Pathogens and Disease Play Havoc on the Host Epiproteome—The “First Line of Response” Role for Proteomic Changes Influenced by Disorder
Abstract
:1. Introduction
2. Review
2.1. Why Use the Broad Interpretation of the Term Epiproteome?
2.2. Marshalling a Diverse Set of Responding Proteins More or Less in Unison
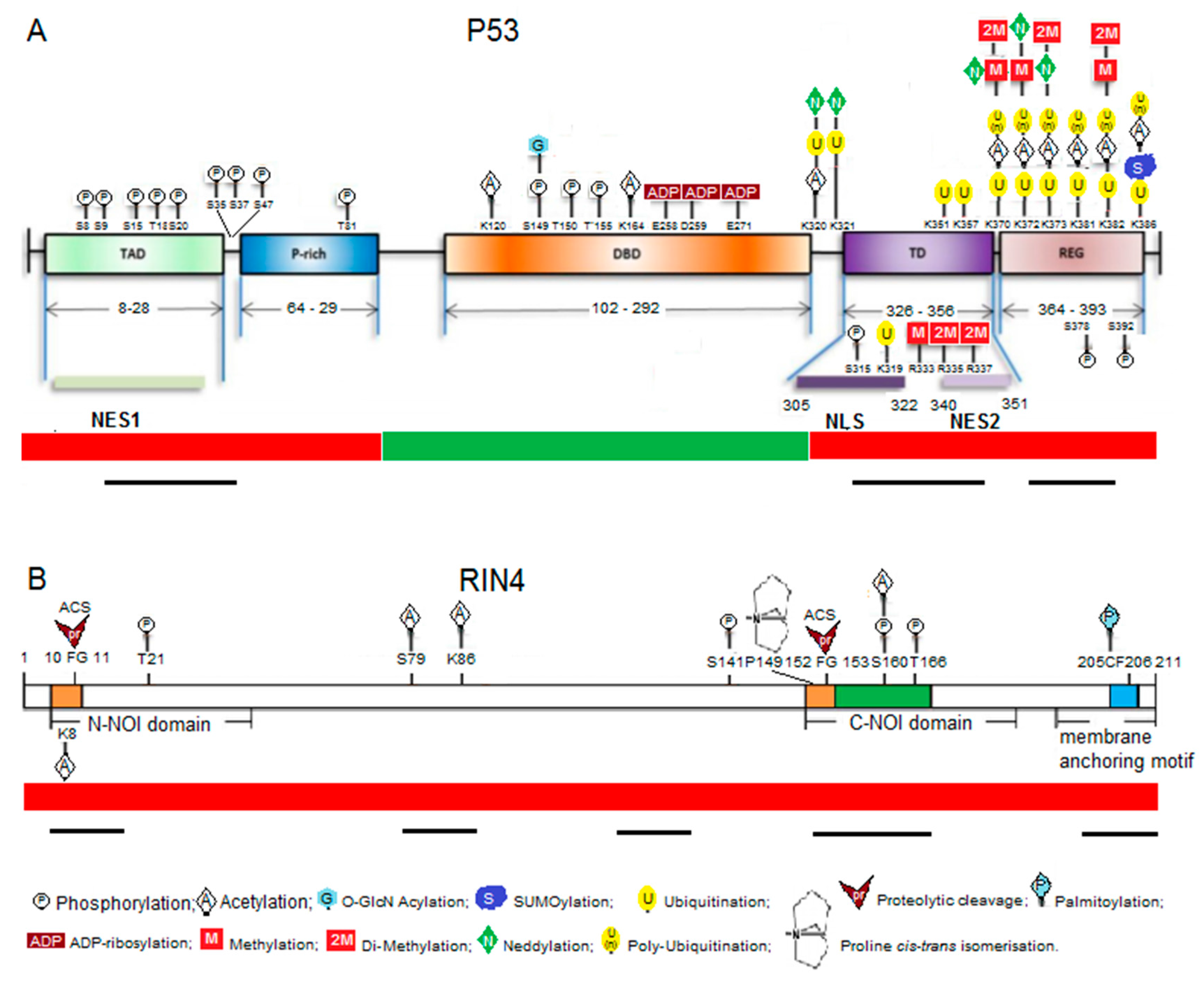
2.3. Animal and Plant Exemplars: p53 and RIN4
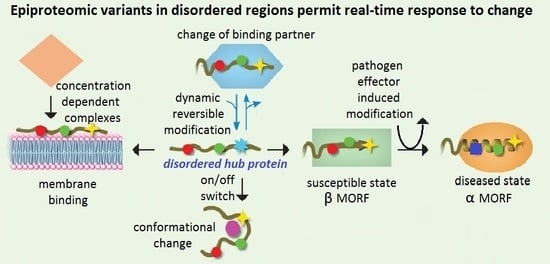
2.4. Characteristics Required for an Integrated Response to Simultaneous Challenges
2.5. How Can Multiple Diverse Signals Be Coordinated in Real-Time?
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Dai, B.; Rasmussen, T.P. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells 2007, 25, 1567–2574. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.M.; Jones, D.P. The redox proteome. J. Biol. Chem. 2013, 288, 26512–26520. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Alfano, J.R. Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011, 14, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Controlled chaos. Science 2008, 322, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.L.; Wagner, A. Mechanisms of mutational robustness in transcriptional regulation. Front. Genet. 2015, 6, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riddihough, G.; Zahn, L.M. What is epigenetics? Science 2010, 330, 611. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Wardenaar, R.; Colomé-Tatché, M.; Gilly, A.; Etcheverry, M.; Labadie, K.; Caillieux, E.; Hospital, F.; Aury, J.M.; Wincker, P.; et al. Mapping the epigenetic basis of complex traits. Science 2014, 343, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Díaz, E.; Jordà, M.; Peinado, M.A.; Rivero, A. Epigenetics of host–pathogen interactions: The road ahead and the road behind. PLoS Pathog. 2012, 8, e1003007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gokhale, N.S.; Horner, S.M. RNA modifications go viral. PLoS Pathog. 2017, 13, e1006188. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Huang, X.; Kelleher, N.L. Epiproteomics: Quantitative analysis of histone marks and codes by mass spectrometry. Curr. Opin. Chem. Biol. 2016, 33, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The cysteine proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Nishi, H.; Hashimoto, K.; Panchenko, A.R. Phosphorylation in protein-protein binding: Effect on stability and function. Structure 2011, 19, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, A.H. Native state proline isomerisation: An intrinsic molecular switch. Biochemistry 2003, 42, 9515–9524. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Covalent lipid modifications of proteins. Curr. Biol. 2013, 23, R431–R435. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, H.; Zou, Y.; Zhang, J.; Long, C.; Li, S.; Chen, S.; Zhou, J.M.; Shao, F. The phosphothreonine lyase activity of a bacterial type III effector family. Science 2007, 315, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Rosebrock, T.R.; Zeng, L.R.; Brady, J.J.; Abramovitch, R.B.; Xiao, F.M.; Martin, G.B. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 2007, 448, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Manning, A.J.; Wolfgeher, D.; Jelenska, J.; Cavanaugh Keri, A.; Xu, H.; Fernandez, S.M.; Michelmore, R.W.; Kron, S.J.; Greenberg, J.T. Acetylation of an NB-LRR plant immune-effector complex suppresses immunity. Cell Rep. 2015, 13, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Higdon, A.; Diers, A.R.; Oh, J.Y.; Landar, A.; Darley-Usmar, V.M. Cell signalling by reactive lipid species: New concepts and molecular mechanisms. Biochem. J. 2012, 442, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Maclaine, N.J.; Hupp, T.R. The regulation of p53 by phosphorylation: A model for how distinct signals integrate into the p53 pathway. Aging 2009, 1, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Zaika, A.I.; Wei, J.; Noto, J.M.; Peek, R.M. Microbial regulation of p53 tumor suppressor. PLoS Pathog. 2015, 11, e1005099. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008, 9 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Rikkerink, E.H.A.; Jones, W.T.; Uversky, V.N. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 2013, 25, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. P53 proteoforms and intrinsic disorder: An illustration of the protein structure–function continuum concept. Int. J. Mol. Sci. 2016, 17, 1874. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Zhu, W.-G. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 2012, 8, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.G.; Oldfield, C.J.; Meng, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Wilcken, R.; Andreeva, A. Tracing the evolution of the p53 tetramerization domain. Structure 2014, 22, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Brown, C.J.; Dunker, A.K.; Uversky, V.N. Intrinsically disordered regions of p53 family are highly diversified in evolution. Biochim. Biophys. Acta 2013, 1834, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Garner, E.; Guilliot, S.; Romero, P.; Albrecht, K.; Hart, J.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E. Protein disorder and the evolution of molecular recognition: Theory, predictions and observations. Pac. Symp. Biocomput. 1998, 3, 473–484. [Google Scholar]
- Brown, C.J.; Johnson, A.K.; Dunker, A.K.; Daughdrill, G.W. Evolution and disorder. Curr. Opin. Struct. Biol. 2011, 21, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Grahn, M.; Wright, A.P. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol. 2011, 12, R65. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Greenwood, D.R.; Templeton, M.D.; Libich, D.S.; McGhie, T.K.; Xue, B.; Yoon, M.; Cui, W.; Kirk, C.A.; Jones, W.T.; et al. The intrinsically disordered structural platform of the plant defence hub protein RIN4 provides insights into its mode of action in the host-pathogen interface and evolution of the NOI protein family. FEBS J. 2014, 281, 3955–3979. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.H.; El-Kasmi, F.; He, Y.; Loehr, A.; Dangl, J.L. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 2014, 16, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Mackey, D.; Holt, B.F.; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef]
- Liu, J.; Elmore, J.M.; Lin, Z.-J.D.; Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011, 9, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Chisholm, S.T.; Dahlbeck, D.; Staskawicz, B.J. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003, 49, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, X.; Chiang, Y.H.; Yadeta, K.A.; Ding, P.; Dong, L.; Zhao, Y.; Li, X.; Yu, Y.; Zhang, L.; et al. Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe 2014, 16, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez, J.D.; Weisbrod, C.R.; Zheng, C.; Eng, J.K.; Bruce, J.E. Protein interactions, post-translational modifications and topologies in human cells. Mol. Cell. Proteom. 2013, 12, 1451–1467. [Google Scholar] [CrossRef] [PubMed]
- Weisbrod, C.R.; Chavez, J.D.; Eng, J.K.; Yang, L.; Zheng, C.; Bruce, J.E. In vivo protein interaction network identified with novel real-time chemical cross-linked peptide identification strategy. J. Proteome Res. 2012, 12, 1569–1579. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Richmond, T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998, 8, 140–146. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, R.; Korolev, N.; Liu, C.F.; Nordenskiöld, L. Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail–H2A acidic patch interaction. J. Mol. Biol. 2017, 429, 2075–2092. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.J.; Buchanan, J.; Kirk, C.; Harvey, D.; Sun, X.; Spagnuolo, J.; Li, S.; Liu, T.; Woods, V.A.; Foster, T.; et al. Inter- and intra-molecular interactions of Arabidopsis thaliana DELLA protein RGL1. Biochem. J. 2011, 435, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Paterson, Y.; Englander, S.W.; Roder, H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science 1990, 249, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Vassall, K.A.; Jenkins, A.D.; Bamm, W.; Haruaz, G. Thermodynamic analysis of the disorder-to-α-helical transition of 18.5-kDa myelin basic protein reveals an equilibrium intermediate representing the most compact conformation. J. Mol. Biol. 2015, 427, 1977–1992. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein intrinsic disorder-base liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Protter, D.S.; Rosen, M.K.; Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Yosef, N.; Regev, A. Impulse control: Temporal dynamics in gene transcription. Cell 2011, 144, 886–896. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rikkerink, E.H.A. Pathogens and Disease Play Havoc on the Host Epiproteome—The “First Line of Response” Role for Proteomic Changes Influenced by Disorder. Int. J. Mol. Sci. 2018, 19, 772. https://doi.org/10.3390/ijms19030772
Rikkerink EHA. Pathogens and Disease Play Havoc on the Host Epiproteome—The “First Line of Response” Role for Proteomic Changes Influenced by Disorder. International Journal of Molecular Sciences. 2018; 19(3):772. https://doi.org/10.3390/ijms19030772
Chicago/Turabian StyleRikkerink, Erik H. A. 2018. "Pathogens and Disease Play Havoc on the Host Epiproteome—The “First Line of Response” Role for Proteomic Changes Influenced by Disorder" International Journal of Molecular Sciences 19, no. 3: 772. https://doi.org/10.3390/ijms19030772