A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response
Abstract
1. Introduction
2. Breast Cancer Treatment According to Histological Subtype
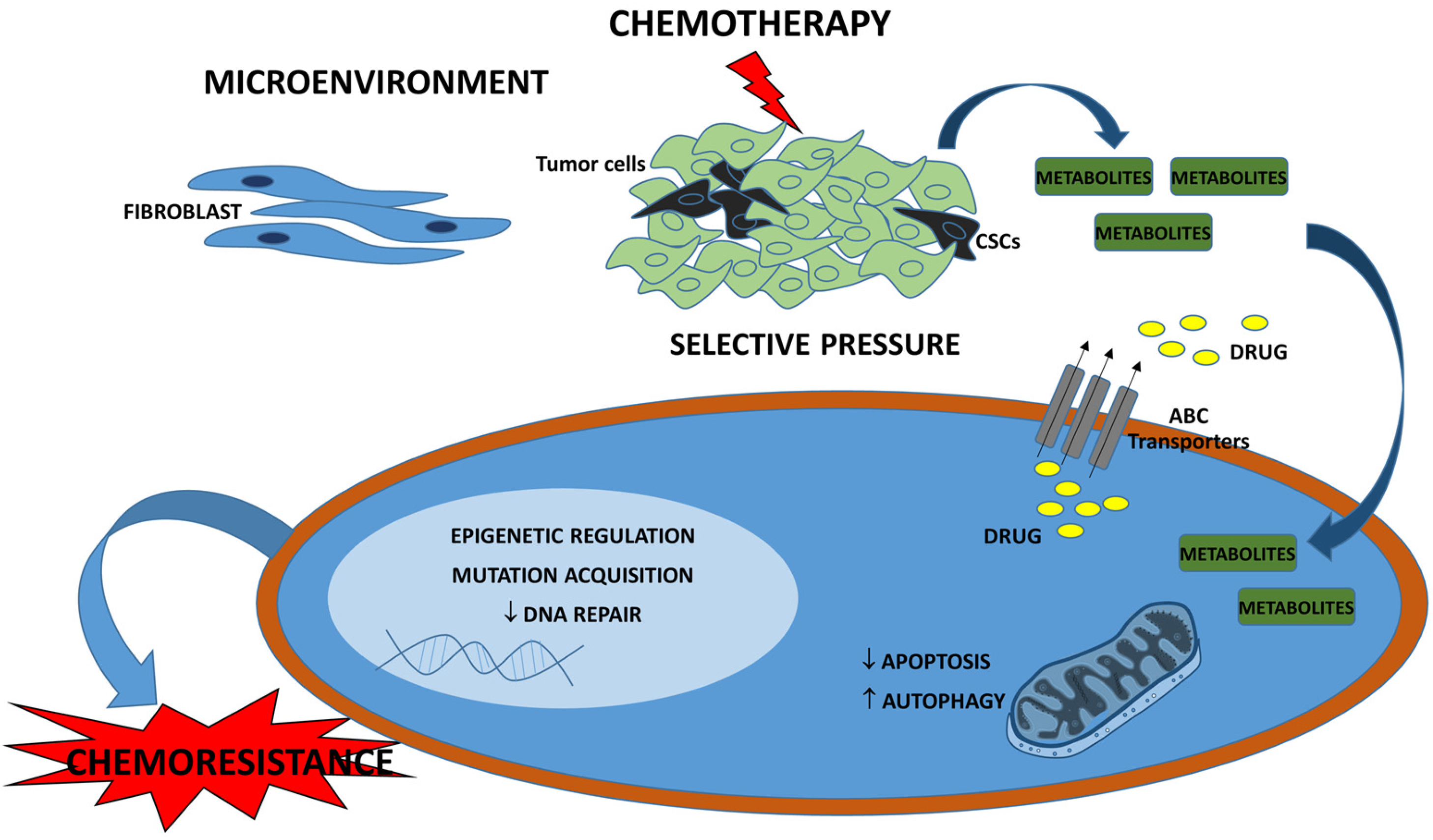
3. Drug Resistance in Breast Cancer
4. Current Metabolomic Approaches
5. Metabolic Profile of Breast Cancer
6. Metabolomic-Based Breast Cancer Chemoresistance
7. Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-tieulent, J.; Jemal, A. Global Cancer Statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Partridge, A.H.; Hughes, M.E.; Warner, E.T.; Ottesen, R.A.; Wong, Y.N.; Edge, S.B.; Theriault, R.L.; Blayney, D.W.; Niland, J.C.; Winer, E.P.; et al. Subtype-Dependent Relationship between Young Age at Diagnosis and Breast Cancer Survival. J. Clin. Oncol. 2016, 34, 3308–3314. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Bray, F.; Ferlay, J.; Lortet-Tieulent, J.; Anderson, B.O.; Jemal, A. International Variation in Female Breast Cancer Incidence and Mortality Rates. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Steding, C.E. Creating chemotherapeutic-resistant breast cancer cell lines: Advances and future perspectives. Future Oncol. 2016, 12, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Luqmani, Y.A. Mechanisms of Drug Resistance in Cancer Chemotherapy. Med. Princ. Pract. 2008, 14, 35–48. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society: Breast Cancer Facts & Figures (2015). Available online: http://www.cancer.org (accessed on 6 August 2017).
- Lee, A.; Djamgoz, M.B.A. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat. Rev. 2018, 62, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Yousefi, B. Breast tumor stroma: A driving force in the development of resistance to therapies. Chem. Biol. Drug Des. 2017, 89, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Yardley, D.A. Drug Resistance and the Role of Combination Chemotherapy in Improving Patient Outcomes. Int. J. Breast Cancer 2013, 2013, 137–414. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Zhang, J.; Yang, W. Activation of the LKB1-SIK1 signaling pathway inhibits the TGF-β-mediated epithelial-mesenchymal transition and apoptosis resistance of ovarian carcinoma cells. Mol. Med. Rep. 2017, 2837–2844. [Google Scholar] [CrossRef] [PubMed]
- Cornelison, R.; Llaneza, D.C.; Landen, C.N. Emerging therapeutics to overcome chemoresistance in epithelial ovarian cancer: A mini-review. Int. J. Mol. Sci. 2017, 18, 2171. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.R.; Jeddi, F.; Soozangar, N.; Somi, M.H.; Samadi, N. The role of Nrf2-Keap1 axis in colorectal cancer, progression, and chemoresistance. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S.; Kobayashi, H.; Graham, C.H. Intrinsic or acquired drug resistance and metastasis: Are they linked phenotypes? J. Cell. Biochem. 1994, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Sauter, E.R. Reliable Biomarkers to Identify New and Recurrent Cancer. Eur. J. Breast Health 2017, 13, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. Metabonomics and its role in drug development and disease diagnosis. Expert Rev. Mol. Diagn. 2004, 4, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bowers, J.; Liu, L.; Wei, S.; Gowda, G.A.N.; Hammoud, Z.; Raftery, D. Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Lanza, I.R.; Zhang, S.; Ward, L.E.; Karakelides, H.; Raftery, D.; Nair, K.S. Quantitative metabolomics by 1H-NMR and LC-MS/MS confirms altered metabolic pathways in diabetes. PLoS ONE 2010, 5, e10538. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Zhang, S.; Gu, H.; Asiago, V.; Shanaiah, N.; Raftery, D. Metabolomics-Based Methods for Early Disease Diagnostics: A Review. Expert Rev. Mol. Diagn. 2008, 8, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ahmed, S. Emerging field of metabolomics: Big promise for cancer biomarker identification and drug discovery. J. Pharm. Biomed. Anal. 2015, 107, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Shajahan-Haq, A.N.; Cheema, M.S.; Clarke, R. Application of metabolomics in drug resistant breast cancer research. Metabolites 2015, 5, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Bucher, E.; Hilvo, M.; Salek, R.; Orešič, M.; Griffin, J.; Brockmöller, S.; Klauschen, F.; Loibl, S.; Barupal, D.K.; et al. Metabolomics of human breast cancer: New approaches for tumor typing and biomarker discovery. Genome Med. 2012, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Esserman, L.J.; Berry, D.A.; DeMichele, A.; Carey, L.; Davis, S.E.; Buxton, M.; Hudis, C.; Gray, J.W.; Perou, C.; Yau, C.; et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: Results from the I-SPY 1 TRIAL—CALGB 150007/150012, ACRIN 6657. J. Clin. Oncol. 2012, 30, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Lamichhane, S.; Krohn, M.; Jernström, S.; Aure, M.R.; Lingjærde, O.C.; Schlichting, E.; Garred, Ø.; et al. Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.S.; Parker, J.S.; Mullins, M.; Cheung, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Kos, Z.; Dabbs, D.J. Biomarker assessment and molecular testing for prognostication in breast cancer. Histopathology 2016, 68, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Morrison, D.H.; Rahardja, D.; King, E.; Peng, Y.; Sarode, V.R. Tumour biomarker expression relative to age and molecular subtypes of invasive breast cancer. Br. J. Cancer 2012, 107, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, Q.; Yu, Q.; Zhang, J.; Liu, Z.; Wang, S.; Lv, S.; Niu, Y. A retrospective study of breast cancer subtypes: The risk of relapse and the relations with treatments. Breast Cancer Res. Treat. 2011, 130, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J. Tailoring therapies—Improving the management of early breast cancer: St GallenInternational Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Cancello, G.; Maisonneuve, P.; Rotmensz, N.; Viale, G.; Mastropasqua, M.G.; Pruneri, G.; Montagna, E.; Iorfida, M.; Mazza, M.; Balduzzi, A.; et al. Progesterone receptor loss identifies Luminal B breast cancer subgroups at higher risk of relapse. Ann. Oncol. 2013, 24, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Yamashita Kashima, Y.; Shu, S.; Yorozu, K.; Moriya, Y.; Harada, N. Mode of action of pertuzumab in combination with trastuzumab plus docetaxel therapy in a HER2 positive breast cancer xenograft model. Oncol. Lett. 2017, 14, 4197–4205. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.A.; Codony-servat, J.; Albanell, J.; Rojo, F.; Baselga, J. Trastuzumab (Herceptin), a Humanized Anti-HER2 Receptor Monoclonal Antibody, Inhibits Basal and Activated HER2 Ectodomain Cleavage in Breast Cancer Cells. Cancer Res. 2001, 61, 4744–4749. [Google Scholar] [PubMed]
- Dey, N.; Williams, C.; Leyland-Jones, B.; de, P. A critical role for HER3 in HER2-amplified and non-amplified breast cancers: Function of a kinase-dead RTK. Am. J. Transl. Res. 2015, 7, 733–750. [Google Scholar] [PubMed]
- Zhang, K.; Sun, J.; Liu, N.; Wen, D.; Chang, D.; Thomason, A.; Yoshinaga, S.K. Transformation of NIH 3T3 cells by HER3 or HER4 receptors requires the presence of HER1 or HER2. J. Biol. Chem. 1996, 271, 3884–3890. [Google Scholar] [CrossRef] [PubMed]
- Lipton, A.; Goodman, L.; Leitzel, K.; Cook, J.; Sperinde, J.; Haddad, M.; Köstler, W.J.; Huang, W.; Weidler, J.M.; Ali, S.; et al. HER3, p95HER2, and HER2 protein expression levels define multiple subtypes of HER2-positive metastatic breast cancer. Breast Cancer Res. Treat. 2013, 141, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, R.; Yan, H.; Zhao, P.; Wu, L.; Wang, H.; Li, T. Prognostic significance of HER3 in patients with malignant solid tumors. Oncotarget 2017, 8, 67140–67151. [Google Scholar] [CrossRef] [PubMed]
- Wuerstlein, R.; Harbeck, N. Neoadjuvant Therapy for HER2-positive Breast Cancer. Rev. Recent Clin. Trials 2017, 12, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Apuri, S. Neoadjuvant and Adjuvant Therapies for Breast Cancer. South Med. J. 2017, 110, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Tevaarwerk, A.J.; Kolesar, J.M. Lapatinib: A small-molecule inhibitor of epidermal growth factor receptor and human epidermal growth factor receptor-2 tyrosine kinases used in the treatment of breast cancer. Clin. Ther. 2009, 31, 2332–2348. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Zheng, X.; Pang, X.; Zhang, Z.; Jing, T.; Xu, W.; Zhang, Q. The potential use of lapatinib-loaded human serum albumin nanoparticles in the treatment of triple-negative breast cancer. Int. J. Pharm. 2015, 484, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M. Molecular stratification of triple-negative breast cancers. Oncologist 2011, 16 (Suppl. 1), 61–70. [Google Scholar] [CrossRef] [PubMed]
- Gerratana, L.; Fanotto, V.; Pelizzari, G.; Agostinetto, E.; Puglisi, F. Do platinum salts fit all triple negative breast cancers? Cancer Treat. Rev. 2016, 48, 34–41. [Google Scholar] [CrossRef] [PubMed]
- La Belle, A.; Khatib, J.; Schiemann, W.P.; Vinayak, S. Role of Platinum in Early-Stage Triple-Negative Breast Cancer. Curr. Treat. Options Oncol. 2017, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xu, S.; Yin, W.; Lin, Y.; Du, Y.; Jiang, Y. Weekly paclitaxel and cisplatin as neoadjuvant chemotherapy with locally advanced breast cancer : A prospective, single arm, phase II study. Oncotarget 2017, 8, 79305–79314. [Google Scholar] [CrossRef] [PubMed]
- Oldridge, D.A.; Wood, A.C.; Weichert-leahey, N.; Crimmins, I.; Winter, C.; Mcdaniel, L.D.; Diamond, M.; Hart, L.S.; Durbin, A.D.; Abraham, B.J.; et al. Associations between sociodemographic and clinicopathological factors, and breast cancer subtypes in a population-based study. Cancer Causes Control 2016, 528, 418–421. [Google Scholar] [CrossRef]
- Germano, S.; O’Driscoll, L. Breast cancer: Uderstanding sensitivity and resistance to chemotherapy and targeted therapies to aid in personalised medicine. Curr. Cancer Drug Targets 2009, 9, 398–418. [Google Scholar] [CrossRef] [PubMed]
- Jeselsohn, R.; Buchwalter, G.; Angelis, C.; Brown, M.; Schiff, M. ESR1 mutations as a mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Groenendijk, F.H.; Bernards, R. Drug resistance to targeted therapies: Déjà vu all over again. Mol. Oncol. 2014, 8, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Garduño, A.M.; Mendoza-Rodríguez, M.G.; Urrutia-Cabrera, D.; Domínguez-Robles, M.C.; Pérez-Yépez, E.A.; Ayala-Sumuano, J.T.; Meza, I. IL-1β induced methylation of the estrogen receptor ERα gene correlates with EMT and chemoresistance in breast cancer cells. Biochem. Biophys. Res. Commun. 2017, 490, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Hong, C.W.; Wang, Y.C.; Huang, W.J.; Yeh, Y.L.; Wang, B.J.; Wang, Y.J.; Chiu, H.W. A novel histone deacetylase inhibitor TMU-35435 enhances etoposide cytotoxicity through the proteasomal degradation of DNA-PKcs in triple-negative breast cancer. Cancer Lett. 2017, 400, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chun, J.; Pan, C.; Alesi, G.N.; Li, D.; Magliocca, K.R.; Kang, Y.; Chen, Z.G.; Shin, D.M.; Khuri, F.R.; et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene 2016, 8, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jung, J.Y.; Choi, S.; Lee, H.; Morales, L.D.; Koh, J.T.; Kim, S.H.; Choi, Y.D.; Choi, C.; Slaga, T.J.; et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy 2017, 13, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ohnuma, S.; Fukuda, M.; Chufan, E.E.; Kudoh, K.; Kanehara, K.; Sugisawa, N.; Ishida, M.; Naitoh, T.; Shibata, H.; et al. Synthetic analogs of curcumin modulate the function of multidrug resistance–linked ATP-binding cassette transporter ABCG2. Drug Metab. Dispos. 2017, 45, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Nickel, S.; Selo, M.A.; Fallack, J.; Clerkin, C.G.; Huwer, H.; Schneider-Daum, N.; Lehr, C.M.; Ehrhardt, C. Expression and Activity of Breast Cancer Resistance Protein (BCRP/ABCG2) in Human Distal Lung Epithelial Cells In Vitro. Pharm. Res. 2017, 34, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Xie, Y.; Baer, M.R.; Ross, D.D. Role of breast cancer resistance protein (BCRP/ABCG2) in cancer drug resistance. Biochem. Pharmacol. 2012, 83, 1084–1103. [Google Scholar] [CrossRef] [PubMed]
- Hölzel, M.; Bovier, A.; Tüting, T. Plasticity of tumour and immune cells: A source of heterogeneity and a cause for therapy resistance? Nat. Rev. Cancer 2013, 13, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Hemann, M.T. DNA damage-mediated induction of a chemoresistant niche. Cell 2010, 143, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Shree, T.; Olson, O.C.; Elie, B.T.; Kester, J.C.; Garfall, A.L.; Simpson, K.; Bell-Mcguinn, K.M.; Zabor, E.C.; Brogi, E.; Joyce, J.A. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011, 25, 2465–2479. [Google Scholar] [CrossRef] [PubMed]
- Chaiwun, B.; Sukhamwang, N.; Trakultivakorn, H.; Saha, B.; Young, L.; Tsao-Wei, D.; Naritoku, W.Y.; Groshen, S.; Taylor, C.R.; Imam, S.A. GSTPi-positive tumour microenvironment-associated fibroblasts are significantly associated with GSTPi-negative cancer cells in paired cases of primary invasive breast cancer and axillary lymph node metastases. Br. J. Cancer 2011, 105, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, H.M.; Koo, J.S. Differential expression of cancer-associated fibroblast-related proteins according to molecular subtype and stromal histology in breast cancer. Breast Cancer Res. Treat. 2015, 149, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Pontiggia, O.; Sampayo, R.; Raffo, D.; Motter, A.; Xu, R.; Bissell, M.J.; De Kier Joffé, E.B.; Simian, M. The tumor microenvironment modulates tamoxifen resistance in breast cancer: A role for soluble stromal factors and fibronectin through β1 integrin. Breast Cancer Res. Treat. 2012, 133, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Utsumi, J.; Toi, M.; Shimizu, K. Characteristic gene expression profiles of human fibroblasts and breast cancer cells in a newly developed bilateral coculture system. BioMed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Opyrchal, M.; Salisbury, J.L.; Iankov, I.; Goetz, M.P.; McCubrey, J.; Gambino, M.W.; Malatino, L.; Puccia, G.; Ingle, J.N.; Galanis, E.; et al. Inhibition of Cdk2 kinase activity selectively targets the CD44 +/CD24-/Low stem-like subpopulation and restores chemosensitivity of SUM149PT triple-negative breast cancer cells. Int. J. Oncol. 2014, 45, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Shima, H.; Yamada, A.; Ishikawa, T.; Endo, I. Are breast cancer stem cells the key to resolving clinical issues in breast cancer therapy? Gland Surg. 2017, 6, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M. Molecular stratification of Triple negative breast cancer. Oncologist 2010, 15, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Seung, J.K.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Mukherjee, S.; Khan, P.; Kajal, K.; Mazumdar, M.; Manna, A.; Mukherjee, S.; De, S.; Jana, D.; Sarkar, D.K.; et al. Aspirin suppresses the acquisition of chemoresistance in breast cancer by disrupting an NFB-IL6 signaling axis responsible for the generation of cancer stem cells. Cancer Res. 2016, 76, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Judes, G.; Rifa, K.; Daures, M.; Dubois, L.; Bignon, Y.J.; Penault-Llorca, F.; Bernard-Gallon, D. High-throughput Omics technologies: New tools for the study of triple-negative breast cancer. Cancer Lett. 2016, 382, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Denkert, C. Tissue-Based Metabolomics to Analyze the Breast Cancer Metabolome. Recent Results Cancer Res. 2016, 207, 157–175. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.L.; Walker, A.K.; Sloan, E.K.; Creek, D.J. Optimized method for untargeted metabolomics analysis of MDA-MB-231 breast cancer cells. Metabolites 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, N.R.; Sharma, U. Breast tissue metabolism by magnetic resonance spectroscopy. Metabolites 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Penkert, J.; Ripperger, T.; Schieck, M.; Schlegelberger, B.; Steinemann, D.; Illig, T. On metabolic reprogramming and tumor biology: A comprehensive survey of metabolism in breast cancer. Oncotarget 2016, 7, 67626–67649. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Rethinking the war on cancer. Lancet 2014, 383, 558–563. [Google Scholar] [CrossRef]
- Hirschey, M.D.; DeBerardinis, R.J.; Diehl, A.M.E.; Drew, J.E.; Frezza, C.; Green, M.F.; Jones, L.W.; Ko, Y.H.; Le, A.; Lea, M.A.; et al. Dysregulated metabolism contributes to oncogenesis. Semin. Cancer Biol. 2015, 35, S129–S150. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Ambs, S. Metabolic signatures of human breast cancer. Mol. Cell. Oncol. 2015, 2, e992217. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.D.; Vignoli, A.; Tenori, L.; Uy, G.L.; Van To, T.; Adebamowo, C.; Hossain, S.M.; Biganzoli, L.; Risi, E.; Love, R.R.; et al. Serum metabolomic profiles identify ER-positive early breast cancer patients at increased risk of disease recurrence in a multicenter population. Clin. Cancer Res. 2017, 23, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Samudio, I.; Fiegl, M.; Andreeff, M. Mitochondrial uncoupling and the Warburg effect: Molecular basis for the reprogramming of cancer cell metabolism. Cancer Res. 2009, 69. [Google Scholar] [CrossRef] [PubMed]
- Otto, W. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Marchiq, I.; Pouysségur, J. Hypoxia, cancer metabolism and the therapeutic benefit of targeting lactate/H+ symporters. J. Mol. Med. 2016, 94, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zhou, B.; Zhao, A.; Qiu, Y.; Zhao, X.; Garmire, L.; Shvetsov, Y.B.; Yu, H.; Yen, Y.; Jia, W. Lowered circulating aspartate is a metabolic feature of human breast cancer. Oncotarget 2015, 6, 33369–33381. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sharma, R.K.; Chagtoo, M.; Agarwal, G.; George, N.; Sinha, N.; Godbole, M.M. 1H NMR metabolomics reveals association of high expression of inositol 1, 4, 5 trisphosphate receptor and metabolites in breast cancer patients. PLoS ONE 2017, 12, e0169330. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.I.; Jamal, Q.; Iqbal, A.; Shaikh, F.; Somroo, S.; Musharraf, S.G. Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci. Rep. 2017, 7, 1715. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Collado, R.; Quiles, J.L.; Sol, J.; Ruiz-sanjuan, M.; Fernandez, M.; De, C.; Pamplona, R. A plasma metabolomic signature discloses human breast cancer. Oncotarget 2017, 8, 19522–19533. [Google Scholar] [CrossRef] [PubMed]
- Flote, V.G.; Vettukattil, R.; Bathen, T.F.; Egeland, T.; McTiernan, A.; Frydenberg, H.; Husøy, A.; Finstad, S.E.; Lømo, J.; Garred, Ø.; et al. Lipoprotein subfractions by nuclear magnetic resonance are associated with tumor characteristics in breast cancer. Lipids Health Dis. 2016, 15, 56. [Google Scholar] [CrossRef] [PubMed]
- Richard, V.; Conotte, R.; Mayne, D.; Colet, J.-M. Does the 1H-NMR plasma metabolome reflect the host-tumor interactions in human breast cancer? Oncotarget 2017, 8, 49915–49930. [Google Scholar] [CrossRef] [PubMed]
- Fuss, T.L.; Cheng, L.L. Evaluation of cancer metabolomics using ex vivo high resolution magic angle spinning (HRMAS) magnetic resonance spectroscopy (MRS). Metabolites 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Haukaas, T.H.; Euceda, L.R.; Giskeødegård, G.F.; Bathen, T.F. Metabolic portraits of breast cancer by HR MAS MR spectroscopy of intact tissue samples. Metabolites 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- El Ansari, R.; McIntyre, A.; Craze, M.L.; Ellis, I.O.; Rakha, E.A.; Green, A.R. Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology 2017. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.S.; Kwak, H.C.; Lee, K.S.; Kang, K.W.; Oh, S.J.; Lee, K.H.; Kim, H.M.; Ma, J.Y.; Kim, S.K. Sulfur amino acid metabolism in doxorubicin-resistant breast cancer cells. Toxicol. Appl. Pharmacol. 2011, 255, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Li, M.; Zha, W.; Zhao, Q.; Gu, R.; Liu, L.; Shi, J.; Zhou, J.; Zhou, F.; Wu, X.; et al. Metabolomic approach to evaluating adriamycin pharmacodynamics and resistance in breast cancer cells. Metabolomics 2013, 9, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Morvan, D. Functional metabolomics uncovers metabolic alterations associated to severe oxidative stress in MCF7 breast cancer cells exposed to ascididemin. Mar. Drugs 2013, 11, 3846–3860. [Google Scholar] [CrossRef] [PubMed]
- Bayet-Robert, M.; Morvan, D. Metabolomics Reveals Metabolic Targets and Biphasic Responses in Breast Cancer Cells Treated by Curcumin Alone and in Association with Docetaxel. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.A.; Winnike, J.H.; McRitchie, S.L.; Clark, R.F.; Pathmasiri, W.W.; Sumner, S.J. Metabolomics Analysis of Hormone-Responsive and Triple-Negative Breast Cancer Cell Responses to Paclitaxel Identify Key Metabolic Differences. J. Proteome Res. 2016, 15, 3225–3240. [Google Scholar] [CrossRef] [PubMed]
- Miolo, G.; Muraro, E.; Caruso, D.; Crivellari, D.; Ash, A.; Scalone, S.; Lombardi, D.; Rizzolio, F.; Giordano, A.; Corona, G. Phamacometabolomics study identifies circulating spermidine and tryptophan as potential biomarkers associated with the complete pathological response to trastuzumab-paclitaxel neoadjuvant therapy in HER-2 positive breast cancer. Oncotarget 2016, 7, 5657–5670. [Google Scholar] [CrossRef] [PubMed]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colnago, L.A. Impact of chemotherapy on metabolic reprogramming: Characterization of the metabolic profile of breast cancer MDA-MB-231 cells using 1H HR-MAS NMR spectroscopy. J. Pharm. Biomed. Anal. 2017, 146, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xiang, L.; Yuan, Z.; Li, S.; Yang, C.; Liu, H.; Jiang, Y.; Cai, Z. Metabolic profiling on the effect of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) in MCF-7 cells. Chemosphere 2018, 192, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Van Asten, J.J.A.; Vettukattil, R.; Buckle, T.; Rottenberg, S.; van Leeuwen, F.; Bathen, T.F.; Heerschap, A. Increased levels of choline metabolites are an early marker of docetaxel treatment response in BRCA1-mutated mouse mammary tumors: An assessment by ex vivo proton magnetic resonance spectroscopy. J. Transl. Med. 2015, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Euceda, L.R.; Haukaas, T.H.; Giskeødegård, G.F.; Vettukattil, R.; Engel, J.; Silwal-Pandit, L.; Lundgren, S.; Borgen, E.; Garred, Ø.; Postma, G.; et al. Evaluation of metabolomic changes during neoadjuvant chemotherapy combined with bevacizumab in breast cancer using MR spectroscopy. Metabolomics 2017, 13, 1–14. [Google Scholar] [CrossRef]
- Wei, S.; Liu, L.; Zhang, J.; Bowers, J.; Gowda, G.A.N.; Seeger, H.; Fehm, T.; Neubauer, H.J.; Vogel, U.; Clare, S.E.; et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 2013, 7, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Moestue, S.; Sitter, B.; Frost Bathen, T.; Tessem, M.-B.; Susann Gribbestad, I. HR MAS MR Spectroscopy in Metabolic Characterization of Cancer. Curr. Top. Med. Chem. 2011, 11, 2–26. [Google Scholar] [CrossRef] [PubMed]
| Biological Materials | Approach | Specific Treatment | Metabolic Pathways Identified | Reference |
|---|---|---|---|---|
| MCF-7 | Immunoblot analyses | Adriamycin | Sulfur amino acid metabolism | [98] |
| MCF-7 | GC-MS | Adriamycin | Increase in glycerol metabolism and decrease in glutathione biosynthesis. | [99] |
| MCF-7 | NMR | Ascididemin | Increase in citrate, gluconate and polyunsaturated fatty acids and decrease in glycerophospho-choline and ethanolamine. | [100] |
| MCF-7 MDA-MB-231 | NMR | curcumin +/− docetaxel (dose- and time-response) | Changes in glutathione metabolism, lipid metabolism, and glucose utilization—some biphasic changes depending on exposure. | [101] |
| BT474 MCF-7 MDA-MB-231 MDA-MB-468 | NMR | Paclitaxel | In luminal A cell lines: lactate and creatine decreased while certain choline metabolites and myo-inositol increased with paclitaxel. In TNBC cell lines: glutamine, glutamate, and glutathione increased, whereas lysine, proline, and valine decreased in the presence of drug. | [102] |
| Human serum samples | LC-MS | Trastuzumab-placlitaxel | Changes in spermidine and tryptophan. | [103] |
| MDA-MB-231 | HR-MAS NMR | Tamoxifen, cisplatin and doxorubicin | Changes in acetate, lactate and phosphocholine. | [104] |
| MCF-7 | UHPLC-MS | Polybrominated diphenyl ethers (PBDEs) | Change in the pentose phosphate pathway. | [105] |
| Tissue samples mouse model | HR-MAS | Docetaxel | In docetaxel-sensitive tumors: increase in choline metabolites. In tumors resistant to docetaxel: metabolites derived from choline did not increase during treatment. | [106] |
| Human breast tumor tissue | HR-MAS | 5-Fluorouracil, epirubicin, cyclophosphamide followed by taxane randomized to bevacizumab | Lower glucose and higher lactate was observed in patients exhibiting a good response compared to those with no response | [107] |
| Human serum samples | LC-MS NMR | Epirubucin and cyclophosphamide followed of doxorubicin in association to trastuzumab in HER2-positive cases | Concentrations significantly different threonine, isoleucine, glutamine and linolenic acid. | [108] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, M.R.; Santos, J.C.; Ribeiro, M.L.; Talarico, M.C.R.; Viana, L.R.; Derchain, S.F.M. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. https://doi.org/10.3390/ijms19020617
Cardoso MR, Santos JC, Ribeiro ML, Talarico MCR, Viana LR, Derchain SFM. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. International Journal of Molecular Sciences. 2018; 19(2):617. https://doi.org/10.3390/ijms19020617
Chicago/Turabian StyleCardoso, Marcella Regina, Juliana Carvalho Santos, Marcelo Lima Ribeiro, Maria Cecília Ramiro Talarico, Lais Rosa Viana, and Sophie Françoise Mauricette Derchain. 2018. "A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response" International Journal of Molecular Sciences 19, no. 2: 617. https://doi.org/10.3390/ijms19020617
APA StyleCardoso, M. R., Santos, J. C., Ribeiro, M. L., Talarico, M. C. R., Viana, L. R., & Derchain, S. F. M. (2018). A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. International Journal of Molecular Sciences, 19(2), 617. https://doi.org/10.3390/ijms19020617





