Prospects of Understanding the Molecular Biology of Disease Resistance in Rice
Abstract
1. Introduction
2. Mechanism of Plant Defense against Biotic Stresses
3. Resistance Gene Architecture and Resistance Hypothesis
4. Signal Molecules and Their Networks Involved in Rice Defense Response
4.1. Mitogen-Associated Protein (MAP) Kinase and Ca2+ Signaling
4.2. Role of ROS and NO in Rice Defense Signaling
4.3. Phytohormone-Mediated Signaling
5. Role of Regulatory Elements in Rice Disease Resistance
6. Breeding Approaches to Control Diseases in Rice
6.1. Introduction of Resistant Exotic Lines
6.2. Introgression of Resistance Genes
6.3. Pyramiding of Resistance Genes
7. Molecular Breeding Approaches for Disease Resistance
7.1. Marker-Assisted Selection and Mapping of Resistance Genes
7.2. Resistance Gene Pyramiding by MAS
7.3. Genome-Wide Association Study of Resistance Genes
7.4. Mutation Breeding for Rice Resistance
8. Transgenic Approaches for Disease Resistance in Rice
8.1. Over-Expression/Functional Complementation Test in Rice Resistance
8.2. Role of RNA Interference (RNAi) in Disease Resistance
8.3. CRISPR/Cas9 Immune System
9. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sharma, T.R.; Rai, A.K.; Gupta, S.K.; Vijayan, J.; Devanna, B.N.; Ray, S. Rice blast management through host-plant resistance, retrospect and prospects. Agric. Res. 2012, 1, 37–52. [Google Scholar] [CrossRef]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Rao, G.J. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H. Rice Diseases, 2nd ed.; Commonwealth Mycological Institute: Kew, UK, 1985; pp. 61–96. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Bray Speth, E.; Lee, Y.N.; He, S.Y. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 2006, 10, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Nurnberger, T.; Kemmerling, B. Pathogen-associated molecular patterns (PAMP) and PAMP-triggered immunity. Annu. Plant Rev. 2009, 34, 16–47. [Google Scholar]
- Chen, X.; Ronald, P.C. Innate immunity in rice. Trends Plant Sci. 2011, 16, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Felix, G. Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 2005, 8, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.C.; Beattie, G.A. An overview of plant defenses against pathogens and herbivores. Plant Health Instruct. 2008, 94. [Google Scholar] [CrossRef]
- Serrano, M.; Coluccia, F.; Torres, M.L.; Haridon, F.; Metraux, J.P. The cuticle and plant defense to pathogens. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Doughari, J. An overview of plant immunity. J. Plant Pathol. Microbiol. 2015, 6, 10–4172. [Google Scholar]
- Nicaise, V.; Roux, M.; Zipfel, C. Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009, 150, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- Tena, G.; Boudsocq, M.; Sheen, J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011, 14, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Chassot, C.; Nawrath, C.; Metraux, J. Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 2007, 49, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Tonnessen, B.W.; Manosalva, P.; Lang, J.M.; Baraoidan, M.; Bordeos, A.; Mauleon, R.; Oard, J.; Hulbert, S.; Leung, H.; Leach, J.E. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015, 87, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Davidson, R.M.; Liu, B.; Zhu, X.; Hulbert, S.H.; Leung, H.; Leach, J.E. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009, 149, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.; Wang, M.; Wang, J.; Liu, B.; Wang, H. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Cayrol, B.; Delteil, A.; Gobbato, E.; Kroj, T.; Morel, J.-B. Three wall-associated kinases required for rice basal immunity from protein complexes in the plasma membrane. Plant Signal. Behav. 2016, 11, e1149676. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Xia, J.; Chen, Z.; Yu, Y.; Li, Q.F.; Zhang, Y.C.; Zhang, J.P.; Wang, C.Y.; Zhu, X.Y.; Zhang, W. Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci. Rep. 2016, 6, 25493. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Kopp, E.; Medzhitov, R. Recognition of microbial infection by Toll-like receptors. Curr. Opin. Immunol. 2003, 15, 396–401. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, Y.; Zhou, Z.; Zhou, J.M. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 2016, 59, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Korneli, C.; Lummer, M.; Navarro, L. Emerging role for RNA based regulation in plant immunity. New Phytol. 2013, 197, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Leipe, D.D.; Koonin, E.V.; Aravind, L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004, 343, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Takken, F.L.W.; Albrecht, M.; Tameling, W.I.L. Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 2006, 9, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.M.; Lin, Z.J.D.; Coaker, G. Plant NB-LRR signaling: Upstreams and downstreams. Curr. Opin. Plant Biol. 2011, 14, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Qi, D.; DeYoung, B.J.; Innes, R.W. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 2012, 158, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; van den Berg, H.A.; Cornelissen, B.J.C.; Takken, F.L.W. Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 2007, 45, 43–72. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chand, S.; Singh, N.K.; Sharma, T.R. Genome-wide distribution, organisation and functional characterization of disease resistance and defence response genes across rice species. PLoS ONE 2015, 10, e0125964. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Bernoux, M.; Ve, T.; Williams, S.; Warren, C.; Hatters, D.; Valkov, E.; Zhang, X.; Ellis, J.G.; Kobe, B.; Dodds, P.N. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 2011, 9, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Van Der Biezen, E.A.; Jones, J.D.G. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- Van der Hoorn, R.A.L.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Bernoux, M.; Moncuquet, P.; Kroj, T.; Dodds, P.N. A novel conserved mechanism for plant NLR protein pairs: The “integrated decoy” hypothesis. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.T.; Monteiro, F.; Dangl, J.L. Treasure your exceptions: Unusual domains in immune receptors reveal host virulence targets. Cell 2015, 161, 957–960. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2015, 33, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Germain, H.; Seguin, A. Innate immunity: Has poplar made its BED? New Phytol. 2011, 189, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Sengupta, S.; Prasad, M.; Ghose, T.K. Genetic diversity of the conserved motifs of six bacterial leaf blight resistance genes in a set of rice landraces. BMC Genet. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AvrPik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors Avr-Pia and Avr1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Kroj, T.; Chanclud, E.; Michel Romiti, C.; Grand, X.; Morel, J. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Johnson, K.C.M.; Zhu, Z.; Huang, Y.; Chen, F.; Zhang, Y.; Li, X. Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front. Plant Sci. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, S.; Jia, Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009, 177, 468–478. [Google Scholar] [CrossRef]
- Yoshimura, S.; Yamanouchi, U.; Katayose, Y.; Toki, S.; Wang, Z.X.; Kono, I.; Kurata, N.; Yano, M.; Iwata, N.; Sasaki, T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 1998, 95, 1663–1668. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Sharma, T.R. Comparative analysis of zinc finger proteins involved in plant disease resistance. PLoS ONE 2012, 7, e42578. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.G.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Saxena, I.; Srikanth, S.; Chen, Z. Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Jaggi, M.; Raghuram, B.; Tuteja, N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011, 6, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Reyna, N.S.; Yang, Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 2006, 19, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Sharma, K.; Mishra, R.S. Elicitor recognition, signal transduction and induced resistance in plants. J. Plant Int. 2012, 7, 95–120. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Oelmuller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defense responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yang, Y.; He, Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant 2013, 3, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhong, S.; Li, Q.; Zhu, Z.; Lou, Y.; Wang, L.; Wang, J.; Wang, M.; Li, Q.; Yang, D.; et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 2007, 5, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Qiu, D.; Yuan, T.; Ding, X.; Li, H.; Duan, L.; Xu, C.; Li, X.; Wang, S. Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 2008, 31, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, A.; Inoue, H.; Goto, S.; Nakayama, A.; Sugano, S.; Hayashi, N.; Takatsuji, H. The nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 2013, 73, 302–313. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, G.L. Plant innate immunity in rice: A defense against pathogen infection. Natl. Sci. Rev. 2016, 3, 295–308. [Google Scholar] [CrossRef]
- Taniguchi, S.; Hosokawa-Shinonaga, Y.; Tamaoki, D.; Yamada, S.; Akimitsu, K.; Gomi, K. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014, 37, 451–461. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Xu, J.; Hofte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Gheysen, G.; Hofte, M. Hormone defense networking in rice: Tales from a different world. Trends Plant Sci. 2013, 18, 555–565. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Seifi, H.S.; Filipe, O.; Haeck, A.; Huu, S.N.; Demeestere, K.; Hofte, M. The DELLA protein SLR1 integrates and amplifies salicylic acid and jasmonic acid dependent innate immunity in rice. Plant Physiol. 2016, 170, 1831–1847. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.J.; Shimono, M.; Sugano, S.; Kojima, M.; Liu, X.; Inoue, H.; Sakakibara, H.; Takatsuji, H. Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant Microbe Interact. 2013, 26, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, H.; Li, Y.; Yu, H.; Li, X.; Xiao, J.; Wang, S. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 2011, 155, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, Y.; Guo, Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Belkhadir, Y.; Jaillais, Y.; Epple, P.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2012, 109, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Asselbergh, B.; De Vleesschauwer, D.; Hofte, M. Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant-pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N. Understanding disease resistance signaling in rice against various pests and pathogens. Austin J. Plant Biol. 2016, 2, 1011. [Google Scholar]
- Iwai, T.; Seo, S.; Mitsuhara, I.; Ohashi, Y. Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol. 2007, 48, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance Regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Boden, E.; Arias, J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 2003, 15, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ding, X.; Yuan, M.; Qiu, D.; Li, X.; Xu, C.; Wang, S. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Mol. Biol. 2006, 60, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.; Jiang, C.J.; Miyazawa, S.I.; Masumoto, C.; Yazawa, K.; Hayashi, N.; Shimono, M.; Nakayama, A.; Miyao, M.; Takatsuji, H. Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 2010, 74, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Chern, M.; Fitzgerald, H.A.; Canlas, P.E.; Navarre, D.A.; Ronald, P.C. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 2005, 18, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in Benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayashi, N.; Matsushita, A.; Xinqiong, L.; Nakayama, A.; Sugano, S.; Jiang, C.J.; Takatsuji, H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. USA 2013, 110, 9577–9582. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes plays opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, H.; Deng, Y.; Xiao, J.; Li, X.; Wang, S. The WRKY45-2WRKY13WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 2015, 167, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Ding, X.; Xiong, M.; Cai, M.; Cao, Y.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Jimmy, J.L.; Babu, S. Role of OsWRKY transcription factors in rice disease resistance. Trop. Plant Pathol. 2015, 40, 355–361. [Google Scholar] [CrossRef]
- Chujo, T.; Takai, R.; Akimoto-Tomiyama, C.; Ando, S.; Minami, E.; Nagamura, Y.; Kaku, H.; Shibuya, N.; Yasuda, M.; Nakashita, H.; et al. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 2007, 1769, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; Takahashi, A.; Nishizawa, Y.; Minami, E.; Nojiri, H.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 2014, 9, e98737. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef] [PubMed]
- Abbruscato, P.; Nepusz, T.; Mizzi, L.; Del Corvo, M.; Morandini, P.; Fumasoni, I.; Michel, C.; Paccanaro, A.; Guiderdoni, E.; Schaffrath, U.; et al. OsWRKY22, a monocot wrky gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 2012, 13, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Miyamoto, K.; Shimogawa, T.; Shimizu, T.; Otake, Y.; Yokotani, N.; Nishizawa, Y.; Shibuya, N.; Nojiri, H.; Yamane, H.; et al. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 2013, 82, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, X.; Wang, X.; Chu, C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, H.; Jang, J.C.; Xiao, T.; He, H.; Jiang, D.; Tang, X. OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Zhao, W.; Meng, X.; Wang, M.; Peng, Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 2007, 172, 120–130. [Google Scholar] [CrossRef]
- Yokotani, N.; Tsuchida-Mayama, T.; Ichikawa, H.; Mitsuda, N.; Ohme-Takagi, M.; Kaku, H.; Minami, E.; Nishizawa, Y. OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol. Plant Microbe Interact. 2014, 27, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhao, W.; Lin, R.; Wang, M.; Peng, Y.-L. Identification of a novel rice bZIP-type transcription factor gene, OsbZIP1, involved in response to infection of Magnaporthe grisea. Plant Mol. Biol. Rep. 2005, 23, 301–302. [Google Scholar] [CrossRef]
- Li, W.; Zhong, S.; Li, G.; Li, Q.; Mao, B.; Deng, Y.; Zhang, H.; Zeng, L.; Song, F.; He, Z. Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res. 2011, 21, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, Q.; Li, Y.; Lei, C.; Zhai, Y.; Sun, X.; Sun, D.; Sun, Y.; Lu, T. A putative leucine-rich repeat receptor kinase, OsBRR1, is involved in rice blast resistance. Planta 2009, 230, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Jisha, V.; Dampanaboina, L.; Vadassery, J.; Mithofer, A.; Kappara, S.; Ramanan, R. Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 2015, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Song, F.; Goodman, R.M.; Zheng, Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 2006, 163, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Liu, H.; Li, Y.; Li, X.; Xu, C.; Long, M.; Wang, S. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 2009, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.Y.; Zeng, N.Y.; Tong, S.W.; Li, W.Y.F.; Xue, Y.; Zhao, K.J.; Wang, C.; Zhang, Q.; Fu, Y.; Sun, Z.; et al. Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytol. 2008, 179, 530–545. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Wong, H.L.; Kawasaki, T.; Hasegawa, M.; Kodama, O.; Shimamoto, K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible Overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Schaffrath, U.; Mauch, F.; Freydl, E.; Schweizer, P.; Dudler, R. Constitutive expression of the defense-related Rir1b gene in transgenic rice plants confers enhanced resistance to the rice blast fungus Magnaporthe grisea. Plant Mol. Biol. 2000, 43, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Kuroda, M.; Yamakawa, H.; Ashizawa, T.; Hirayae, K.; Kurimoto, L.; Shinya, T.; Shibuya, N. Suppression of a phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 2009, 150, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Sawada, K.; Hasegawa, M.; Tokuda, L.; Kameyama, J.; Kodama, O.; Kohchi, T.; Yoshida, K.; Shinmyo, A. Enhanced resistance to blast fungus and bacterial blight in transgenic rice constitutively expressing OsSBP, a rice homologue of mammalian selenium-binding proteins. Biosci. Biotechnol. Biochem. 2004, 68, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Akamatsu, A.; Hayashi, K.; Housen, Y.; Okuda, J.; Yao, A.; Nakashima, A.; Takahashi, H.; Yoshida, H.; Wong, H.L. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 2010, 7, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Domingo, C.; Andres, F.; Tharreau, D.; Iglesias, D.J.; Talon, M. Constitutive Expression of OsGH3.1 Reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 2009, 22, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Brar, D.S.; Dalmacio, R.; Elloran, R.; Aggarwal, R.; Angeles, R.; Khush, G.S. Gene transfer and molecular characterization of introgression from wild Oryza species into rice. In Rice Genetics III; Khush, G.S., Ed.; International Rice Research Institute: Manila, Philippines, 1996; pp. 477–486. [Google Scholar]
- Jena, K.K.; Khush, G.S. Introgression of genes from Oryza officinalis Well ex Watt to cultivated rice, O. sativa L. Theor. Appl. Genet. 1990, 80, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Multani, D.S.; Jena, K.K.; Brar, D.S.; Delos Reyes, B.G.; Angeles, E.R.; Khush, G.S. Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin to cultivated rice, O. sativa L. Theor. Appl. Genet. 1994, 88, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Mackill, D.J.; Bonman, J.M.; McCouch, S.R.; Champoux, M.C.; Nelson, R.J. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 1994, 136, 1421–1434. [Google Scholar] [PubMed]
- Young, N.D. QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 1996, 34, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. Can the strategic use of disease resistant hosts protect their inherent durability? In Durability of Disease Resistance; Jacobs, T., Parleviet, J.E., Eds.; Kluwer: Wageningen, The Netherlands, 1993; pp. 83–96. [Google Scholar]
- Simmonds, N.W. Introgression and incorporation. Strategies for the use of crop genetic resources. Biol. Rev. 1993, 68, 539–562. [Google Scholar] [CrossRef]
- Thippeswamy, S.; Chandramohan, Y.; Zameema, S.; Srinivas, B.; Padmaja, D.; Pravalika, K. Identification of blast resistant rice (Oryza sativa L.) genotypes in indigenous and exotic germplasm and validation of pi gene linked molecular markers. Electron. J. Plant Breed. 2016, 7, 21–29. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.H.; Xu, C.G.; Zhang, Q. Improvement of bacterial blight resistance of Minghui63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection. Crop Sci. 2000, 40, 239–244. [Google Scholar] [CrossRef]
- Bonman, J.M. Durable resistance to rice blast disease-environmental influences. Euphytica 1992, 63, 115–123. [Google Scholar] [CrossRef]
- Tan, G.X.; Ren, X.; Weng, Q.M.; Shi, Z.Y.; Zhu, L.L.; He, G.C. Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta Genet. Sin. 2004, 31, 724–729. [Google Scholar] [PubMed]
- Young, N.D.; Tanksley, S.D. RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 1989, 77, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B. Modernization in plant breeding approaches for improving biotic stress resistance in crop plants. Turk. J. Agric. For. 2015, 39, 1–16. [Google Scholar] [CrossRef]
- Steffan, P.; Borgen, A.; Lazzaro, M.; Backes, G.; Torp, A.M.; Rasmussen, S.K. Marker assisted breeding and mass selection of wheat composite cross populations. In Proceedings of the 10 Year’s Anniversary Conference Organic Plant Breeding: What Makes the Difference, Frankfurt, Germany, 3–4 November 2011. [Google Scholar]
- McDonald, B.A. Using dynamic diversity to achieve durable disease resistance in agricultural ecosystems. Trop. Plant Pathol. 2014, 39, 191–196. [Google Scholar] [CrossRef]
- Keneni, G.; Bekele, E.; Imtiaz, M.; Dagne, K. Genetic vulnerability of modern crop cultivars: Causes, mechanism and remedies. Int. J. Plant Res. 2012, 2, 69–79. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Sattari, A.; Fakheri, B.; Hassan, F.S.C.; Noroozi, M. Blast resistance in rice: A review of breeding and biotechnology. Int. J. Agric. Crop Sci. 2014, 7, 329–333. [Google Scholar]
- Ashizawa, T.; Zenbayashi, K.; Koizumi, S. Development of a simulation model for forecasting rice blast epidemics in multiline. Jpn. J. Phytopathol. 2001, 67, 194–197. [Google Scholar]
- Ishizaki, K.; Hoshi, T.; Abe, S.; Sasaki, Y.; Kobayashi, K.; Kasaneyama, H.; Matsui, T.; Azuma, S. Breeding of blast resistant lines in rice variety “Koshihikari” and evaluation of their characters. Breed. Sci. 2005, 55, 371–377. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Jiang, G.; Xu, Y.; He, Y.Q. Pyramiding of Xa7 and Xa21 for the improvement of disease resistance to bacterial blight in hybrid rice. Plant Breed. 2006, 125, 600–605. [Google Scholar] [CrossRef]
- Petit-Houdenot, Y.; Fudal, I. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 2017, 8, 1072. [Google Scholar] [CrossRef] [PubMed]
- Langridge, P.; Lagudah, E.S.; Holton, T.A.; Appels, R.; Sharp, P.J.; Chalmers, K.J. Trends in genetic and genome analyses in wheat: A review. Aust. J. Agric. Res. 2001, 52, 1043–1077. [Google Scholar] [CrossRef]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Barman, S.R.; Gowda, M.; Venu, R.C.; Chattoo, B.B. Identification of a major blast resistance gene in the rice cultivar “Tetep”. Plant Breed 2004, 123, 300–302. [Google Scholar] [CrossRef]
- Lin, F.; Chen, S.; Que, Z.; Wang, L.; Liu, X.; Pan, Q. The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 2007, 177, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Koizumi, S.; La, T.N.; Zenbayashi, K.S.; Ashizawa, T.; Yasuda, N.; Imazaki, I.; Miyasaka, A. Pi35(t), a new gene conferring partial resistance to leaf blast in the rice cultivar Hokkai 188. Theor. Appl. Genet. 2006, 113, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Sallaud, C.; Lorieux, M.; Roumen, E.; Tharreau, D.; Berruyer, R.; Svestasrani, P.; Garsmeur, O.; Ghesquiere, A.; Notteghem, J.L. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor. Appl. Genet. 2003, 106, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang, L.; Pan, Q. Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology 2004, 94, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Fukuta, Y.; Yanoria, M.J.T.; Mercado-Escueta, D.; Ebron, L.A.; Fujita, Y.; Araki, E.; Khush, G.S. Quantitative trait loci (QTL) reactions to rice blast isolates from Japan and the Philippines. In Rice Blast: Interaction with Rice and Control; Springer: Berlin, Germany, 2004; pp. 113–121. [Google Scholar]
- Mir, G.N.; Khush, G.S. Genetics of resistance to bacterial blight in rice cultivar DV 86. Crop Res. 1990, 3, 194–198. [Google Scholar]
- Wu, X.; Li, X.; Xu, C.; Wang, S. Fine genetic mapping of xa24, a recessive gene for resistance against Xanthomonas oryzae pv.oryzae in rice. Theor. Appl. Genet. 2008, 118, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Li, S.G.; Xu, J.C.; Zhai, W.X.; Ling, Z.Z.; Ma, B.T.; Wang, Y.P.; Wang, W.M.; Cao, G.; Ma, Y.Q. Identification of two blast resistance genes in a rice variety, Digu. J. Phytopathol. 2004, 152, 77–85. [Google Scholar] [CrossRef]
- Lei, C.; Hao, K.; Yang, Y.; Ma, J.; Wang, S.; Wang, J.; Cheng, Z.; Zhao, S.; Zhang, X.; Guo, X. Identification and fine mapping of two blast resistance genes in rice cultivar 93-11. Crop J. 2013, 1, 2–14. [Google Scholar] [CrossRef]
- Zhou, J.H.; Wang, J.L.; Xu, J.C.; Lei, C.L.; Ling, Z.Z. Identification and mapping of a rice blast resistance gene Pi-g(t) in the cultivar Guangchangzhan. Plant Pathol. 2004, 53, 191–196. [Google Scholar] [CrossRef]
- Tabien, R.E.; Li, Z.; Paterson, A.H.; Marchetti, M.A.; Stansel, J.W.; Pinson, S.R.M.; Park, W.D. Mapping of four major rice blast resistance genes from ‘Lemont’ and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 2000, 101, 1215–1225. [Google Scholar] [CrossRef]
- Pan, Q.; Wang, L.; Ikehashi, H.; Tanisaka, T. Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 1996, 86, 1071–1075. [Google Scholar] [CrossRef]
- Pan, Q. H.; Wang, L.; Ikehashi, H.; Yamagata, H.; Tanisaka, T. Identification of two new genes conferring resistance to rice blast in the Chinese native cultivar “Maowangu”. Plant Breed. 1998, 117, 27–31. [Google Scholar] [CrossRef]
- Ogawa, T.; Yamamoto, T. Inheritance of resistance to bacterial blight in rice. In Rice Genetics I: (In 2 Parts), Proceedings of the International Rice Genetics Symposium, Manila, Philippines, 27–31 May 1985; Banta, S.J., Ed.; World Scientific: Singapore, 2008; pp. 471–479. [Google Scholar]
- Goto, T.; Matsumoto, T.; Furuya, N.; Tsuchiya, K.; Yoshimura, A. Mapping of bacterial blight resistance gene Xa11 on rice chromosome 3. Jpn. Agric. Res. Quart. 2009, 43, 221–225. [Google Scholar] [CrossRef]
- Busungu, C.; Taura, S.; Sakagami, J.I.; Ichitani, K. Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 2016, 66, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wang, L.; Pan, Q. A new recessive gene conferring resistance against rice blast. Rice 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S. Linkage studies on the resistance to bacterial leaf blight, Xanthomonas oryzae (Uyeda et Ishiyama) Dowson, in rice. Bull. Natl. Inst. Agric. Sci. Ser. D 1967, 16, 1–18. [Google Scholar]
- Yoshimura, S.; Umehara, Y.; Kurata, N.; Nagamura, Y.; Sasaki, T.; Minobe, Y.; Iwata, N. Identification of a YAC clone carrying the Xa1 allele, a bacterial blight resistance gene in rice. Theor. Appl. Genet. 1996, 93, 117–122. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Li, D.; Zhu, Y.; Tan, M.; Zhang, D.; Lin, X. Fine mapping of Xa2, a bacterial blight resistance gene in rice. Mol. Breed. 2006, 17, 1–6. [Google Scholar] [CrossRef]
- Ogawa, T.; Morinaka, T.; Fujii, K.; Kimura, T. Inheritance of resistance of rice varieties Kogyoku and Java 14 to bacterial group V of Xanthomonas oryzae. Jpn. J. Phytopathol. 1978, 44, 137–141. [Google Scholar] [CrossRef]
- Taura, S.; Ogawa, T.; Tabien, R.E.; Khush, G.S.; Yoshimura, A.; Omura, T. The specific reaction of Taichung Native 1 to Philippine races of bacterial blight and inheritance of resistance to race 5 (PXO 112). Rice Genet. Newsl. 1987, 4, 101–102. [Google Scholar]
- Tan, Z.; Zhang, Q.; Zhu, L.; Wang, C. RFLP Mapping of a Rice bacterial blight resistance gene X_ (# alpha#-14). Hereditas 1998, 20, 30–33. [Google Scholar]
- Wang, C.; Wen, G.; Lin, X.; Liu, X.; Zhang, D. Identification and fine mapping of the new bacterial blight resistance gene, Xa31(t) in rice. Eur. J. Plant Pathol. 2009, 123, 235–240. [Google Scholar] [CrossRef]
- Terashima, T.; Fukuoka, S.; Saka, N.; Kudo, S. Mapping of a blast field resistance gene Pi39(t) of elite rice strain Chubu 111. Plant Breed. 2008, 127, 485–489. [Google Scholar] [CrossRef]
- Fukuoka, S.; Okuno, K.; Kawase, M. Rice Blast Disease Gene Pi21, Resistance Gene pi21 and Utilization Thereof. U.S. Patent WO/2007/000880, 4 January 2007. [Google Scholar]
- Shinoda, H.; Toriyama, K.; Yunoki, T.; Ezuka, A.; Sakurai, Y. Studies on the varietal resistance of rice to blast 6. Linkage relationship of blast resistance genes. Jpn. Chugoku Nogyo Shikengo Fukuyama Bull. Ser. A 1971, 20, 1–25. [Google Scholar]
- Causse, M.A.; Fulton, T.M.; Cho, Y.G.; Ahn, S.N.; Chunwongse, J.; Wu, K.; Xiao, J.; Yu, Z.; Ronald, P.C.; Harrington, S.E. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 1994, 138, 1251–1274. [Google Scholar] [PubMed]
- Iyer, A.S.; McCouch, S.R. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 2004, 17, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Garris, A.J.; Iyer, A.S.; Chapman, B.; Kresovich, S.; McCouch, S.R. High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 2003, 107, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.N.; Kim, Y.K.; Han, S.S.; Choi, H.C.; Moon, H.P.; McCouch, S.R. Molecular mapping of a gene for resistance to a Korean isolate of rice blast. Rice Genet Newsl. 1996, 13, 74–76. [Google Scholar]
- Naqvi, N.I.; Chattoo, B.B. Development of a sequence characterized amplified region (SCAR) based indirect selection method for a dominant blast-resistance gene in rice. Genome 1996, 39, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, G.S.; Khush, G.S.; Mew, T.W. Genetic analysis of bacterial blight resistance in seventy-four cultivars of rice, Oryza sativa L. Theor. Appl. Genet. 1978, 53, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Kaji, R.; Ogawa, T. Identification of the located chromosome of the resistance gene, Xa7, to bacterial leaf blight in rice. Breed. Sci. 1995, 45, 79. [Google Scholar]
- Porter, B.W.; Chittoor, J.M.; Yano, M.; Sasaki, T.; White, F.F. Development and mapping of markers linked to the rice bacterial blight resistance gene. Crop Sci. 2003, 43, 1484–1492. [Google Scholar] [CrossRef]
- Chen, S.; Huang, Z.; Zeng, L.; Yang, J.; Liu, Q.; Zhu, X. High-resolution mapping and gene prediction of Xanthomonas oryzae pv.oryzae resistance gene Xa7. Mol. Breed. 2008, 22, 433–441. [Google Scholar] [CrossRef]
- Gu, K.; Tian, D.; Yang, F.; Wu, L.; Sreekala, C.; Wang, D.; Wang, G.-L.; Yin, Z. High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 2004, 108, 800–807. [Google Scholar] [PubMed]
- Korinsak, S.; Sriprakhon, S.; Sirithanya, P.; Jairin, J.; Vanavichit, A.; Toojinda, T. Identification of microsatellite markers (SSR) linked to a new bacterial blight resistance gene xa33(t) in rice cultivar ‘Ba7’. Maejo Int. J. Sci. Technol. 2009, 3, 235–247. [Google Scholar]
- Deng, Y.; Zhu, X.; Shen, Y.; He, Z. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 2006, 113, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G.-L. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Jeung, J.U.; Kim, B.R.; Cho, Y.C.; Han, S.S.; Moon, H.P.; Lee, Y.T.; Jena, K.K. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 2007, 115, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Koide, Y.; Telebanco-Yanoria, M.J.; Fukuta, Y.; Kobayashi, N. Detection of novel blast resistance genes, Pi58(t) and Pi59(t), in a Myanmar rice landrace based on a standard differential system. Mol. Breed. 2013, 32, 241–252. [Google Scholar] [CrossRef]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G.-L. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.; Deng, X.; Liu, J.; Sun, P.; Liu, Y.; Huang, H.; Jiang, N.; Kang, H.; Ning, Y. Molecular mapping of the blast resistance genes Pi2-1 and Pi51(t) in the durably resistant rice “Tianjingyeshengdao”. Phytopathology 2012, 102, 779–786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G. AB lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-L.; Fan, Y.-Y.; Li, D.-B.; Zheng, K.-L.; Leung, H.; Zhuang, J.-Y. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor. Appl. Genet. 2005, 111, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Fjellstrom, R.; Conaway-Bormans, C.A.; McClung, A.M.; Marchetti, M.A.; Shank, A.R.; Park, W.D. Development of DNA markers suitable for marker assisted selection of three genes conferring resistance to multiple pathotypes. Crop Sci. 2004, 44, 1790–1798. [Google Scholar] [CrossRef]
- Hayasaka, H. Mapping genes conferring rice blast resistance in rice variety Kasalath using RFLP markers. II. Linkage analysis of the resistance gene on chromosome 6. Breed. Sci. 1995, 45, 168. [Google Scholar]
- Ballini, E.; Morel, J.; Droc, G.; Price, A.; Courtois, B.; Notteghem, J.; Tharreau, D.A. Genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 2008, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Li, Z.; Wu, J.; Wang, Y.; Wu, L.; Wang, S.; Wang, D.; Wen, T.; Liang, Y.; Sun, P. Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice 2012, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, S.; Yang, J.; Zhou, S.; Zeng, L.; Han, J.; Su, J.; Wang, L.; Pan, Q. The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theor. Appl. Genet. 2012, 124, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Vikal, Y.; Chawla, H.; Sharma, R.; Lore, J.S.; Singh, K. Mapping of bacterial blight resistance gene xa8 in rice (Oryza sativa L.). Indian J. Genet. Plant Breed. 2014, 74, 589–595. [Google Scholar] [CrossRef]
- Pan, Q.H.; Tanisaka, T.; Ikehashi, H. Studies on the genetics and breeding of blast resistance in rice IV. Gene analysis for the blast resistance of a indica variety Kasalath. Breed. Sci. 1995, 45, 170. [Google Scholar]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z.; Sanchez, A.; Park, Y.J.; Bennetzen, J.L.; Zhang, Q.; Wang, S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2006, 112, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Lin, L.; Tabien, R.E.; Khush, G.S. A new recessive gene for resistance to bacterial blight of rice. Rice Genet. Newsl. 1987, 4, 98–100. [Google Scholar]
- Sanchez, A.C.; Ilag, L.L.; Yang, D.; Brar, D.S.; Ausubel, F.; Khush, G.S.; Yano, M.; Sasaki, T.; Li, Z.; Huang, N. Genetic and physical mapping of xa13, a recessive bacterial blight resistance gene in rice. Theor. Appl. Genet. 1999, 98, 1022–1028. [Google Scholar] [CrossRef]
- Zhang, G.; Angeles, E.R.; Abenes, M.L.P.; Khush, G.S.; Huang, N. RAPD and RFLP mapping of the bacterial blight resistance gene xa-13 in rice. Theor. Appl. Genet. 1996, 93, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed]
- Berruyer, R.; Adreit, H.; Milazzo, J.; Gaillard, S.; Berger, A.; Dioh, W.; Lebrun, M.-H.; Tharreau, D. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-H. Construction of a molecular map of rice and gene mapping using a double-haploid population of a cross between indica and japonica varieties. Rice Genet. Newsl. 1993, 10, 132–135. [Google Scholar]
- Liu, X.Q.; Wang, L.; Chen, S.; Lin, F.; Pan, Q.H. Genetic and physical mapping of Pi36(t), a novel rice blast resistance gene located on rice chromosome 8. Mol. Genet. Genom. 2005, 274, 394–401. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, X.; Wang, L.; Wang, L.; Lin, F.; Cheng, Y.; Chen, Z.; Liao, Y.; Pan, Q. Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci. China Life Sci. 2012, 55, 141–149. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, B.; Zhang, S.; Zhu, X.; Yang, Q.; Wu, S.; Mei, M.; Mauleon, R.; Leach, J.; Mew, T.; Leung, H. Candidate defense genes as predictors of quantitative blast resistance in rice. Mol. Plant Microbe Interact. 2004, 17, 1146–1152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pan, Q.-H.; Hu, Z.-D.; Takatoshi, T.; Wang, L. Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. Acta Bot. Sin. 2003, 24, 871–877. [Google Scholar]
- Kinoshita, T.; Kiyosawa, S. Some considerations on linkage relationships between Pii and Piz in the blast resistance of rice. Rice Genet. Newsl. 1997, 14, 57–59. [Google Scholar]
- Mackill, D.J.; Bonman, J.M. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 1992, 82, 746–749. [Google Scholar] [CrossRef]
- Jeon, J.-S.; Chen, D.; Yi, G.-H.; Wang, G.L.; Ronald, P.C. Genetic and physical mapping of Pi5(t), a locus associated with broad-spectrum resistance to rice blast. Mol. Genet. Genom. 2003, 269, 280–289. [Google Scholar]
- Liu, Y.; Liu, B.; Zhu, X.; Yang, J.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Cao, Y.; Xu, C.; Li, X.; Wang, S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor. Appl. Genet. 2006, 113, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Ezuka, A.; Horino, O.; Toriyama, K. Inheritance of resistance of rice variety Wase Aikoku 3 to Zanthomonas oryzae. Bulletin 1975, 28, 124–130. [Google Scholar]
- Yoshimura, S.; Yoshimura, A.; Iwata, N.; McCouch, S.R.; Abenes, M.L.; Baraoidan, M.R.; Mew, T.W.; Nelson, R.J. Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol. Breed. 1995, 1, 375–387. [Google Scholar] [CrossRef]
- Sun, X.; Yang, Z.; Wang, S.; Zhang, Q. Identification of a 47-kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor. Appl. Genet. 2003, 106, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Mew, T.W.; Cruz, V.; Reyes, R.C. Interaction of Xanthomonas campestris pv. oryzae and a resistant rice cultivar. Phytopathology 1982, 72, 786–789. [Google Scholar] [CrossRef]
- Yoshimura, A.; Mew, T.W.; Khush, G.S.; Omura, T. Inheritance of resistance to bacterial blight in rice cultivar Cas 209. Phytopathology 1983, 73, 1409–1412. [Google Scholar] [CrossRef]
- Gu, K.; Sangha, J.S.; Li, Y.; Yin, Z. High-resolution genetic mapping of bacterial blight resistance gene Xa10. Theor. Appl. Genet. 2008, 116, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Wang, G.-L.; Chen, L.-L.; Kim, H.-S.; Pi, L.-Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.-X.; Zhu, L.-H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S.; Bacalangco, E.; Ogawa, T. A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newsl. 1991, 7, 121–122. [Google Scholar]
- Ronald, P.C.; Albano, B.; Tabien, R.; Abenes, L.; Wu, K.; McCouch, S.; Tanksley, S.D. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Gen. Genet. 1992, 236, 113–120. [Google Scholar] [PubMed]
- Lin, X.H.; Zhang, D.P.; Xie, Y.F.; Gao, H.P.; Zhang, Q. Identifying and mapping a new gene for bacterial blight resistance in rice based on RFLP markers. Phytopathology 1996, 86, 1156–1159. [Google Scholar] [CrossRef]
- Wang, C.; Tan, M.; Xu, X.; Wen, G.; Zhang, D.; Lin, X. Localizing the bacterial blight resistance gene, Xa22(t), to a 100-kilobase bacterial artificial chromosome. Phytopathology 2003, 93, 1258–1262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, Q.; Lin, S.C.; Zhao, B.Y.; Wang, C.L.; Yang, W.C.; Zhou, Y.L.; Li, D.Y.; Chen, C.B.; Zhu, L.H. Identification and tagging a new gene for resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) from O. rufipogon. Rice Genet. Newsl. 1998, 15, 138–142. [Google Scholar]
- Jin, X.W.; Wang, C.L.; Yang, Q.; Jiang, Q.X.; Fan, Y.L.; Liu, G.C.; Zhao, K.J. Breeding of near-isogenic line CBB30 and molecular mapping of Xa30(t), a new resistance gene to bacterial blight in rice. Sci. Agric. Sin. 2007, 40, 1094–1100. [Google Scholar]
- Zheng, C.-K.; Chun-Lian, W.; Yuan-Jie, Y.U.; Liang, Y.-T.; Kai-Jun, Z. Identification and molecular mapping of Xa32(t), a novel resistance gene for bacterial blight (Xanthomonas oryzae pv. oryzae) in rice. Acta Agron. Sin. 2009, 35, 1173–1180. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, D.; Lin, X. Identification and mapping of a novel bacterial blight resistance gene Xa35(t) originated from Oryza minuta. Sci. Agric. Sin. 2010, 43, 2611–2618. [Google Scholar]
- Zhang, F.; Zhuo, D.; Huang, L.; Wang, W.; Xu, J.; Vera Cruz, C.; Li, Z.; Zhou, Y. Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 2015, 64, 568–575. [Google Scholar] [CrossRef]
- Kim, S.-M.; Suh, J.-P.; Qin, Y.; Noh, T.-H.; Reinke, R.F.; Jena, K.K. Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 2015, 128, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Bao, Y.; Xie, L.; Su, Y.; Chu, R.; He, W.; Huang, J.; Wang, J.; Zhang, H. Fine mapping and identification of blast resistance gene Pi-hk1 in a broad-spectrum resistant japonica rice landrace. Phytopathology 2013, 103, 1162–1168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V.; Pandit, A.; Singh, A.; Gaikwad, K.; Upreti, H.C. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genomics 2005, 274, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, J.L.; Correa-Victoria, F.J.; Escobar, F.; Prado, G.; Aricapa, G.; Duque, M.C.; Tohme, J. Identification of microsatellite markers linked to the blast resistance gene Pi-1(t) in rice. Euphytica 2008, 160, 295–304. [Google Scholar] [CrossRef]
- Fujii, K.; Hayano-Saito, Y.; Saito, K.; Sugiura, N.; Hayashi, N.; Tsuji, T.; Izawa, T.; Iwasaki, M. Identification of a RFLP marker tightly linked to the panicle blast resistance gene, Pb1, in rice. Breed. Sci. 2000, 50, 183–188. [Google Scholar] [CrossRef]
- Goto, I. Genetic studies on the resistance of rice plant to the blast fungus. I. Inheritance of resistance in crosses Sensho X H-79 and Imochi-shirazu X H-79. Ann. Phytopathol. Soc. Jpn. 1970, 36, 304–312. [Google Scholar] [CrossRef]
- Goto, I.; Jaw, Y.-L.; Baluch, A.A. Genetic studies on resistance of rice plant to blast fungus. Jpn. J. Phytopathol. 1981, 47, 252–254. [Google Scholar] [CrossRef][Green Version]
- Gowda, M.; Roy-Barman, S.; Chattoo, B.B. Molecular mapping of a novel blast resistance gene Pi38 in rice using SSLP and AFLP markers. Plant Breed. 2006, 125, 596–599. [Google Scholar] [CrossRef]
- Yunoki, T.; Ezuka, A.; Morinaka, T.; Sakurai, Y.; Shinoda, H.; Toriyama, K. Studies on the varietal resistance to Rice blast. 4. Variation of field resistance due to fungus strains. Bull. Chugoku Agric. Exp. Stn. Ser. A 1970, 21–41. [Google Scholar]
- Zenbayashi-Sawata, K.; Ashizawa, T.; Koizumi, S. Pi34-AvrPi34: A new gene-for-gene interaction for partial resistance in rice to blast caused by Magnaporthe grisea. J. Gen. Plant Pathol. 2005, 71, 395–401. [Google Scholar] [CrossRef]
- Kwon, S.-W.; Cho, Y.-C.; Kim, Y.-G.; Suh, J.-P.; Jeung, J.-U.; Roh, J.-H.; Lee, S.-K.; Jeon, J.-S.; Yang, S.-J.; Lee, Y.-T. Development of near-isogenic japonica rice lines with enhanced resistance to Magnaporthe grisea. Mol. Cells 2008, 25, 407–416. [Google Scholar] [PubMed]
- Chauhan, R.; Farman, M.; Zhang, H.-B.; Leong, S. Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene Avr1-CO39 of Magnaporthe grisea. Mol. Genet. Genom. 2002, 267, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-H.; Dela Vina, M.; Inukai, T.; Mackill, D.J.; Ronald, P.C.; Nelson, R.J. Molecular mapping of the blast resistance gene, Pi44(t), in a line derived from a durably resistant rice cultivar. Theor. Appl. Genet. 1999, 98, 1046–1053. [Google Scholar] [CrossRef]
- Sun, P.; Liu, J.; Wang, Y.; Jiang, N.; Wang, S.; Dai, Y.; Gao, J.; Li, Z.; Pan, S.; Wang, D. Molecular mapping of the blast resistance gene Pi49 in the durably resistant rice cultivar Mowanggu. Euphytica 2013, 192, 45–54. [Google Scholar] [CrossRef]
- Kaji, R.; Ogawa, T. RFLP Mapping of Blast Resistance Gene Pi-km in Rice; International Rice Research Notes; Food and Agriculture Organization: Rome, Italy, 1996. [Google Scholar]
- Ahn, S.-N.; Kim, Y.-K.; Hong, H.-C.; Han, S.-S.; Kwon, S.-J.; Choi, H.-C.; Moon, H.-P.; McCouch, S.R. Molecular mapping of a new gene for resistance to rice blast (Pyricularia grisea Sacc.). Euphytica 2000, 116, 17–22. [Google Scholar] [CrossRef]
- Huang, H.; Huang, L.; Feng, G.; Wang, S.; Wang, Y.; Liu, J.; Jiang, N.; Yan, W.; Xu, L.; Sun, P. Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi 3150. Phytopathology 2011, 101, 620–626. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prasad, M.S.; Kanthi, B.A.; Balachandran, S.M.; Seshumadhav, M.; Mohan, K.M.; Viraktamath, B.C. Molecular mapping of rice blast resistance gene Pi-1(t) in the elite indica variety Samba mahsuri. World J. Microbiol. Biotechnol. 2009, 25, 1765–1769. [Google Scholar] [CrossRef]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Rasabandith, S.; Angeles, E.R.; Khush, G.S. Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathology 2003, 93, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.-Y.; Ma, W.-B.; Wu, J.-L.; Chai, R.-Y.; Lu, J.; Fan, Y.-Y.; Jin, M.-Z.; Leung, H.; Zheng, K.-L. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 2002, 128, 363–370. [Google Scholar] [CrossRef]
- Kumar, P.; Pathania, S.; Katoch, P.; Sharma, T.R.; Plaha, P.; Rathour, R. Genetic and physical mapping of blast resistance gene Pi42(t) on the short arm of rice chromosome 12. Mol. Breed. 2010, 25, 217–228. [Google Scholar] [CrossRef]
- Wu, K.S.; Martinez, C.; Lentini, Z.; Tohme, J.; Chumley, F.G.; Scolnik, P.A.; Valent, B. Cloning a blast resistance gene by chromosome walking. In Rice Genetics III: (In 2 Parts), Proceedings of the Third International Rice Genetics Symposium, Manila, Philippines, 16–20 October 1995; World Scientific: Singapore, 2008; pp. 669–674. [Google Scholar]
- Yu, Z.H.; Mackill, D.J.; Bonman, J.M.; Tanksley, S.D. Tagging genes for blast resistance in rice via linkage to RFLP markers. Theor. Appl. Genet. 1991, 81, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Inukai, T.; Nelson, R. Mapping for blast resistance gene H-3 derived from rice cuhivar Pai-Kan-Tao. Rep. Hokkaido Br. CropSol. See Jap. Soc. Breed. 1994, 35, 54–55. [Google Scholar]
- Hayashi, K.; Yoshida, H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009, 57, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Bryan, G.T.; Wu, K.-S.; Farrall, L.; Jia, Y.; Hershey, H.P.; McAdams, S.A.; Faulk, K.N.; Donaldson, G.K.; Tarchini, R.; Valent, B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lin, F.; Wang, L.; Pan, Q. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 2007, 176, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Y.; Wang, L.; Jing, J.-X.; Li, Z.-Q.; Lin, F.; Huang, L.-F.; Pan, Q.-H. The Pikm gene, conferring stable resistance to isolates of Magnaporthe oryzae, was finely mapped in a crossover-cold region on rice chromosome 11. Mol. Breed. 2007, 20, 179–188. [Google Scholar] [CrossRef]
- Talukder, Z.I.; McDonald, A.J.S.; Price, A.H. Loci controlling partial resistance to rice blast do not show marked QTL× environment interaction when plant nitrogen status alters disease severity. New Phytol. 2005, 168, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, J.; Ling, Z.; Zhu, L. Analysis of rice blast resistance genes by QTL mapping. Chin. Sci. Bull. 2004, 49, 337–342. [Google Scholar] [CrossRef]
- Albar, L.; Lorieux, M.; Ahmadi, N.; Rimbault, I.; Pinel, A.; Sy, A.A.; Fargette, D.; Ghesquiere, A. Genetic basis and mapping of the resistance to rice yellow mottle virus. QTLs identification and relationship between resistance and plant morphology. Theor. Appl. Genet. 1998, 97, 1145–1154. [Google Scholar] [CrossRef]
- Djedatin, G.; Ndjiondjop, M.N.; Sanni, A.; Lorieux, M.; Verdier, V.; Ghesquiere, A. Identification of novel major and minor QTLs associated with Xanthomonas oryzae pv.oryzae (African strains) resistance in rice (Oryza sativa L.). Rice 2016, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.D.; Zeng, D.L.; Ma, L.Y.; Ji, Z.J.; Guo, L.B.; Li, X.M.; Qian, Q. Mapping QTLs for bacterial blight resistance in a DH population from japonica/indica cross of rice (Oryzae sativa). Chin. J. Rice Sci. 2006, 20, 102–104. [Google Scholar]
- Fiyaz, R.A.; Yadav, A.K.; Krishnan, S.G.; Ellur, R.K.; Bashyal, B.M.; Grover, N.; Bhowmick, P.K.; Nagarajan, M.; Vinod, K.K.; Singh, N.K.; et al. Mapping quantitative trait loci responsible for resistance to Bakanae disease in rice. Rice 2016, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Volante, A.; Tondelli, A.; Aragona, M.; Valente, M.T.; Biselli, C.; Desiderio, F.; Bagnaresi, P.; Matic, S.; Gullino, M.L.; Infantino, A.; et al. Identification of bakanae disease resistance loci in japonica rice through genome wide association study. Rice 2017, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Tabien, R.; Li, Z.; Paterson, A.; Marchetti, M.; Stansel, J.; Pinson, S. Mapping QTLs for field resistance to the rice blast pathogen and evaluating their individual and combined utility in improved varieties. Theor. Appl. Genet. 2002, 105, 313–324. [Google Scholar] [PubMed]
- Tang, D.; Wu, W.; Li, W.; Lu, H.; Worland, A.J. Mapping of QTLs conferring resistance to bacterial leaf streak in rice. Theor. Appl. Genet. 2000, 101, 286–0291. [Google Scholar] [CrossRef]
- Loan, L.C.; Du, P.V.; Li, Z. Molecular dissection of quantitative resistance of sheath blight in rice (Oryza sativa L.). Omonrice 2004, 12, 1–12. [Google Scholar]
- Sato, H.; Matsumoto, K.; Ota, C.; Yamakawa, T.; Kihara, J.; Mizobuchi, R. Confirming a major QTL and finding additional loci responsible for field resistance to brown spot (Bipolaris oryzae) in rice. Breed. Sci. 2015, 65, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Bagali, P.G.; Hittalmani, S.; Shashidhar, S.Y.; Shashidhar, H.E. Identification of DNA markers linked to partial resistance for blast disease in rice across four locations. In Advances in Rice Blast Research. Developments in Plant Pathology; Tharreau, D., Lebrun, M.H., Talbot, N.J., Notteghem, J.L., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 34–42. [Google Scholar]
- Li, Y.S.; Zhang, Y.D.; Zhu, Z.; Zhao, L.; Wang, C.L. QTL analysis for resistance to rice false smut by using recombinant inbred lines in rice. Chin. J. Rice Sci. 2008, 22, 472–476. [Google Scholar]
- Lopez-Gerena, J. Mapping QTL Controlling Durable Resistance to Rice Blast in the Cultivar Oryzica Llanos 5. Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2006. [Google Scholar]
- Chen, H.; Wang, S.; Xing, Y.; Xu, C.; Hayes, P.M.; Zhang, Q. Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc. Natl. Acad. Sci. USA 2003, 100, 2544–2549. [Google Scholar] [CrossRef] [PubMed]
- Pinson, S.R.M.; Capdevielle, F.M.; Oard, J.H. Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 2005, 45, 503–510. [Google Scholar] [CrossRef]
- Loan, L.C.; Du, P.; Li, Z. Identification of genes conferring resistance to some Philippine and Vietnamese races of blast. Omonrice 2003, 11, 49–62. [Google Scholar]
- Liu, G.; Jia, Y.; Correa-Victoria, F.J.; Prado, G.A.; Yeater, K.M.; Mcclung, A.; Correll, J.C. Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology 2009, 99, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, L.; Zhang, Y.; Sun, D.-Z.; Zhai, H.-Q.; Wan, J.-M. Detection and analysis of QTL for resistance to stripe disease in rice, using backcross inbred lines. Acta Agron. Sin. 2005, 31, 1041–1046. [Google Scholar]
- Katara, J.L.; Sonah, H.; Deshmukh, R.K.; Chaurasia, R.; Kotasthane, A.S. Molecular analysis of QTLs associated with resistance to brown spot in rice (Oryza sativa L.). Indian J. Genet. 2010, 70, 17–21. [Google Scholar]
- Sato, H.; Ando, I.; Hirabayashi, H.; Takeuchi, Y.; Arase, S.; Kihara, J.; Kato, H.; Imbe, T.; Nemoto, H. QTL analysis of brown spot resistance in rice (Oryza sativa L.). Breed. Sci. 2008, 58, 93–96. [Google Scholar] [CrossRef]
- Zhou, Y.L.; Xie, X.W.; Zhang, F.; Wang, S.; Liu, X.Z.; Zhu, L.H.; Xu, J.; Gao, Y.M.; Li, Z. Detection of quantitative resistance loci associated with resistance to rice false smut (Ustilaginoidea virens) using introgression lines. Plant Pathol. 2014, 63, 365–372. [Google Scholar] [CrossRef]
- Chen, C.-H.; Zheng, W.; Huang, X.-M.; Zhang, D.-P.; Lin, X.-H. Major QTL conferring resistance to rice bacterial leaf streak. Agric. Sci. China 2006, 5, 216–220. [Google Scholar] [CrossRef]
- Kunihiro, Y.; Qian, Q.; Sato, H.; Teng, S.; Zeng, D.-L.; Fujimoto, K.; Zhu, L.H. QTL analysis of sheath blight resistance in rice (Oryza sativa L.). Acta Genet. Sin. 2002, 29, 50–55. [Google Scholar] [PubMed]
- Zou, J.H.; Pan, X.B.; Chen, Z.X.; Xu, J.Y.; Lu, J.F.; Zhai, W.X.; Zhu, L.H. Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor. Appl. Genet. 2000, 101, 569–573. [Google Scholar] [CrossRef]
- Xu, Q.; Yuan, X.; Yu, H.; Wang, Y.; Tang, S.; Wei, X. Mapping quantitative trait loci for sheath blight resistance in rice using double haploid population. Plant Breed. 2011, 130, 404–406. [Google Scholar] [CrossRef]
- Wang, B.; Jiang, L.; Chen, L.; Lu, B.; Wang, Q.; Quang, T.L.; Fan, J.; Cheng, X.; Zhai, H.; Xu, D. Screening of rice resources against rice black-streaked dwarf virus and mapping of resistant QTL. Acta Agron. Sin. 2010, 36, 1258–1264. [Google Scholar] [CrossRef]
- Pan, X.; Zou, J.; Chen, Z.; Lu, J.; Yu, H.; Li, H.; Wang, Z.; Pan, X.; Rush, M.C.; Zhu, L. Tagging major quantitative trait loci for sheath blight resistance in rice variety, Jasmine 85. Chinese Sci. Bull. 1999, 44, 1783–1789. [Google Scholar] [CrossRef]
- Yu, Y.C.; Teng, S.; Zeng, D.L.; Dong, G.J.; Qian, Q.; Huang, D.N.; Zhu, L.W. Analysis of QTLs for resistance to rice bacterial blight. Chin. J. Rice Sci. 2003, 17, 315–318. [Google Scholar]
- Sato, H.; Takeuchi, Y.; Hirabayashi, H.; Nemoto, H.; Hirayama, M.; Kato, H.; Imbe, T.; Ando, I. Mapping QTLs for field resistance to rice blast in the Japanese upland rice variety Norin 12. Breed. Sci. 2006, 56, 415–418. [Google Scholar] [CrossRef][Green Version]
- Zhou, T.; Du, L.; Wang, L.; Wang, Y.; Gao, C.; Lan, Y.; Sun, F.; Fan, Y.; Wang, G.; Zhou, Y. Genetic analysis and molecular mapping of QTLs for resistance to rice black-streaked dwarf disease in rice. Sci. Rep. 2015, 5, 10509. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pinson, S.R.M.; Marchetti, M.A.; Stansel, J.W.; Park, W.D. Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor. Appl. Genet. 1995, 91, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-J.; Zhong, H.; Zhou, Y.; Zuo, H.; Zhou, L.-H.; Zhu, J.-Y.; Ji, C.-Q.; Gu, S.-L.; Gu, M.-H.; Liang, G.-H. Identification of QTLs for the resistance to rice stripe virus in the indica rice variety Dular. Euphytica 2009, 165, 557–565. [Google Scholar] [CrossRef]
- Wang, C.; Su, C.; Zhai, H.; Wan, J. Identification of QTLs underlying resistance to a virulent strain of Xanthomonas oryzae pv.oryzae in rice cultivar DV85.F. Crop Res. 2005, 91, 337–343. [Google Scholar] [CrossRef]
- Fukuoka, S.; Okuno, K. QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor. Appl. Genet. 2001, 103, 185–190. [Google Scholar] [CrossRef]
- Miyamoto, M.; Yano, M.; Hirasawa, H. Mapping of quantitative trait loci conferring blast field resistance in the Japanese upland rice variety Kahei. Breed. Sci. 2001, 51, 257–261. [Google Scholar] [CrossRef]
- Han, Y.P.; Xing, Y.Z.; Chen, Z.X.; Gu, S.L.; Pan, X.B.; Chen, X.L.; Zhang, Q.F. Mapping QTLs for horizontal resistance to sheath blight in an elite rice restorer line, Minghui 63. Acta Genet. Sin. 2002, 29, 622–626. [Google Scholar] [PubMed]
- Sun, D.-Z.; Jiang, L.; Zhang, Y.-X.; Cheng, X.-N.; Zhai, H.-Q.; Wan, J.-M. Quantitative trait loci for resistance to stripe disease in rice (Oryza sativa). Rice Sci. 2007, 14, 157–160. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, X.Y.; Zhang, S.; Bernardo, M.; Edwards, J.; Galbraith, D.W.; Leach, J.; Zhang, G.; Liu, B.; Leung, H. Dissecting quantitative resistance against blast disease using heterogeneous inbred family lines in rice. Theor. Appl. Genet. 2011, 122, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.-X.; Ji, X.-M.; Yang, Y.; Pan, X.-Y.; Zuo, S.-M.; Zhang, Y.-F.; Zou, J.-H.; Chen, Z.-X.; Zhu, L.-H.; Pan, X.-B. Identification and marker-assisted selection of two major quantitative genes controlling rice sheath blight resistance in backcross generations. Acta Genet. Sin. 2005, 32, 399–405. [Google Scholar] [PubMed]
- .Xiu-Lan, D.; Ling, J.; Shi-Jia, L.; Chun-Ming, W.; Liang-Ming, C.; Zhao-Bang, C.; Yong-Jian, F.; Yi-Jun, Z.; Jian-Min, W. QTL analysis for rice stripe disease resistance gene using recombinant inbred lines (RILs) derived from crossing of Kinmaze and DV85. Acta Genet. Sin. 2004, 31, 287–292. [Google Scholar]
- Ashkani, S.; Rafil, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular breeding strategy and challenges towards the improvement of blast disease resistance in rice crop. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Gupta, Y.K.; Singh, P.K.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Rana, J.C.; et al. Molecular diversity in rice blast resistance gene Pi-ta makes it highly effective against dynamic population of Magnaporthe oryzae. Funct. Integr. Genom. 2013, 13, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Singh, P.K.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Singh, N.K.; Sharma, T.R. Positive selection pressure on rice blast resistance allele Piz-t makes it divergent in Indian landraces. J. Plant Int. 2013, 8, 34–44. [Google Scholar]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Variar, M.; Prashanthi, S.K.; Singh, A.K.; Singh, U.D.; Chand, D.; Singh, N.K.; et al. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, G.; Costanzo, S.; Lee, S.; Dai, Y. Current progress on genetic interactions of rice with rice blast and sheath blight fungi. Front. Agric. China 2009, 3, 231–239. [Google Scholar] [CrossRef]
- Barnwal, M.K.; Kotasthane, A.; Magculia, N.; Mukherjee, P.K.; Savary, S.; Sharma, A.K.; Singh, H.B.; Singh, U.S.; Sparks, A.H.; Variar, M.; et al. A review on crop losses, epidemiology and disease management of rice brown spot to identify research priorities and knowledge gaps. Eur. J. Plant Pathol. 2013, 136, 443–457. [Google Scholar] [CrossRef]
- Mizobuchi, R.; Fukuoka, S.; Tsushima, S.; Yano, M.; Sato, H. QTLs for resistance to major rice diseases exacerbated by global warming: Brown spot, bacterial seedling rot and bacterial grain rot. Rice 2016, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.D.; Guo, L.B.; Li, X.M.; Ji, Z.J.; Ma, L.Y.; Qian, Q. Analysis of QTLs for resistance to rice bakanae disease. Chin. J. Rice Sci. 2006, 6, 657–659. [Google Scholar]
- Hur, Y.J.; Lee, S.B.; Kim, T.H.; Kwon, T.; Lee, J.H.; Shin, D.J.; Park, S.K.; Hwang, U.H.; Cho, J.H.; Yoon, Y.N.; et al. Mapping of qBK1, a major QTL for bakanae disease resistance in rice. Mol. Breed. 2015, 35, 78. [Google Scholar] [CrossRef]
- Guo, X.; Li, Y.; Fan, J.; Li, L.; Huang, F.; Wang, W. Progress in the study of false smut disease in rice. J. Agric. Sci. Technol. A 2012, 2, 1211–1217. [Google Scholar]
- Andargie, M.; Li, J. Arabidopsis thaliana: A model host plant to study plant-pathogen interaction using rice false smut isolates of Ustilaginoidea virens. Front. Plant Sci. 2016, 7, 192. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.; Wang, S. Toward an understanding of the molecular basis of quantitative disease resistance in rice. J. Biotechnol. 2012, 159, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Nino-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Triplett, L.R.; Cohen, S.P.; Heffelfinger, C.; Schmidt, C.L.; Huerta, A.I.; Tekete, C.; Verdier, V.; Bogdanove, A.J.; Leach, J.E. A resistance locus in the American heirloom rice variety Carolina Gold Select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv. oryzicola. Plant J. 2016, 87, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Ham, J.H.; Melanson, R.A.; Rush, M.C. Burkholderia glumae: Next major pathogen of rice? Mol. Plant Pathol. 2011, 12, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Hayano-Saito, Y.; Saito, K.; Nakamura, S.; Kawasaki, S.; Iwasaki, M. Fine physical mapping of the rice stripe resistance gene locus, Stvb-i. Theor. Appl. Genet. 2000, 101, 59–63. [Google Scholar] [CrossRef]
- Albar, L.; Ndjiondjop, M.-N.; Esshak, Z.; Berger, A.; Pinel, A.; Jones, M.; Fargette, D.; Ghesquiere, A. Fine genetic mapping of a gene required for Rice yellow mottle virus cell-to-cell movement. Theor. Appl. Genet. 2003, 107, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Thiemele, D.; Boisnard, A.; Ndjiondjop, M.N.; Cheron, S.; Sere, Y.; Ake, S.; Ghesquiere, A.; Albar, L. Identification of a second major resistance gene to rice yellow mottle virus, RYMV2, in the African cultivated rice species, O. glaberrima. Theor. Appl. Genet. 2010, 121, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Muhsin, M.; Atienza, G.A.; Kwak, D.-Y.; Kim, S.-M.; De Leon, T.B.; Angeles, E.R.; Coloquio, E.; Kondoh, H.; Satoh, K.; et al. Single nucleotide polymorphisms in a gene for translation initiation factor (eIF4G) of rice (Oryza sativa) associated with resistance to rice tungro spherical virus. Mol. Plant Microbe Interact. 2010, 23, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, C.J.; Huang, J.; Yang, S.H.; Tian, D.C.; Zhang, X.H. Allele mining of rice blast resistance genes at AC134922 locus. Biochem. Biophy. Res. Commun. 2014, 446, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H.; Ishikawa, K.; Omura, T.; Cabauatan, P.Q.; Koganezawa, H. Characterization of rice tungro bacilliform and rice tungro spherical viruses. Phytopathology 1991, 81, 1130–1132. [Google Scholar] [CrossRef]
- Hibino, H.; Daquioag, R.D.; Mesina, E.M.; Aguiero, V.M. Resistances in rice to tungro-associated viruses. Plant Dis. 1990, 74, 923–926. [Google Scholar] [CrossRef]
- Chen, J.; Huang, D.-R.; Wang, L.; Liu, G.J.; Zhuang, J.Y. Identificatiion of quantitative trait loci for resistance to whitebacked planthopper, Sogatella furcifera, from an interspecific cross Oryza sativa × O. rufipogon. Breed. Sci. 2010, 60, 153–159. [Google Scholar] [CrossRef]
- Hittalmani, S.; Parco, A.; Mew, T.V.; Zeigler, R.S.; Huang, N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar] [CrossRef]
- Liu, S.P.; Li, X.; Wang, C.Y.; Li, X.H.; He, Y.Q. Improvement of resistance to rice blast in Zhenshan 97 by molecular marker aided selection. Acta Bot. Sin. 2002, 45, 1346–1350. [Google Scholar]
- Narayanan, N.N.; Baisakh, N.; Cruz, C.M.V.; Gnanamanickam, S.S.; Datta, K.; Datta, S.K. Molecular breeding for the development of blast and bacterial blight resistance in rice cv. IR50. Crop Sci. 2002, 42, 2072–2079. [Google Scholar] [CrossRef]
- Lee, S.; Costanzo, S.; Jia, Y.; Olsen, K.M.; Caicedo, A.L. Evolutionary dynamics of the genomic region around the blast resistance gene Pi-ta in AA genome Oryza species. Genetics 2009, 183, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, V.K.; Singh, S.P.; Pandian, R.T.P.; Ellur, R.K.; Singh, D.; Bhowmick, P.K.; Gopala Krishnan, S.; Nagarajan, M.; Vinod, K.K.; et al. Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.Y.; Wu, T.; Liu, W.G.; Wang, F.; Li, J.H.; Zhu, X.Y.; Huang, H.J.; Liu, Z.R.; Liao, Y.L.; Zhu, M.S.; et al. Genetic improvement of resistance to blast and bacterial blight of the elite maintainer line Rongfeng B in hybrid rice (Oryza sativa L.) by using marker-assisted selection. Afr. J. Biotechnol. 2012, 11, 13104–13114. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Mizukami, Y.; Koga, H.; Yamanouchi, U.; Yosuke Yoshioka, Y.; Hayashi, N.; Ebana, K.; Mizobuchi, R.; Yano, M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015, 5, 7773. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.Z.; Zia, M.; Rashid, H.; Chaudhary, M.F.; Chaudhry, Z. Comparison of transgenic plant production for bacterial blight resistance in Pakistani local rice (Oryza sativa L.) cultivars. Afr. J. Biotechnol. 2010, 9, 1892–1904. [Google Scholar]
- Qiu, X.; Pang, Y.; Yuan, Z.; Xing, D.; Xu, J.; Dingkuhn, M.; Li, Z.; Ye, G. Genome-wide association study of grain appearance and milling quality in a worldwide collection of indica rice germplasm. PLoS ONE 2015, 10, e0145577. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R.; Wright, M.H.; Tung, C.W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 10532. [Google Scholar] [CrossRef] [PubMed]
- Raboin, L.-M.; Ballini, E.; Tharreau, D.; Ramanantsoanirina, A.; Frouin, J.; Courtois, B.; Ahmadi, N. Association mapping of resistance to rice blast in upland field conditions. Rice 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically-important loci in rice from mutant populations. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xie, W.; Li, J.; Zhou, F.; Zhang, Q. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 2014, 12, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Mgonja, E.M.; Park, C.H.; Kang, H.; Balimponya, E.G.; Opiyo, S.; Bellizzi, M.; Mutiga, S.K.; Rotich, F.; Ganeshan, V.D.; Mabagala, R.; et al. Genotyping-by-sequencing-based genetic analysis of African rice cultivars and association mapping of blast resistance genes against Magnaporthe oryzae populations in Africa. Phytopathology 2017, 107, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.X.; Xu, J.L.; Luo, R.T.; Shi, D.; Li, Z.K. Genetic analysis and breeding use of blast resistance in a japonica rice mutant R917. Euphytica 2003, 130, 71–76. [Google Scholar] [CrossRef]
- Ahloowalia, B.S.; Maluszynski, M.; Nichterlein, K. Global impact of mutation-derived varieties. Euphytica 2004, 135, 187–204. [Google Scholar] [CrossRef]
- Shu, G.Y. Induced Plant Mutations in the Genomics Era; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009; pp. 425–427. [Google Scholar]
- Alonso, J.M.; Ecker, J.R. moving forward in reverse genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 2006, 7, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Gurr, S.J.; Rushton, P.J. Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol. 2005, 23, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Nordlee, J.A.; Taylor, S.L.; Townsend, J.A.; Thomas, L.A.; Bush, R.K. Identification of a Brazil-nut allergen in transgenic soybeans. N. Engl. J. Med. 1996, 334, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Marvier, M.; McCreedy, C.; Regetz, J.; Kareiva, P. A meta-analysis of effects of Bt cotton and maize on nontarget invertebrates. Science 2007, 316, 1475–1477. [Google Scholar] [CrossRef] [PubMed]
- Radosevich, S.R.; Ghersa, C.M.; Comstock, G. Concerns a weed scientist might have about herbicide-tolerant crops. Weed Technol. 1992, 6, 635–639. [Google Scholar] [CrossRef]
- Jepson, P.C.; Croft, B.A.; Pratt, G.E. Test systems to determine the ecological risks posed by toxin release from Bacillus thuringiensis genes in crop plants. Mol. Ecol. 1994, 3, 81–99. [Google Scholar] [CrossRef]
- Xiao, N.; Wu, Y.; Pan, C.; Yu, L.; Chen, Y.; Liu, G.; Li, Y.; Zhang, X.; Wang, Z.; Dai, Z. Improving of rice blast resistances in japonica by pyramiding major R genes. Front. Plant Sci. 2017, 7, 1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine rich repeat class of plant disease resistance genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-K.; Song, M.-Y.; Seo, Y.-S.; Kim, H.-K.; Ko, S.; Cao, P.-J.; Suh, J.-P.; Yi, G.; Roh, J.-H.; Lee, S. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes. Genetics 2009, 181, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Kumar, S.P.; Gupta, S.K.; Gautam, N.; Singh, N.K.; Sharma, T.R. Functional complementation of rice blast resistance gene Pi-kh (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J. Plant Biochem. Biotechnol. 2011, 20, 55–65. [Google Scholar] [CrossRef]
- Zhai, C.; Lin, F.; Dong, Z.; He, X.; Yuan, B.; Zeng, X.; Wang, L.; Pan, Q. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 2011, 189, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two adjacent nucleotide-binding site–leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Zhai, C.; Wang, W.; Zeng, X.; Xu, X.; Hu, H.; Lin, F.; Wang, L.; Pan, Q. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 2011, 122, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Rai, A.K.; Devanna, B.N.; Singh, P.K.; Kapoor, R.; Rajashekara, H.; Prakash, G.; Sharma, V.; Sharma, T.R. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 2017, 36, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Quilis, J.; LopezGarcia, B.; Meynard, D.; Guiderdoni, E.; San Segundo, B. Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant Biotechnol. J. 2014, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Sripriya, R.; Parameswari, C.; Veluthambi, K. Enhancement of sheath blight tolerance in transgenic rice by combined expression of tobacco osmotin (ap24) and rice chitinase (chi11) genes. In Vitro Cell. Dev. Biol. 2017, 53, 12–21. [Google Scholar] [CrossRef]
- Li, H.; Zhou, S.-Y.; Zhao, W.-S.; Su, S.-C.; Peng, Y.-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.; Tank, H.G.; Prasad, B.D.; Chattoo, B.B. Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res. 2009, 18, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Transgenic rice with inducible ethylene production exhibits broad spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 2013, 11, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Karmakar, S.; Chanda, P.K.; Sarkar, S.N.; Datta, S.K.; Datta, K. Tissue-specific expression of ArabidopsisNPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci. 2016, 250, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Richa, K.; Tiwari, I.M.; Devanna, B.N.; Botella, J.R.; Sharma, V.; Sharma, T.R. Novel chitinase gene LOC_Os11g47510 from indica rice Tetep provides enhanced resistance against sheath blight pathogen Rhizoctonia solani in rice. Front. Plant Sci. 2017, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Velazhahan, R.; Oliva, N.; Ona, I.; Mew, T.; Khush, G.S.; Muthukrishnan, S.; Datta, S.K. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor. Appl. Genet. 1999, 98, 1138–1145. [Google Scholar] [CrossRef]
- Mao, B.; Liu, X.; Hu, D.; Li, D. Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnorpathe oryzae and reveals impact on seed germination. World J. Microbiol. Biotechnol. 2014, 30, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Bundo, M.; Coca, M. Enhancing blast disease resistance by overexpression of the calcium dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 2016, 14, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Bundo, M.; Coca, M. Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 2017, 2963–2975. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, Y.-G.; Shi, Y.; Wu, L.; Xu, Y.-J.; Huang, F.; Guo, X.-Y.; Zhang, Y.; Fan, J.; Zhao, J.-Q. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, S.-L.; Li, J.-L.; Hu, X.-H.; Wang, H.; Cao, X.-L.; Xu, Y.-J.; Zhao, Z.-X.; Xiao, Z.-Y.; Yang, N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Peris Peris, C.; Sire, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, L.; Pan, X.; Hu, Z.; Ling, F.; Yan, Y.; Liu, Y.; Lin, Y. Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 2015, 87, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Hayashi, N.; Sasaya, T.; Mori, M. Overexpression of BSR1 confers broad-spectrum resistance against two bacterial diseases and two major fungal diseases in rice. Breed. Sci. 2016, 66, 396–406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xue, X.; Cao, Z.X.; Zhang, X.T.; Wang, Y.; Zhang, Y.F.; Chen, Z.X.; Pan, X.B.; Zuo, S.M. Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis. 2016, 100, 1634–1642. [Google Scholar] [CrossRef]
- Chen, X.J.; Chen, Y.; Zhang, L.N.; Xu, B.; Zhang, J.H.; Chen, Z.X.; Tong, Y.H.; Zuo, S.M.; Xu, J.Y. Overexpression of OsPGIP1 enhances rice resistance to sheath blight. Plant Dis. 2016, 100, 388–395. [Google Scholar] [CrossRef]
- Luan, Z.H.; Zhou, D.W. Screening of rice (Oryza sativa L.) OsPR1b-interacting factors and their roles in resisting bacterial blight. Genet. Mol. Res. 2015, 14, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Hong, W.; Wu, J.; Wang, Y.; Ji, S.; Zhu, S.; Wei, C.; Zhang, J.; Li, Y. A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. eLife 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, M.; Tang, M.; Dong, B.; Wu, D.; Zhang, Z.; Zhou, B. Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 2015, 237, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Han, Q.; Zi, Q.; Lv, S.; Qiu, D.; Zeng, H. Enhanced disease resistance and drought tolerance in transgenic rice plants overexpressing protein elicitors from Magnaporthe oryzae. PLoS ONE 2017, 12, e0175734. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 2017, 7, 41037. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, I.M.; Jesuraj, A.; Kamboj, R.; Devanna, B.N.; Botella, J.R.; Sharma, T.R. Host delivered RNAi, an efficient approach to increase rice resistance to sheath blight pathogen (Rhizoctonia solani). Sci. Rep. 2017, 7, 7521. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, T.; Maruthasalam, S.; Kalpana, K.; Poovannan, K.; Kumar, K.K.; Kokiladevi, E.; Sudhakar, D.; Samiyappan, R.; Balasubramanian, P. Stability of sheath blight resistance in transgenic ASD16 rice lines expressing a rice chi11 gene encoding chitinase. Biol. Plant 2016, 60, 749–756. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef] [PubMed]
- Sprague, S.J.; Balesdent, M.-H.; Brun, H.; Hayden, H.L.; Marcroft, S.J.; Pinochet, X.; Rouxel, T.; Howlett, B.J. Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia. Eur. J. Plant Pathol. 2006, 114, 33–40. [Google Scholar] [CrossRef]
- Kumari, M.; Devanna, B.N.; Singh, P.K.; Rajashekara, H.; Sharma, V.; Sharma, T.R. Stacking of blast resistance orthologue genes in susceptible indica rice line improves resistance against Magnaporthe oryzae. 3 Biotech 2018, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.A.; Bannenberg, G.; Castresana, C. Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Curr. Opin. Plant Biol. 2008, 11, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Jones, J.D.G. Hormone (dis) harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Delteil, A.; Zhang, J.; Lessard, P.; Morel, J.-B. Potential candidate genes for improving rice disease resistance. Rice 2010, 3, 56–71. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Wagh, S.G.; Alam, M.M.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Toriba, T.; Hirano, H.-Y.; Nishiguchi, M. Analysis of rice RNA-dependent RNA polymerase 6 (OsRDR6) gene in response to viral, bacterial and fungal pathogens. J. Gen. Plant Pathol. 2016, 82, 12–17. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, J.; Liu, Z.; Wang, Z.; Zhou, C.; Wang, H. Host-induced gene silencing of rice blast fungus Magnaporthe oryzae pathogenicity genes mediated by the brome mosaic virus. Genes 2017, 8, 241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, F.; Li, X.; Baller, J.A.; Qi, Y.; Starker, C.G.; Bogdanove, A.J.; Voytas, D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed]
- Wolt, J.D.; Wang, K.; Sashital, D.; Lawrence-Dill, C.J. Achieving plant CRISPR targeting that limits off-target effects. Plant Genome 2016, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wolt, J.D. Risk associated with off-target plant genome editing and methods for its limitation. Emerg. Top. Life Sci. 2017, 2, 231–240. [Google Scholar] [CrossRef]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Chen, J.; Zeng, L.; Goh, M.; Leung, H. Characterizing rice lesion mimic mutants and identifying a mutant with broad spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000, 13, 869–876. [Google Scholar] [CrossRef] [PubMed]

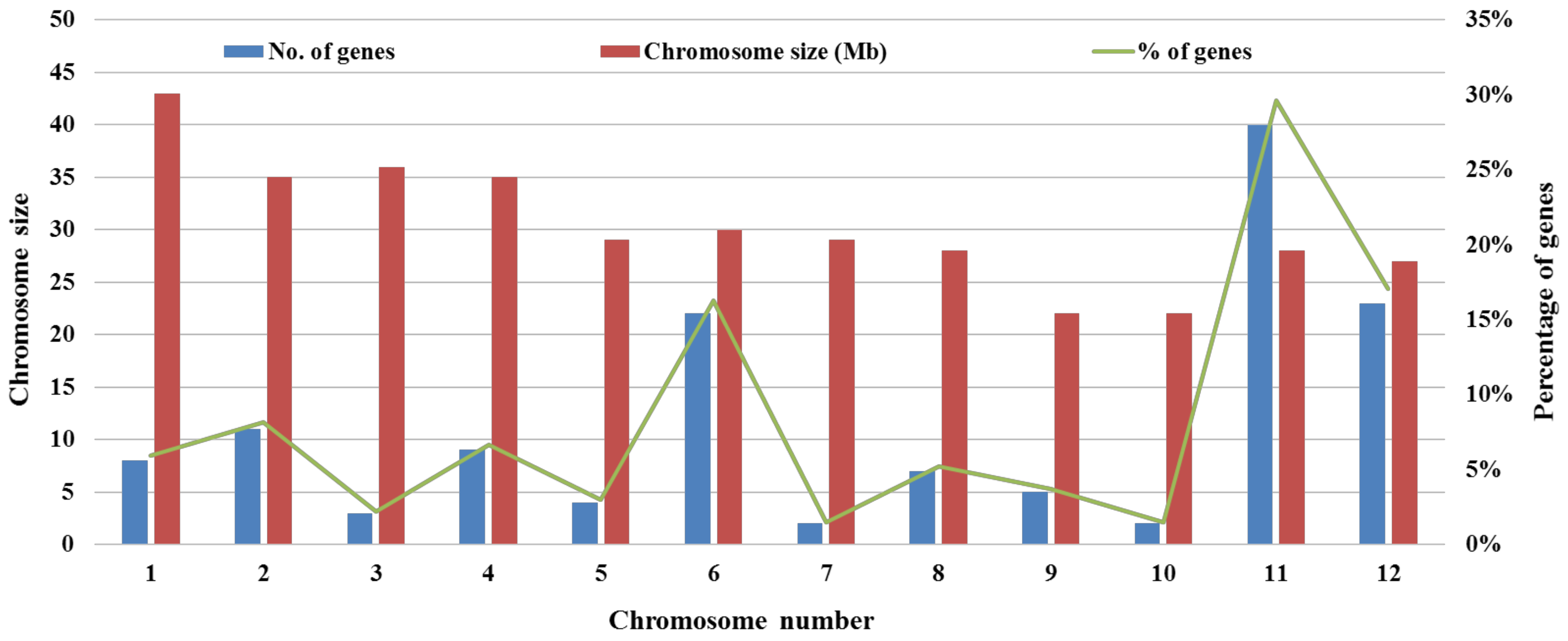

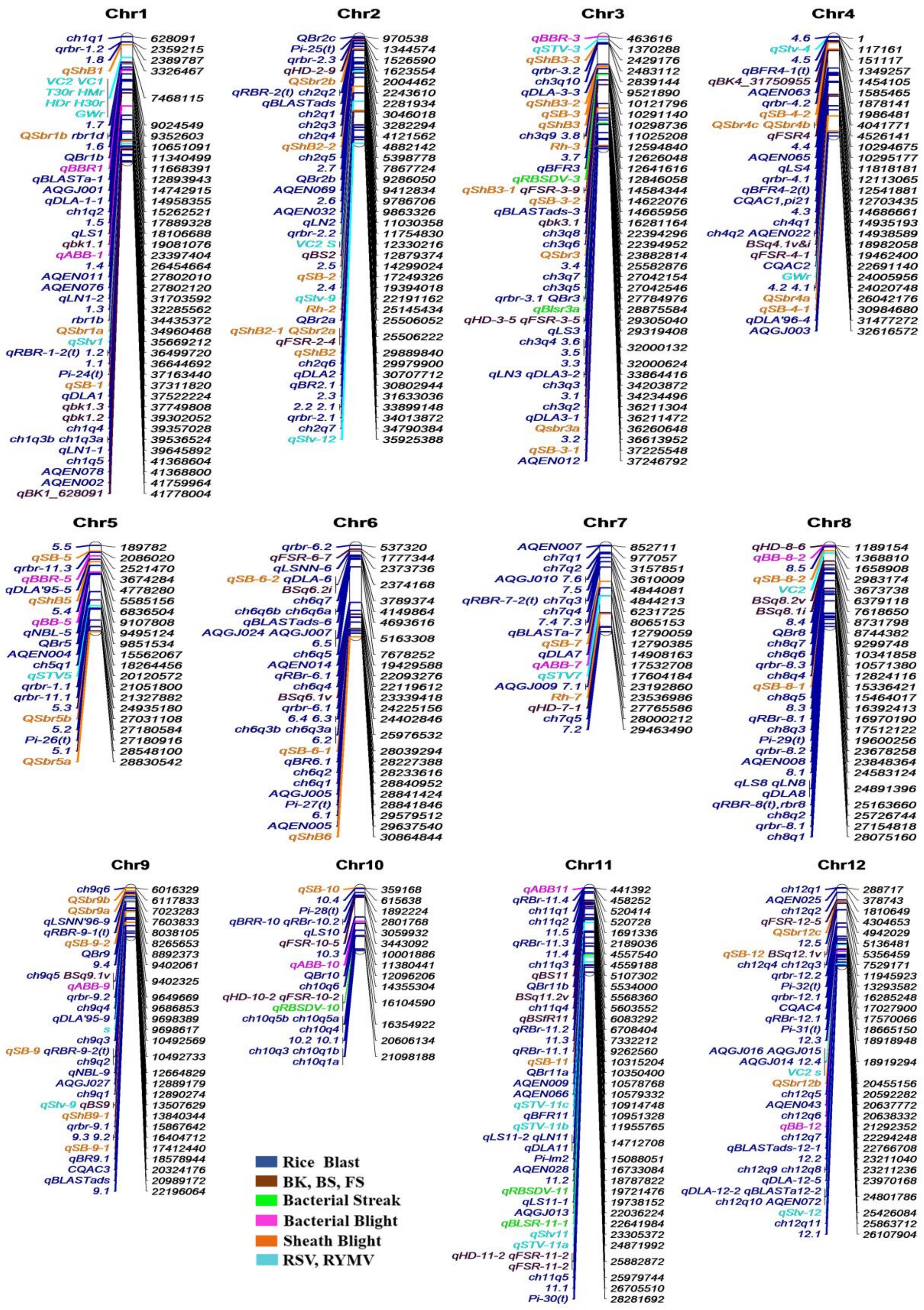
| Chromosome | Genes | References |
|---|---|---|
| 1 | (Xa29(t)), (Pit, Pitp(t), Pi37, Pi35(t), Pi24(t), Pi27, Pish) | [136,149,150,151,152,153,154,155] |
| 2 | (xa24(t)), (Pi-b, Pi25(t)) (Pid1(t), Pi-Da(t)), (Pi-y1(t), Piy2(t)), (Pig(t), Pi-tq5, Pi14(t), Pi16(t)) | [149,153,156,157,158,159,160,161,162,163] |
| 3 | (Xa11), (xa42), (Pi66(t)) | [164,165,166,167] |
| 4 | (Xa1), (Xa2), (Xa12), (Xa14), (xa31(t)), (Pi39(t), pi21, Pikur1, Pi(t)) | [58,168,169,170,171,172,173,174,175,176,177,178] |
| 5 | (xa5), (Pi26(t), Pi23, Pi10) | [153,179,180,181,182] |
| 6 | (Xa7), (Xa27), (xa33(t)), (Pi27(t), Pi-tq1, Pi8, Pi13(t), Pi22(t), Pigm(t)), (Piz-5, Piz-t), (Pi40(t), Pi59(t), Pi9, Pi2-1, Pid2), (Pi25(t), Pi26),(Piz), (Pi13), (Pi2-2, Pi50(t)) | [153,161,162,163,181,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201] |
| 7 | (xa8), (Pi17(t)) | [202,203] |
| 8 | (xa13), (Pi29(t), Pi33, Pizh, Pi36, pi55(t), PiGD-1(t)) | [153,204,205,206,207,208,209,210,211,212,213] |
| 9 | (Pi15, Pi2(t), Pi3(t), Pi5(t), Pi56(t)) | [214,215,216,217,218] |
| 10 | (Pi28(t), PiGD-2(t)) | [153,213] |
| 11 | (Xa3/Xa26), (Xa4), (Xa6, xa9), (Xa10), (Xa21), (Xa22), (Xa23, Xa30(t), Xa32(t), Xa35(t), Xa39, Xa40(t)), (Pi7(t)), (Pik, Pik-p), (Pi30(t), Pi60(t), Pilm2, Pikg),(Pik-h, Pik-s), (Pi-hk1), (Pi54), (Pi-1(t), (Pb1, Pise1, Pikur2, Pi38, Pif, Pi34, Pia, PiCO39(t), Pi44(t), Pi49, Pik-m, Pi18(t), Pi47), (Pi1) | [129,149,153,159,161,162,183,197,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254] |
| 12 | (xa25/Xa25(t)), (Pita-2), (Pi31(t), Pi32(t)), Pi61(t), Pi-tq6, (Ipi(t), IPi3(t)), (Pi21(t), Pi58(t), Pi51(t), Pi-GD-3(t), Pi48,Pi24(t), Pi-42(t), Pi62(t)), (Pi6(t), Pi4(t)), (Pi12(t), Pi19(t), Pita, Pi39(t), Pi20(t)) | [149,153,159,161,178,181,192,194,213,252,255,256,257,258,259,260,261,262,263,264] |
| Chromosome | QTLs | References |
|---|---|---|
| 1 | (1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8),(AQEN002, AQEN011, AQEN076, AQEN078), AQGJ001,( ch1q1, ch1q2, ch1q3a, ch1q3b, h1q4, ch1q5),(GWr, H30r, HDr, HMr, T30r, VC1,VC2), Pi-24(t), qABB-1, qBBR1,(qbk1.1, qbk1.2, qbk1.3), qBK1_628091, qBLASTa-1, qBlsr1,(QBr1a, QBr1b), qBSfR1,(qDLA1, qLN1-1, qLN1-2, qLS1), qDLA-1-1, qFSR1, qrbr-1.2,(qRBR-1-1(t), rbr1a, qRBR-1-2(t), rbr1b, qRBR-1-4(t), rbr1d), qSB-1,(QSbr1a, QSbr1b), qShB1, qStv1, | [129,153,196,199,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283] |
| 2 | (2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7),(AQEN032, AQEN069),(BSq2.1v&i, BSq2.2v&i),(ch2q1, ch2q2, ch2q3, ch2q4, ch2q5, ch2q6, ch2q7), Pi-25(t), qBLASTads, qBlsr2, qBR2.1,(QBr2a, QBr2b, QBr2c), qBS2,(qDLA2, qLN2), qFSR-2-4,,(qRBR-2(t), rbr2),(qrbr-2.2, qrbr-2.3), qSB-2,(QSbr2a, QSbr2b), qShB2,(qShB2-1, qShB2-2),(qStv-12,qStv-9), Rh-2,(S, VC2) | [129,153,196,199,266,267,272,273,274,278,280,281,282,284,285,286,287,288,289,290,291,292] |
| 3 | (3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, qFSR-3-5, qFSR-3-9), AQEN012,(ch3q10, ch3q2, ch3q3, ch3q4, ch3q5, ch3q6, ch3q7, ch3q8, ch3q9), qBBR-3, qBBR3-1, qBFR3, qbk3.1, qBLASTads-3,(qBlsr3a, qBlsr3b, qBlsr3c, qBlsr3d), QBr3,(qDLA3-1, DLA3-2, qLN3, qLS3), qDLA-3-3 ,(qrbr-3.1, qrbr-3.2), qRBSDV-3, qSB-3,(qSB-3-1, qSB-3-2), QSbr3, Qsbr3a, qShB3,(qShB3-1, qShB3-2, qShB3-3), qSTV-3, Qxa-3, Rh-3 | [129,196,199,266,269,270,272,273,274,276,278,280,281,282,288,290,292,293,294,295,296,297,298] |
| 4 | (4.1, 4.2, 4.3, 4.4, 4.5, 4.6), qFSR-4-1,( AQEN022, AQEN063, AQEN065), AQGJ003, BSq4.1v&i,(ch4q1, ch4q2),(CQAC1, pi21, CQAC2), GWr, qBB-4, qBBR-4,(qBFR4-1(t), qBFR4-2(t)), qBK4_31750955,(qBlsr4a, qBlsr4b), QBr4, qBSfR4, qDLA’96-4, qFSR4, qLS4,(qrbr-4.1, qrbr-4.2),(qSB-4-1, qSB-4-2),(QSbr4a, QSbr4b, QSbr4c), qStv-4 | [129,196,199,265,266,267,268,271,273,274,275,276,277,278,280,281,284,286,291,293,299,300] |
| 5 | (5.1, 5.2, 5.3, 5.4, 5.5), AQEN004, ch5q1, Pi-26(t), qBB-5, qBBR-5,(qBlsr5a, qBlsr5b), QBr5,(qDLA’95-5, qNBL-5),(qrbr-1.1, qrbr-11.1, qrbr-11.3, qrbr-2.1), qSB-5,(QSbr5a, QSbr5b), qShB5, qSTV5, xa5 | [129,153,199,266,268,273,274,276,278,281,282,290,293,298,301,302] |
| 6 | (6.1, 6.2, 6.3, 6.4, 6.5), qFSR-6-7,(AQEN005, AQEN014),(AQGJ005, AQGJ007, AQGJ024),(BSq6.1v, BSq6.2i),(ch6q1, ch6q2, ch6q3a, ch6q3b, ch6q4, ch6q5, ch6q6a, ch6q6b, ch6q7), Pi-27(t), qBBR-6,(qBLASTads-6, Pi-tq1), qBR6.1,(qDLA-6, qLSNN-6),(qrbr-6.1, qrbr-6.2),(qSB-6-1, qSB-6-2), qShB6, xa7 | [129,153,199,265,266,272,276,278,280,282,284,286,293,298,303] |
| 7 | (7.1, 7.2, 7.3, 7.4, 7.5, 7.6), AQEN007,(AQGJ009, AQGJ010),(ch7q1, ch7q2, ch7q3, ch7q4, ch7q5), qABB-7, qBBR7, qBLASTa-7, QBr7, qDLA7,(qRBR-7-2(t), rbr7b), qSB-7, qStv7, Rh-7 | [129,196,199,265,266,268,269,272,274,279,288,292,302] |
| 8 | (8.1, 8.2, 8.3, 8.4, 8.5), qFSR-8-3, AQEN008,(BSq8.1i, BSq8.2v),(ch8q1, ch8q2, ch8q3, ch8q4, ch8q5, ch8q6, ch8q7), Pi-29(t),qBB-8-2, QBr8,(qDLA8, qLN8, qLS8),(qRBR-8(t), rbr8),(qrbr-8.1, qrbr-8.2, qrbr-8.3),(qSB-8-1, qSB-8-2), Qsbr8a, VC2 | [129,153,196,199,266,267,268,274,278,279,280,284,286,296] |
| 9 | (9.1, 9.2, 9.3, 9.4), AQGJ027, BSq9.1v,(ch9q1, ch9q2, ch9q3, ch9q4, ch9q5, ch9q6), CQAC3, qABB-9, qBLASTads, QBr9, qBR9.1, qBS9,(qDLA’95-9, qLSNN’96-9, qNBL-9),(qrbr-9.1, qrbr-9.2),(qRBR-9-1(t), rbr9a, qRBR-9-2(t), rbr9b), qSB-9,(qSB-9-1, qSB-9-2),(QSbr9a,QSbr9b), qShB9-1, qStv-9, s | [199,213,265,266,267,268,272,274,276,278,279,280,281,282,284,285,289,291,299,301] |
| 10 | (10.1, 10.2, 10.3, 10.4),(qFSR-10-2, qFSR-10-5),(ch10q1a, ch10q1b, ch10q2, ch10q3, ch10q4, ch10q5a, ch10q5b, ch10q6), Pi-28(t), qABB-10, QBr10, qFSR10, qLS10, qRBSDV-10, qSB-10 | [153,196,199,266,268,277,280,281,286,295] |
| 11 | (11.1, 11.2, 11.3, 11.4, 11.5), qFSR-11-2,(AQEN009, AQEN028, AQEN066), AQGJ013,(BSq11.1v&i, BSq11.2v),(ch11q1, ch11q2, ch11q3, ch11q4, ch11q5), Pi-30(t), Pi-lm2, qABB11, qBFR11, qBlsr11, QBr11a, QBr11b, qBS11, qBSfR11,(qDLA11, qLN11, qLS11-1, qLS11-2), qRBSDV-11, qSB-11, qStv11,(qSTV-11a, qSTV-11b, qSTV-11c) | [129,153,196,199,265,266,268,274,277,281,284,286,294,297,304,305] |
| 12 | (12.1, 12.2, 12.3, 12.4, 12.5), qFSR-12-5,(AQEN025, AQEN043, AQEN072),(AQGJ014, AQGJ015, AQGJ016), BSq12.1v,(ch12q1,, ch12q2, ch12q3, ch12q4, ch12q5, ch12q6, ch12q7, ch12q8, ch12q9, ch12q10, ch12q11), CQAC4,(Pi-31(t), Pi-32(t)), qBB-12,(qBLASTa12-2, qBLASTads-12-1, Pi-tq6), (qDLA-12-2, qDLA-12-5), qFSR12,(qrbr-12.1, qrbr-12.2), qSB-12,(QSbr12a, QSbr12b, QSbr12c), qStv-12,(s, VC2) | [129,153,199,265,266,267,268,272,276,277,278,280,281,284,286,291,299] |
| Gene | Type | Method | Pathogen | Reference |
|---|---|---|---|---|
| Pib | NBS-LRR | OE | M. oryzae | [355] |
| Pi-ta | NBS-LRR | OE | M. oryzae | [262] |
| Pi9 | NBS-LRR | OE | M. oryzae | [193] |
| Pi-2 | NBS-LRR | OE | M. oryzae | [190] |
| Pi5 | CC-NB-LRR | OE | M. oryzae | [356] |
| Pi21 | Proline-rich protein | RNAi | M. oryzae | [357] |
| Pi36 | CC-NB-LRR | OE | M. oryzae | [263] |
| Pi37 | NBS-LRR | OE | M. oryzae | [151] |
| Pi54 | NBS-LRR | OE | M. oryzae | [358] |
| pi-d2 | B-lectin domain | OE | M. oryzae | [195] |
| Pik | CC-NBS-LRR | OE & RNAi | M. oryzae | [359] |
| Pikm | NBS-LRR | OE | M. oryzae | [360] |
| Pik-p | CC-NBS-LRR | OE & RNAi | M. oryzae | [361] |
| Pi54rh | NBS-LRR | OE | M. oryzae | [307] |
| Pi54of | CC-LRR | OE | M. oryzae | [311] |
| Pi54 and Pi54rh | NBS-LRR | OE | M. oryzae | [362] |
| mpi and pci | Proteinase inhibitors | OE | M. oryzae& Insect pests | [363] |
| chi11 and ap24 | Rice chitinase & Tobacco osmotin | OE | R. solani | [364] |
| OsBRR1 | LRR-RLK | OE | M. oryzae | [112] |
| OsWAK1 | Protein kinase | OE | M. oryzae | [365] |
| Dm-AMP1 | Antifungal plant defensin | OE | M. oryzae&R. solani | [366] |
| OsACS2 | Ethylene biosynthetic gene | OE | M. oryzae&R. solani | [367] |
| At. NPR1 | Defense gene | OE | R. solani | [368] |
| (Loc_Os11g47510) | Chitinase | OE | R. solani | [369] |
| PR-5 | Thaumatin-like protein | OE | R. solani | [370] |
| RCH10 and AGLU1 | Chitinase & Alfalfa β-1,3-glucanase gene | OE | M. oryzae&R. solani | [371] |
| OsCPK4 | Protein kinase | OE | M. oryzae&R. solani | [372] |
| OsCPK10 | Protein kinase | OE | M. oryzae&R. solani | [373] |
| miR160a and miR398b | miRNA | OE | M. oryzae | [374] |
| miR169a | miRNA | OE | M. oryzae | [375] |
| osa-miR7695 | miRNA | OE | M. oryzae | [376] |
| OsPGIP1 | Polygalacturonase inhibiting proteins | OE | R. solani | [377] |
| BSR1 | Receptor like kinase | OE | Xoo, M. oryzae, Burkholderiaglumae, Cochliobolusmiyabeans, RSV | [378] |
| OsOSM1 | Osmotin protein (PR) | OE | R. solani | [379] |
| OsPGIP1 | Polygalacturonase inhibiting proteins | OE | R. solani | [380] |
| OsPR1b | PR | OE | Xoo | [381] |
| OsSAMS1 | S-adenosyl-l-methionine synthetase | RNAi | RDV | [382] |
| OsDCL1 | Dicer-like | RNAi | M. oryzae | [383] |
| miR528 | miRNA | OE | RDV | [384] |
| miR168 | miRNA | RNAi | RDV, RSV | [385] |
| ALS | Acetolactate Synthase | CRISPR/Cas9 | Herbicide | [386] |
| OsSWEET13 | Sucrose transport | TALENs | Xoo | [387] |
| MoHrip1 and MoHrip2 | Effector protein | OE | M. oryzae | [388] |
| MoSM1 | Secreted protein | OE | M. oryzae&Xoo | [389] |
| RPMK1-1 and RPMK1-2 | Kinases | RNAi | R. solani | [390] |
| chi11 | Chitinase | OE | R. solani | [391] |
| OsSWEET11, OsSWEET14 | Sucrose transporter | CRISPR/Cas9 | Xoo | [392] |
| OsSWEET14 | Sucrose transporter | TALEN | Xoo | [393] |
| OsERF922 | ERF | CRISPR/Cas9 | M. oryzae | [394] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.K.; Nag, A.; Arya, P.; Kapoor, R.; Singh, A.; Jaswal, R.; Sharma, T.R. Prospects of Understanding the Molecular Biology of Disease Resistance in Rice. Int. J. Mol. Sci. 2018, 19, 1141. https://doi.org/10.3390/ijms19041141
Singh PK, Nag A, Arya P, Kapoor R, Singh A, Jaswal R, Sharma TR. Prospects of Understanding the Molecular Biology of Disease Resistance in Rice. International Journal of Molecular Sciences. 2018; 19(4):1141. https://doi.org/10.3390/ijms19041141
Chicago/Turabian StyleSingh, Pankaj Kumar, Akshay Nag, Preeti Arya, Ritu Kapoor, Akshay Singh, Rajdeep Jaswal, and Tilak Raj Sharma. 2018. "Prospects of Understanding the Molecular Biology of Disease Resistance in Rice" International Journal of Molecular Sciences 19, no. 4: 1141. https://doi.org/10.3390/ijms19041141
APA StyleSingh, P. K., Nag, A., Arya, P., Kapoor, R., Singh, A., Jaswal, R., & Sharma, T. R. (2018). Prospects of Understanding the Molecular Biology of Disease Resistance in Rice. International Journal of Molecular Sciences, 19(4), 1141. https://doi.org/10.3390/ijms19041141