprotein 3-alpha-l-fucosyltransferase A |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sim, J.-S.; Kesawat, M.S.; Kumar, M.; Kim, S.-Y.; Mani, V.; Subramanian, P.; Park, S.; Lee, C.-M.; Kim, S.-R.; Hahn, B.-S. Lack of the α1,3-Fucosyltransferase Gene (Osfuct) Affects Anther Development and Pollen Viability in Rice. Int. J. Mol. Sci. 2018, 19, 1225. https://doi.org/10.3390/ijms19041225
Sim J-S, Kesawat MS, Kumar M, Kim S-Y, Mani V, Subramanian P, Park S, Lee C-M, Kim S-R, Hahn B-S. Lack of the α1,3-Fucosyltransferase Gene (Osfuct) Affects Anther Development and Pollen Viability in Rice. International Journal of Molecular Sciences. 2018; 19(4):1225. https://doi.org/10.3390/ijms19041225
Chicago/Turabian StyleSim, Joon-Soo, Mahipal Singh Kesawat, Manu Kumar, Su-Yeon Kim, Vimalraj Mani, Parthiban Subramanian, Soyoung Park, Chang-Muk Lee, Seong-Ryong Kim, and Bum-Soo Hahn. 2018. "Lack of the α1,3-Fucosyltransferase Gene (Osfuct) Affects Anther Development and Pollen Viability in Rice" International Journal of Molecular Sciences 19, no. 4: 1225. https://doi.org/10.3390/ijms19041225
APA StyleSim, J.-S., Kesawat, M. S., Kumar, M., Kim, S.-Y., Mani, V., Subramanian, P., Park, S., Lee, C.-M., Kim, S.-R., & Hahn, B.-S. (2018). Lack of the α1,3-Fucosyltransferase Gene (Osfuct) Affects Anther Development and Pollen Viability in Rice. International Journal of Molecular Sciences, 19(4), 1225. https://doi.org/10.3390/ijms19041225








