DNA Methylation and Histone Modification in Hypertension
Abstract
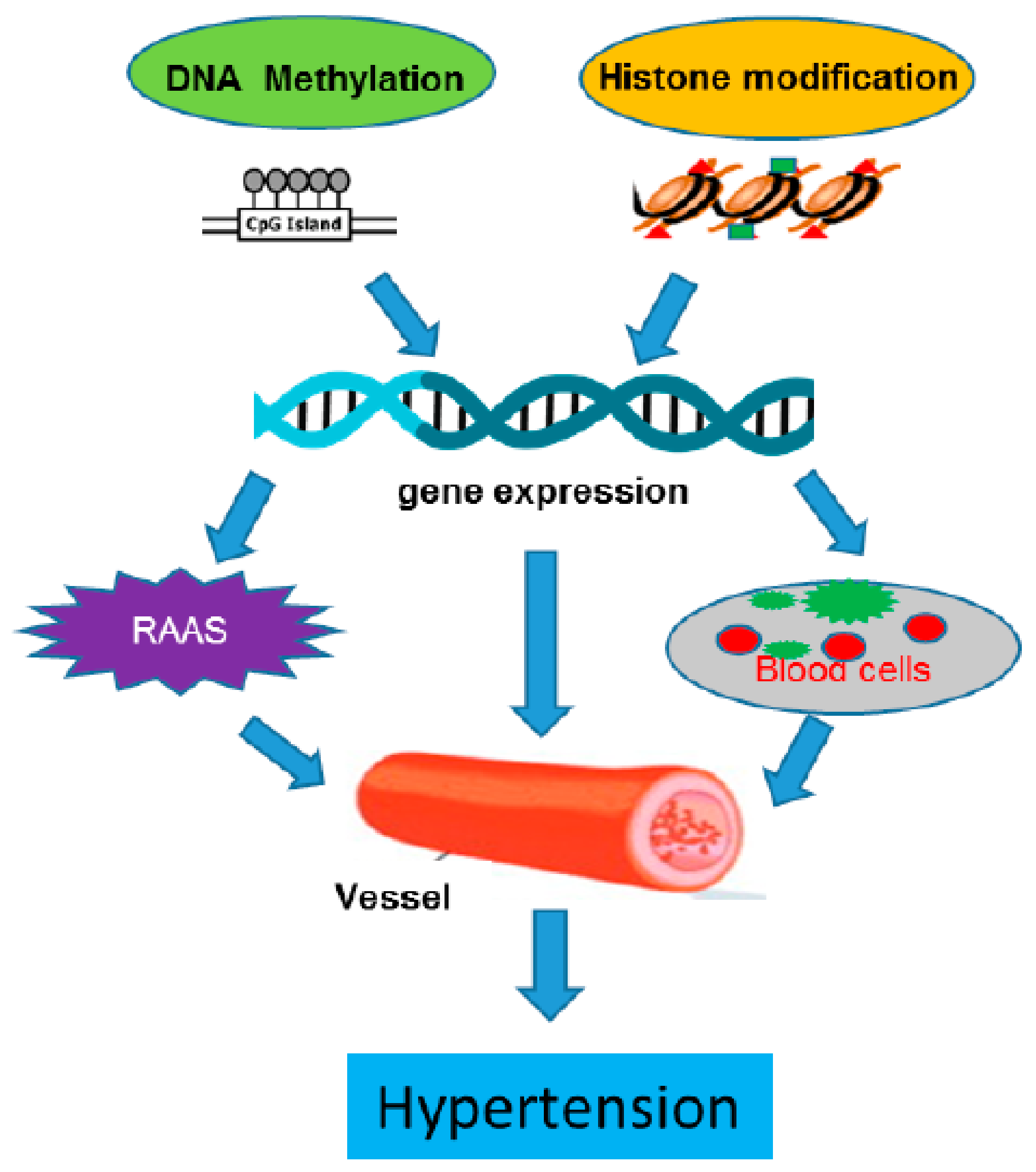
1. Introduction
2. Discovery, Development, and Detection
2.1. DNA Methylation
2.2. Histone Modification
3. Epigenomic Regulation in Hypertensive Vasculature
3.1. Epigenomic Regulation in the Whole Vessel
3.2. Epigenomic Regulation in Vascular Smooth Muscle Cells (VSMCs)
3.3. Epigenomic Regulation in Endothelial Cell Dysregulation
4. Clinical Application of Epigenomic Studies in Human Systemic Hypertension
5. Mechanisms Underlying Epigenetic Alterations in Hypertension
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 acc/aha/aapa/abc/acpm/ags/apha/ash/aspc/nma/pcna guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: Executive summary: A report of the american college of cardiology/american heart association task force on clinical practice guidelines. Hypertension 2017. [Google Scholar] [CrossRef]
- Liang, M.; Cowley, A.W., Jr.; Mattson, D.L.; Kotchen, T.A.; Liu, Y. Epigenomics of hypertension. Semin. Nephrol. 2013, 33, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Merai, R.; Siegel, C.; Rakotz, M.; Basch, P.; Wright, J.; Wong, B.; Dhsc; Thorpe, P. Cdc grand rounds: A public health approach to detect and control hypertension. Morb. Mortal. Wkly. Rep. 2016, 65, 1261–1264. [Google Scholar] [CrossRef] [PubMed]
- Wise, I.A.; Charchar, F.J. Epigenetic modifications in essential hypertension. Int. J. Mol. Sci. 2016, 17, 451. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Owens, G.K. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012, 74, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Pei, F.; Wang, X.; Yue, R.; Chen, C.; Huang, J.; Huang, J.; Li, X.; Zeng, C. Differential expression and DNA methylation of angiotensin type 1a receptors in vascular tissues during genetic hypertension development. Mol. Cell. Biochem. 2015, 402, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bogdarina, I.; Welham, S.; King, P.J.; Burns, S.P.; Clark, A.J. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ. Res. 2007, 100, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.; Goyal, D.; Leitzke, A.; Gheorghe, C.P.; Longo, L.D. Brain renin-angiotensin system: Fetal epigenetic programming by maternal protein restriction during pregnancy. Reprod. Sci. 2010, 17, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Rivière, G.; Lienhard, D.; Andrieu, T.; Vieau, D.; Frey, B.M.; Frey, F.J. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics 2014, 6, 478–489. [Google Scholar] [CrossRef]
- Friso, S.; Pizzolo, F.; Choi, S.W.; Guarini, P.; Castagna, A.; Ravagnani, V.; Carletto, A.; Pattini, P.; Corrocher, R.; Olivieri, O. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis 2008, 199, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.A.; Baek, I.; Seok, Y.M.; Yang, E.; Cho, H.M.; Lee, D.Y.; Hong, S.H.; Kim, I.K. Promoter hypomethylation upregulates na+-k+-2cl-cotransporter 1 in spontaneously hypertensive rats. Biochem. Biophys. Res. Commun. 2010, 396, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, C.; Chen, M.; Zhang, H.; Yang, S.; Zhang, L. Chronic hypoxia during gestation causes epigenetic repression of the estrogen receptor-α gene in ovine uterine arteries via heightened promoter methylation. Hypertension 2012, 60, 697–704. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, R.; Jin, Y.; Tang, W.H.; Qin, L.; Zhang, X.; Tellides, G.; Hwa, J.; Yu, J.; Martin, K.A. Ten-eleven translocation-2 (tet2) is a master regulator of smooth muscle cell plasticity. Circulation 2013, 128, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.N.; Liu, P.P.; Wang, L.; Yuan, F.; Xu, L.; Xin, Y.; Fei, L.J.; Zhong, Q.L.; Huang, Y.; Xu, L.; et al. Lower add1 gene promoter DNA methylation increases the risk of essential hypertension. PLoS ONE 2013, 8, e63455. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Q.; Fan, R.; Gu, T.L.; Zhong, Q.L.; Gong, M.L.; Dong, C.Z.; Hao, L.M.; Zhang, L.N. Hypermethylation of scnn1a gene-body increases the risk of essential hypertension. Int. J. Clin. Exp. Pathol. 2016, 9, 8047–8056. [Google Scholar]
- Zhong, Q.; Liu, C.; Fan, R.; Duan, S.; Xu, X.; Zhao, J.; Mao, S.; Zhu, W.; Hao, L.; Yin, F.; et al. Association of scnn1b promoter methylation with essential hypertension. Mol. Med. Rep. 2016, 14, 5422–5428. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Gu, T.; Zhong, F.; Fan, R.; Zhu, F.; Ren, P.; Yin, F.; Zhang, L. Hypomethylation of the toll-like receptor-2 gene increases the risk of essential hypertension. Mol. Med. Rep. 2017, 16, 964–970. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bostrom, A.E.; Mwinyi, J.; Voisin, S.; Wu, W.; Schultes, B.; Zhang, K.; Schioth, H.B. Longitudinal genome-wide methylation study of roux-en-y gastric bypass patients reveals novel CpG sites associated with essential hypertension. BMC Med. Genom. 2016, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.A.; Cho, H.M.; Lee, D.Y.; Kim, K.C.; Han, H.S.; Kim, I.K. Tissue-specific upregulation of angiotensin-converting enzyme 1 in spontaneously hypertensive rats through histone code modifications. Hypertension 2012, 59, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Manabe, I.; Owens, G.K. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel p19-derived in vitro smooth muscle differentiation system. Circ. Res. 2001, 88, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Ren, X.S.; Xiong, X.Q.; Chen, Y.Z.; Zhao, M.X.; Wang, J.J.; Zhou, Y.B.; Han, Y.; Chen, Q.; Li, Y.H.; et al. Nlrp3 inflammasome activation contributes to vsmc phenotypic transformation and proliferation in hypertension. Cell Death Dis. 2017, 8, e3074. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Matouk, C.C.; Rachlis, A.; Lin, S.; Tai, S.C.; D’Abreo, C.; Marsden, P.A. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J. Biol. Chem. 2005, 280, 24824–24838. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.M.; Lee, D.Y.; Kim, H.Y.; Lee, H.A.; Seok, Y.M.; Kim, I.K. Upregulation of the na(+)-k(+)-2cl(-) cotransporter 1 via histone modification in the aortas of angiotensin ii-induced hypertensive rats. Hypertens. Res. 2012, 35, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G. A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6, a018200. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Kaplan, W.D.; Kinosita, R. Formation of the sex chromatin by a single x-chromosome in liver cells of rattus norvegicus. Exp. Cell Res. 1959, 18, 415–418. [Google Scholar] [CrossRef]
- Lyon, M.F. Gene action in the x-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Riggs, A.D. X inactivation, differentiation, and DNA methylation. Cytogenet. Genome Res. 1975, 14, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Doskočil, J.; Šorm, F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim. Biophys. Acta 1962, 55, 953–959. [Google Scholar] [CrossRef]
- Bird, A.P.; Southern, E.M. Use of restriction enzymes to study eukaryotic DNA methylation. J. Mol. Biol. 1978, 118, 27–47. [Google Scholar] [CrossRef]
- Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Gorlach, A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharmacol. 2017, 174, 1533–1554. [Google Scholar] [CrossRef] [PubMed]
- Branco, M.R.; Ficz, G.; Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2011, 13, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Gilsbach, R.; Preissl, S.; Gruning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Wurch, A.; Bonisch, U.; Gunther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 2014, 5, 5288. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schubeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, G.; Stegle, O.; Reik, W. Single-cell epigenomics: Recording the past and predicting the future. Science 2017, 358, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Helin, K. Role of tet enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Millis, R.M. Epigenetics and hypertension. Curr. Hypertens. Rep. 2011, 13, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone h3 lysine 9 creates a binding site for hp1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Canas, B.; Pappin, D.; Kouzarides, T. Histone h3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (nurd) repressor complex. J. Biol. Chem. 2002, 277, 11621–11624. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998, 12, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Collas, P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010, 45, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 2013, 58, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, H.; Wu, S.; Wang, C. Single-cell sequencing technologies for cardiac stem cell studies. Stem Cells Dev. 2017, 26, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Wu, X.; Li, X.; Wen, L.; Tang, F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013, 23, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Tang, Q.; Wan, M.; Cui, K.; Zhang, Y.; Ren, G.; Ni, B.; Sklar, J.; Przytycka, T.M.; Childs, R.; et al. Genome-wide detection of dnase i hypersensitive sites in single cells and ffpe tissue samples. Nature 2015, 528, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; de Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; de Graaf, C.A.; Amendola, M.; et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, C.; Wu, S. Single-cell sequencing for drug discovery and drug development. Curr. Top. Med. Chem. 2017, 17, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Mulqueen, R.M.; Pokholok, D.; Norberg, S.; Fields, A.J.; Sun, D.; Torkenczy, K.A.; Shendure, J.; Trapnell, C.; O’Roak, B.J.; Xia, Z.; et al. Scalable and efficient single-cell DNA methylation sequencing by combinatorial indexing. bioRxiv 2017. [Google Scholar] [CrossRef]
- Kim, J.D.; Lee, A.; Choi, J.; Park, Y.; Kang, H.; Chang, W.; Lee, M.S.; Kim, J. Epigenetic modulation as a therapeutic approach for pulmonary arterial hypertension. Exp. Mol. Med. 2015, 47, e175. [Google Scholar] [CrossRef] [PubMed]
- Chelladurai, P.; Seeger, W.; Pullamsetti, S.S. Epigenetic mechanisms in pulmonary arterial hypertension: The need for global perspectives. Eur. Respir. Rev. 2016, 25, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Saco, T.V.; Parthasarathy, P.T.; Cho, Y.; Lockey, R.F.; Kolliputi, N. Role of epigenetics in pulmonary hypertension. Am. J. Physiol. Cell Physiol. 2014, 306, C1101–C1105. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Ryan, J.J.; Marsboom, G.; Archer, S.L. Epigenetic mechanisms of pulmonary hypertension. Pulm. Circ. 2011, 1, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Raftopoulos, L.; Katsi, V.; Makris, T.; Tousoulis, D.; Stefanadis, C.; Kallikazaros, I. Epigenetics, the missing link in hypertension. Life Sci. 2015, 129, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Funder, J.; Pearce, P.; Smith, R.; Smith, A. Mineralocorticoid action: Target tissue specificity is enzyme, not receptor, mediated. Science 1988, 242, 583–585. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, P.; Sansonnens, A.; Dick, B.; Frey, F.J. In vivo 11 -hsd-2 activity: Variability, salt-sensitivity, and effect of licorice. Hypertension 2001, 38, 1330–1336. [Google Scholar] [CrossRef] [PubMed]
- Pizzolo, F.; Friso, S.; Morandini, F.; Antoniazzi, F.; Zaltron, C.; Udali, S.; Gandini, A.; Cavarzere, P.; Salvagno, G.; Giorgetti, A.; et al. Apparent mineralocorticoid excess by a novel mutation and epigenetic modulation by HSD11B2 promoter methylation. J. Clin. Endocrinol. Metab. 2015, 100, E1234–E1241. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.C.; Lytle, C.; Zhu, T.T.; Payne, J.A.; Benz, E., Jr.; Forbush, B., 3rd. Molecular cloning and functional expression of the bumetanide-sensitive Na-K-Cl cotransporter. Proc. Natl. Acad. Sci. USA 1994, 91, 2201–2205. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.W.; Flagella, M.; Sutliff, R.L.; Lorenz, J.N.; Nieman, M.L.; Weber, C.S.; Paul, R.J.; Shull, G.E. Decreased blood pressure and vascular smooth muscle tone in mice lacking basolateral na(+)-k(+)-2cl(-) cotransporter. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H1846–H1855. [Google Scholar] [CrossRef] [PubMed]
- Garg, P.; Martin, C.F.; Elms, S.C.; Gordon, F.J.; Wall, S.M.; Garland, C.J.; Sutliff, R.L.; O’Neill, W.C. Effect of the Na-K-2Cl cotransporter NKCC1 on systemic blood pressure and smooth muscle tone. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H2100–H2105. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, M.E.; Karas, R.H. The protective effects of estrogen on the cardiovascular system. N. Engl. J. Med. 1999, 340, 1801–1811. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gao, P.; Zhang, H.; Chen, H.; Zheng, W.; Lv, X.; Xu, T.; Wei, Y.; Liu, D.; Liang, C. Sirt1 inhibits angiotensin ii-induced vascular smooth muscle cell hypertrophy. Acta Biochim. Biophys. Sin. 2011, 43, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xu, T.T.; Lu, J.; Li, L.; Xu, J.; Hao, D.L.; Chen, H.Z.; Liu, D.P. Overexpression of sirt1 in vascular smooth muscle cells attenuates angiotensin ii-induced vascular remodeling and hypertension in mice. J. Mol. Med. 2014, 92, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Brozovich, F.V.; Nicholson, C.J.; Degen, C.V.; Gao, Y.Z.; Aggarwal, M.; Morgan, K.G. Mechanisms of vascular smooth muscle contraction and the basis for pharmacologic treatment of smooth muscle disorders. Pharmacol. Rev. 2016, 68, 476–532. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Leslie, K.L.; Martin, K.A. Epigenetic regulation of smooth muscle cell plasticity. Biochim. Biophys. Acta 2015, 1849, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Levy, E.; Spahis, S.; Bigras, J.L.; Delvin, E.; Borys, J.M. The epigenetic machinery in vascular dysfunction and hypertension. Curr. Hypertens. Rep. 2017, 19, 52. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Li, L. Histone acetylation and recruitment of serum responsive factor and creb-binding protein onto sm22 promoter during sm22 gene expression. Circ. Res. 2002, 90, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, C.; Tang, R.; Chen, H.; Zhang, Z.; Tatsuguchi, M.; Wang, D.Z. Acetylation of myocardin is required for the activation of cardiac and smooth muscle genes. J. Biol. Chem. 2012, 287, 38495–38504. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, Z.; Zhang, C.L.; Oh, J.; Xing, W.; Li, S.; Richardson, J.A.; Wang, D.Z.; Olson, E.N. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol. Cell. Biol. 2005, 25, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.J.; Martin, K.A. Coordinating regulation of gene expression in cardiovascular disease: Interactions between chromatin modifiers and transcription factors. Front. Cardiovasc. Med. 2017, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Lee, J.J.; Stoll, S.; Ma, B.; Wiener, R.; Wang, C.; Costa, K.D.; Qiu, H. Inhibition of SRF/myocardin reduces aortic stiffness by targeting vascular smooth muscle cell stiffening in hypertension. Cardiovasc. Res. 2017, 113, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Lee, J.J.; Stoll, S.; Ma, B.; Costa, K.D.; Qiu, H. Rho kinase regulates aortic vascular smooth muscle cell stiffness via actin/SRF/myocardin in hypertension. Cell. Physiol. Biochem. 2017, 44, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Siti, H.N.; Kamisah, Y.; Kamsiah, J. The role of oxidative stress, antioxidants and vascular inflammation in cardiovascular disease (a review). Vasc. Pharmacol. 2015, 71, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.B.; Yang, S.H.; Toth, B.; Kovalenko, A.; Wallach, D. Activation of the NLRP3 inflammasome by proteins that signal for necroptosis. Methods Enzymol. 2014, 545, 67–81. [Google Scholar] [PubMed]
- Krishnan, S.M.; Sobey, C.G.; Latz, E.; Mansell, A.; Drummond, G.R. Il-1β and il-18: Inflammatory markers or mediators of hypertension? Br. J. Pharmacol. 2014, 171, 5589–5602. [Google Scholar] [CrossRef] [PubMed]
- Sandoo, A.; van Zanten, J.J.; Metsios, G.S.; Carroll, D.; Kitas, G.D. The endothelium and its role in regulating vascular tone. Open Cardiovasc. Med. J. 2010, 4, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Nadar, S.; Blann, A.; Lip, G. Endothelial dysfunction: Methods of assessment and application to hypertension. Curr. Pharm. Des. 2004, 10, 3591–3605. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Uludag, M.O.; Usanmaz, S.E.; Ayaloglu-Butun, F.; Akcali, K.C.; Demirel-Yilmaz, E. Resveratrol affects histone 3 lysine 27 methylation of vessels and blood biomarkers in doca salt-induced hypertension. Mol. Biol. Rep. 2015, 42, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Smolarek, I.; Wyszko, E.; Barciszewska, A.M.; Nowak, S.; Gawronska, I.; Jablecka, A.; Barciszewska, M.Z. Global DNA methylation changes in blood of patients with essential hypertension. Med. Sci. Monit. 2010, 16, CR149–CR155. [Google Scholar] [PubMed]
- Kato, N.; Loh, M.; Takeuchi, F.; Verweij, N.; Wang, X.; Zhang, W.; Kelly, T.N.; Saleheen, D.; Lehne, B.; Leach, I.M.; et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 2015, 47, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Staessen, J.A.; Bianchi, G. Adducin and hypertension. Pharmacogenomics 2005, 6, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.E.; Campbell, R.D.; Sanderson, C.M. Novel NG36/G9a gene products encoded within the human and mouse mhc class iii regions. Mamm. Genome 2014, 12, 916–924. [Google Scholar] [CrossRef]
- Lehnertz, B.; Northrop, J.P.; Antignano, F.; Burrows, K.; Hadidi, S.; Mullaly, S.C.; Rossi, F.M.; Zaph, C. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J. Exp. Med. 2010, 207, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.C.; Langley-Evans, S.C. Early administration of angiotensin-converting enzyme inhibitor captopril, prevents the development of hypertension programmed by intrauterine exposure to a maternal low-protein diet in the rat. Clin. Sci. 1998, 94, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.A.; Dunn, R.L.; Marchand, M.C.; Langley-Evans, S.C. Increased systolic blood pressure in rats induced by a maternal low-protein diet is reversed by dietary supplementation with glycine. Clin. Sci. 2002, 103, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Lillycrop, K.A.; Slater-Jefferies, J.L.; Hanson, M.A.; Godfrey, K.M.; Jackson, A.A.; Burdge, G.C. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br. J. Nutr. 2007, 97, 1064–1073. [Google Scholar] [PubMed]
- Alexander, B.T.; Ojeda, N.B. Prenatal inflammation and the early origins of hypertension. Clin. Exp. Pharmacol. Physiol. 2008, 35, 1403–1404. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, A.M.; Alexanderson, C.; Molne, J.; Haraldsson, B.; Hansell, P.; Holmang, A. Prenatal exposure to interleukin-6 results in hypertension and alterations in the renin-angiotensin system of the rat. J. Physiol. 2006, 575, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.Q.; Zhang, H.G.; Li, S.H.; Jia, Y.; Liu, Y.; Zhou, J.Z.; Wei, Y.L.; Hao, L.Y.; Tang, Y.; Su, M.; et al. Prenatal exposure to inflammation induced by zymosan results in activation of intrarenal renin-angiotensin system in adult offspring rats. Inflammation 2010, 33, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Wei, Y.; Yu, C.; Zhou, J.; Li, S.; Pang, Y.; Li, G.; Li, X. Prenatal exposure to zymosan results in hypertension in adult offspring rats. Clin. Exp. Pharmacol. Physiol. 2008, 35, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.Q.; Zhang, H.G.; Yuan, Z.B.; Yang, D.L.; Hao, L.Y.; Li, X.H. Prenatal exposure to lipopolysaccharide alters the intrarenal renin-angiotensin system and renal damage in offspring rats. Hypertens. Res. 2010, 33, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.L.; Li, X.H.; Zhou, J.Z. Prenatal exposure to lipopolysaccharide results in increases in blood pressure and body weight in rats. Acta Pharmacol. Sin. 2007, 28, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yin, N.; Deng, Y.; Wei, Y.; Huang, Y.; Pu, X.; Li, L.; Zheng, Y.; Guo, J.; Yu, J.; et al. Ascorbic acid protects against hypertension through downregulation of ace1 gene expression mediated by histone deacetylation in prenatal inflammation-induced offspring. Sci. Rep. 2016, 6, 39469. [Google Scholar] [CrossRef] [PubMed]
- Kuzawa, C.W.; Sweet, E. Epigenetics and the embodiment of race: Developmental origins of us racial disparities in cardiovascular health. Am. J. Hum. Biol. 2009, 21, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Jirtle, R.L.; Skinner, M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007, 8, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, L. Epigenetic mechanisms in developmental programming of adult disease. Drug Discov. Today 2011, 16, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kondo, T.; Takada, I.; Youn, M.Y.; Yamamoto, Y.; Takahashi, S.; Matsumoto, T.; Fujiyama, S.; Shirode, Y.; Yamaoka, I.; et al. DNA demethylation in hormone-induced transcriptional derepression. Nature 2009, 461, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Wang, J.; Nawaz, Z.; Liu, J.M.; Qin, J.; Wong, J. Both corepressor proteins smrt and n-cor exist in large protein complexes containing hdac3. EMBO J. 2000, 19, 4342–4350. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.Y.; Downes, M.; Ordentlich, P.; Evans, R.M. Isolation of a novel histone deacetylase reveals that class i and class ii deacetylases promote smrt-mediated repression. Gene Dev. 2000, 14, 55–66. [Google Scholar] [PubMed]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of jmjc domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Metzger, E.; Wissmann, M.; Yin, N.; Muller, J.M.; Schneider, R.; Peters, A.H.; Gunther, T.; Buettner, R.; Schule, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Toumazou, C.; Tsukada, Y.; Erdjument-Bromage, H.; Tempst, P.; Wong, J.; Zhang, Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006, 125, 483–495. [Google Scholar] [CrossRef] [PubMed]
| Genes | Mark | Status | Species | Models | Tissues/Cells | Function | Ref |
|---|---|---|---|---|---|---|---|
| DNA methylation | |||||||
| Atgr1α | 5mC | Hypo | Rat | SHR | Aorta and mesentery artery | Increased expression of receptor and effect of RAAS | [6] |
| Atgr1β | 5mC | Hypo | Rat | Maternal low protein rat | RAAS | [7] | |
| Ace-1 | 5mC | Hypo | Mice | Maternal protein deficient mice | RAAS | [8] | |
| ACE-1 | 5mC | Hyper | Human | Human PBMCs; cell culture (HepG2, HT29, HMEC-1, SUT) | RAAS | [9] | |
| HSD11B2 | 5mC | Hyper | Human | Glucocorticoid treatment | Human PBMCs | Renal sodium balance | [10] |
| Sslc12a2 (NKCC1) | 5mC | Hypo | Rat | SHR | Aorta and heart | Ionic balance | [11] |
| ESR1 (ERα) | 5mC | Hyper | Sheep | Uterine artery | vasodilation | [12] | |
| SRF, MYOCD, MYH11 | 5mC | Hyper | Human | Human coronary artery SMCs | contraction phenotype | [13] | |
| ADD1 | 5mC | Hypo | Human | Human PBMCs | Ionic balance | [14] | |
| SCNN1A | 5mC | Hyper | Human | Human PBMCs | Ionic balance | [15] | |
| SCNN1B | 5mC | CpG1 Hyper, CpG2 Hypo | Human | Human PBMCs | Ionic balance | [16] | |
| TLR2 | 5mC | Hypo | Human | Human PBMCs | Chronic inflammation | [17] | |
| EHMT2 | 5mC | Hypo | Human | Human PBMCs | Chronic inflammation | [18] | |
| Histone modification | |||||||
| Ace1 | H3Ac, H3K4me3, H3K9me2 | Hyper, Hyper, Hypo | Rat | SHR | Heart, kidney | RAAS | [19] |
| SM22 | H3Ac | Hyper | Mouse | 10T1/2 cells | Contractile phenotype | [20] | |
| Nlrp3 | H3K9Ac | Hyper | Rat | SHR | VSMCs | Chronic inflammation | [21] |
| NOS3 (eNOS) | H3K9Ac, H4K12 H3K4 me2, H3K4me3 | Hyper | Human | Cell culture; HUVEC, HMVEC, VSMC, HEPG2, HeLa, JEG-3 | Vasodilation in endothelial cells | [22] | |
| Slc12a2 (NKCC1) | H3Ac H3K27me3 | Hyper, Hypo | Rat | Angiotensin II delivery | Aorta | Ionic balance | [23] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stoll, S.; Wang, C.; Qiu, H. DNA Methylation and Histone Modification in Hypertension. Int. J. Mol. Sci. 2018, 19, 1174. https://doi.org/10.3390/ijms19041174
Stoll S, Wang C, Qiu H. DNA Methylation and Histone Modification in Hypertension. International Journal of Molecular Sciences. 2018; 19(4):1174. https://doi.org/10.3390/ijms19041174
Chicago/Turabian StyleStoll, Shaunrick, Charles Wang, and Hongyu Qiu. 2018. "DNA Methylation and Histone Modification in Hypertension" International Journal of Molecular Sciences 19, no. 4: 1174. https://doi.org/10.3390/ijms19041174
APA StyleStoll, S., Wang, C., & Qiu, H. (2018). DNA Methylation and Histone Modification in Hypertension. International Journal of Molecular Sciences, 19(4), 1174. https://doi.org/10.3390/ijms19041174




