Functional Association between Regulatory RNAs and the Annexins
Abstract
:1. Introduction
2. Annexin Expression Changes by Intracellular Stress
3. MicroRNAs—Important Stress-Induced Regulators of Gene Expression
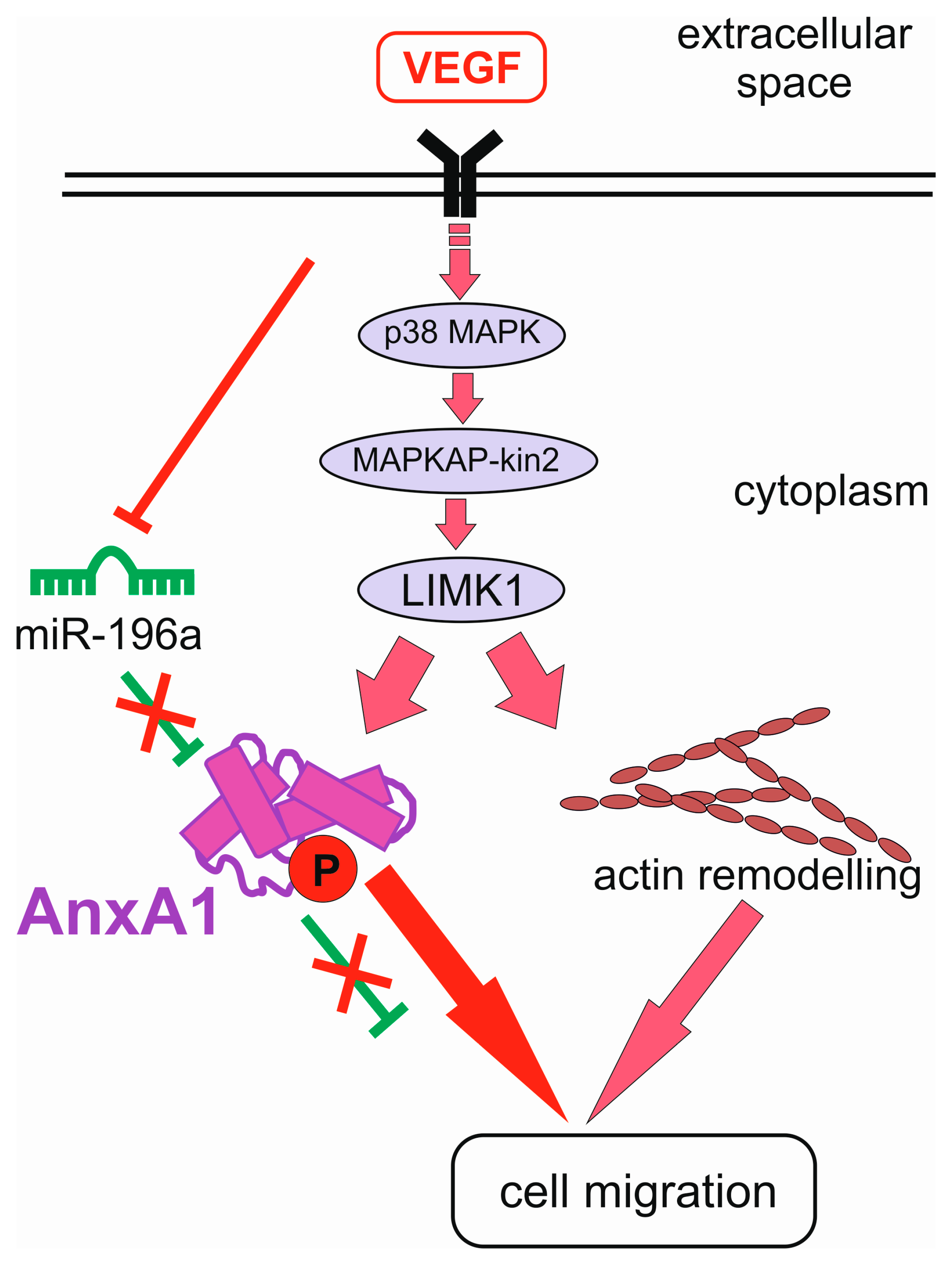
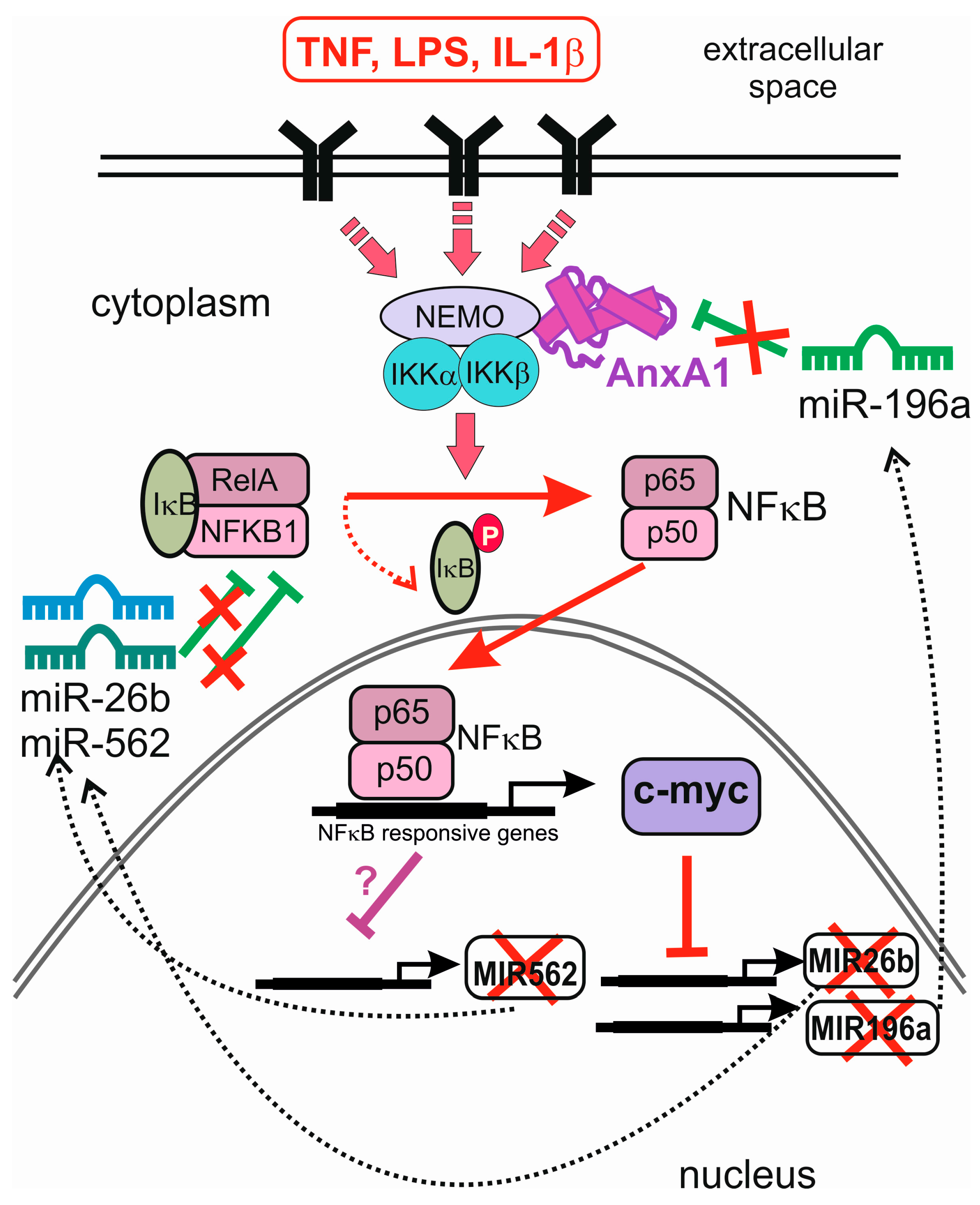
4. Feedback and Feed-Forward Loops between miRNAs and Annexins
5. RNA Binding by Annexins: Implications for Localisation and Function
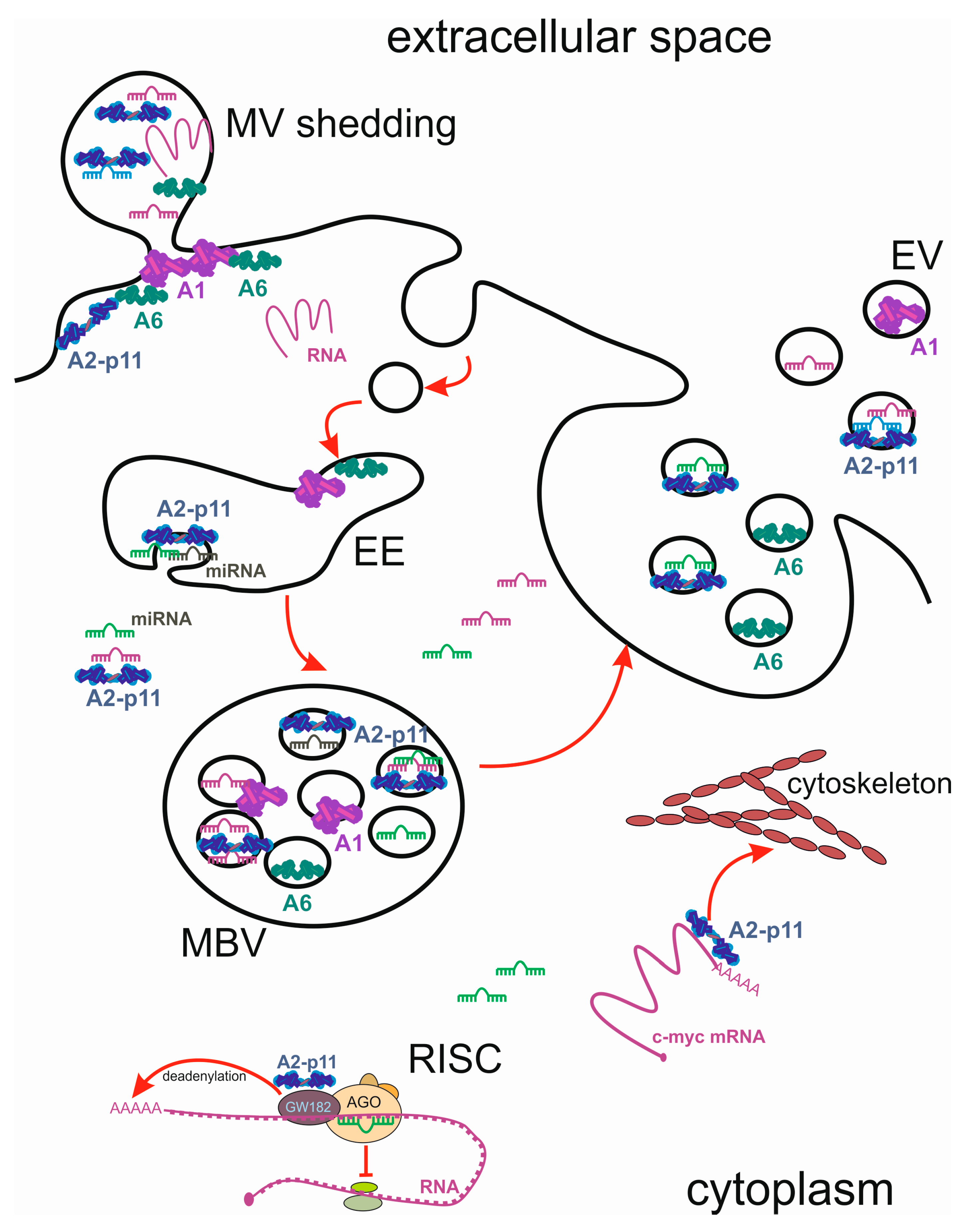
6. Intercellular Transport of miRNA and Annexins by Extracellular Vesicles (EVs)
6.1. Exosome Biogenesis, Release, and Uptake
6.2. miRNA Packaging and Transfer via EVs
7. Annexins as Stress-Induced Cargo of EVs
7.1. Annexins in Exosomes
7.2. Annexins in Shed Microvesicles
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Friedlander, M.R.; Lizano, E.; Houben, A.J.; Bezdan, D.; Banez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; Gonzalez, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Sharp, P.A. microRNAs: A safeguard against turmoil? Cell 2007, 130, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskaya, K.; Babiychuk, E.B.; Draeger, A. The annexins: Spatial and temporal coordination of signaling events during cellular stress. Cell. Mol. Life Sci. 2009, 66, 2623–2642. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskaya, K.; Babiychuk, E.B.; Hostettler, A.; Rescher, U.; Draeger, A. Annexins as intracellular calcium sensors. Cell Calcium 2007, 41, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskaya, K.; Tschumi, F.; Babiychuk, E.B.; Stroka, D.; Draeger, A. Annexins sense changes in intracellular pH during hypoxia. Biochem. J. 2008, 409, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Konopka-Postupolska, D.; Clark, G. Annexins as overlooked regulators of membrane trafficking in plant cells. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, E.J.; Yang, E.J.; Lee, J.E.; Park, A.R.; Song, W.H.; Park, O.K. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 2004, 16, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Gorecka, K.M.; Thouverey, C.; Buchet, R.; Pikula, S. Potential role of annexin AnnAt1 from Arabidopsis thaliana in pH-mediated cellular response to environmental stimuli. Plant Cell Physiol. 2007, 48, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Dihazi, H.; Asif, A.R.; Agarwal, N.K.; Doncheva, Y.; Muller, G.A. Proteomic analysis of cellular response to osmotic stress in thick ascending limb of Henle’s loop (TALH) cells. Mol. Cell. Proteom. 2005, 4, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Grossi, A.; Lametsch, R.; Karlsson, A.H.; Lawson, M.A. Mechanical stimuli on C2C12 myoblasts affect myoblast differentiation, focal adhesion kinase phosphorylation and galectin-1 expression: A proteomic approach. Cell Biol. Int. 2011, 35, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Monastyrskaya, K.; Babiychuk, E.B.; Draeger, A.; Burkhard, F.C. Down-regulation of annexin A1 in the urothelium decreases cell survival after bacterial toxin exposure. J. Urol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.J.; Moss, S.E. Annexins and disease. Biochem. Biophys. Res. Commun. 2004, 322, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Mussunoor, S.; Murray, G.I. The role of annexins in tumour development and progression. J. Pathol. 2008, 216, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Rhodes, D.R.; Ingold, C.; Chinnaiyan, A.M.; Rubin, M.A. Dysregulation of the annexin family protein family is associated with prostate cancer progression. Am. J. Pathol. 2003, 162, 255–261. [Google Scholar] [CrossRef]
- Srivastava, M.; Torosyan, Y.; Raffeld, M.; Eidelman, O.; Pollard, H.B.; Bubendorf, L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int. J. Cancer 2007, 121, 2628–2636. [Google Scholar] [CrossRef] [PubMed]
- Kollermann, J.; Schlomm, T.; Bang, H.; Schwall, G.P.; von Eichel-Streiber, C.; Simon, R.; Schostak, M.; Huland, H.; Berg, W.; Sauter, G.; et al. Expression and prognostic relevance of annexin A3 in prostate cancer. Eur. Urol. 2008, 54, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Langbein, S.; Lehmann, J.; Harder, A.; Steidler, A.; Michel, M.S.; Alken, P.; Badawi, J.K. Protein profiling of bladder cancer using the 2D-PAGE and SELDI-TOF-MS technique. Technol. Cancer Res. Treat. 2006, 5, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Shen, K.H.; Huang, L.C.; Huang, H.Y.; Wang, Y.H.; Wu, T.F. Annexin-I overexpression is associated with tumour progression and independently predicts inferior disease-specific and metastasis-free survival in urinary bladder urothelial carcinoma. Pathology 2010, 42, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Babiychuk, E.B.; Monastyrskaya, K.; Potez, S.; Draeger, A. Blebbing confers resistance against cell lysis. Cell Death Differ. 2011, 18, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.V.; Hogdall, C.K.; Jochumsen, K.M.; Hogdall, E.V.S. Annexin A2 and cancer: A systematic review. Int. J. Oncol. 2018, 52, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rothermund, C.A.; Ayala-Sanmartin, J.; Vishwanatha, J.K. Nuclear annexin II negatively regulates growth of LNCaP cells and substitution of ser 11 and 25 to glu prevents nucleo-cytoplasmic shuttling of annexin II. BMC Biochem. 2003, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, H.; Liu, S.; Guo, C.; Wang, J.; Greenaway, F.T.; Sun, M.Z. Role of annexin A6 in cancer. Oncol. Lett. 2015, 10, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Grewal, T.; Enrich, C. Molecular mechanisms involved in Ras inactivation: The annexin A6-p120GAP complex. Bioessays 2006, 28, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Spitzer, J.I.; Morozov, P.; Tuschl, T. miRNAs in human cancer. J. Pathol. 2011, 223, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schuttler, C.G.; Fehr, C.; Junemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wu, S.; Ding, J.; Lin, J.; Wei, L.; Gu, J.; He, X. MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res. 2010, 38, 7211–7218. [Google Scholar] [CrossRef] [PubMed]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. Rna 2008, 14, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Ma, Z.; Huo, Y.; Xiao, Z.; Li, Y.; Wang, Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. Rna 2011, 17, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Catto, J.W.; Alcaraz, A.; Bjartell, A.S.; De Vere White, R.; Evans, C.P.; Fussel, S.; Hamdy, F.C.; Kallioniemi, O.; Mengual, L.; Schlomm, T.; et al. MicroRNA in prostate, bladder, and kidney cancer: A systematic review. Eur. Urol. 2011, 59, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Gheinani, A.H.; Burkhard, F.C.; Monastyrskaya, K. Deciphering microRNA code in pain and inflammation: Lessons from bladder pain syndrome. Cell. Mol. Life Sci. 2013, 70, 3773–3789. [Google Scholar] [CrossRef] [PubMed]
- Solito, E.; de Coupade, C.; Canaider, S.; Goulding, N.J.; Perretti, M. Transfection of annexin 1 in monocytic cells produces a high degree of spontaneous and stimulated apoptosis associated with caspase-3 activation. Br. J. Pharmacol. 2001, 133, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Luthra, R.; Singh, R.R.; Luthra, M.G.; Li, Y.X.; Hannah, C.; Romans, A.M.; Barkoh, B.A.; Chen, S.S.; Ensor, J.; Maru, D.M.; et al. MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 2008, 27, 6667–6678. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.E.; Raulf, N.; Gaken, J.; Lawler, K.; Urbano, T.G.; Bullenkamp, J.; Gobeil, S.; Huot, J.; Odell, E.; Tavassoli, M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int. J. Cancer 2015, 137, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Pin, A.L.; Houle, F.; Fournier, P.; Guillonneau, M.; Paquet, E.R.; Simard, M.J.; Royal, I.; Huot, J. Annexin-1-mediated endothelial cell migration and angiogenesis are regulated by vascular endothelial growth factor (VEGF)-induced inhibition of miR-196a expression. J. Biol. Chem. 2012, 287, 30541–30551. [Google Scholar] [CrossRef] [PubMed]
- Grewal, T.; Koese, M.; Rentero, C.; Enrich, C. Annexin A6-regulator of the EGFR/Ras signalling pathway and cholesterol homeostasis. Int. J. Biochem. Cell Biol. 2010, 42, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Grewal, T.; Hoque, M.; Conway, J.R.W.; Reverter, M.; Wahba, M.; Beevi, S.S.; Timpson, P.; Enrich, C. Annexin A6-A multifunctional scaffold in cell motility. Cell Adh. Migr. 2017, 11, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Poeter, M.; Radke, S.; Koese, M.; Hessner, F.; Hegemann, A.; Musiol, A.; Gerke, V.; Grewal, T.; Rescher, U. Disruption of the annexin A1/S100A11 complex increases the migration and clonogenic growth by dysregulating epithelial growth factor (EGF) signaling. Biochim. Biophys. Acta 2013, 1833, 1700–1711. [Google Scholar] [CrossRef] [PubMed]
- Koese, M.; Rentero, C.; Kota, B.P.; Hoque, M.; Cairns, R.; Wood, P.; Vila de Muga, S.; Reverter, M.; Alvarez-Guaita, A.; Monastyrskaya, K.; et al. Annexin A6 is a scaffold for PKCα to promote EGFR inactivation. Oncogene 2013, 32, 2858–2872. [Google Scholar] [CrossRef] [PubMed]
- Bist, P.; Leow, S.C.; Phua, Q.H.; Shu, S.; Zhuang, Q.; Loh, W.T.; Nguyen, T.H.; Zhou, J.B.; Hooi, S.C.; Lim, L.H. Annexin-1 interacts with NEMO and RIP1 to constitutively activate IKK complex and NF-κB: Implication in breast cancer metastasis. Oncogene 2011, 30, 3174–3185. [Google Scholar] [CrossRef] [PubMed]
- Anbalagan, D.; Yap, G.; Yuan, Y.; Pandey, V.K.; Lau, W.H.; Arora, S.; Bist, P.; Wong, J.S.; Sethi, G.; Nissom, P.M.; et al. Annexin-A1 regulates microRNA-26b* and microRNA-562 to directly target NF-κB and angiogenesis in breast cancer cells. PLoS ONE 2014, 9, e114507. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Anbalagan, D.; Lee, L.H.; Samy, R.P.; Shanmugam, M.K.; Kumar, A.P.; Sethi, G.; Lobie, P.E.; Lim, L.H. ANXA1 inhibits miRNA-196a in a negative feedback loop through NF-kB and c-Myc to reduce breast cancer proliferation. Oncotarget 2016, 7, 27007–27020. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.S.; Cui, J.; Ning, J.N.; Wang, G.S.; Qian, G.S.; Lu, K.Z.; Yi, B. MiR-206 controls the phenotypic modulation of pulmonary arterial smooth muscle cells induced by serum from rats with hepatopulmonary syndrome by regulating the target gene, annexin A2. Cell. Physiol. Biochem. 2014, 34, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- Bronisz, A.; Wang, Y.; Nowicki, M.O.; Peruzzi, P.; Ansari, K.; Ogawa, D.; Balaj, L.; De Rienzo, G.; Mineo, M.; Nakano, I.; et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014, 74, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.K.; Chen, Q.; Chen, Y.B.; Gu, M.; Zheng, D.C.; Zhou, J.; Wang, Z. microRNA-155 promotes the proliferation of prostate cancer cells by targeting annexin 7. Mol. Med. Rep. 2015, 11, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.; Hirata, F. Lipocortin (Annexin) I heterotetramer binds to purine RNA and pyrimidine DNA. Biochem. Biophys. Res. Commun. 1999, 265, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.; Hirata, F. DNA chain unwinding and annealing reactions of lipocortin (annexin) I heterotetramer: Regulation by Ca(2+) and Mg(2+). Biochem. Biophys. Res. Commun. 2002, 291, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, A.P.; Darlix, J.L.; Spahr, P.F. A cellular protein phosphorylated by the avian sarcoma virus transforming gene product is associated with ribonucleoprotein particles. EMBO J. 1983, 2, 309–315. [Google Scholar] [PubMed]
- Filipenko, N.R.; MacLeod, T.J.; Yoon, C.S.; Waisman, D.M. Annexin A2 is a novel RNA-binding protein. J. Biol. Chem. 2004, 279, 8723–8731. [Google Scholar] [CrossRef] [PubMed]
- Mickleburgh, I.; Burtle, B.; Hollas, H.; Campbell, G.; Chrzanowska-Lightowlers, Z.; Vedeler, A.; Hesketh, J. Annexin A2 binds to the localization signal in the 3′ untranslated region of c-myc mRNA. FEBS J. 2005, 272, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Hollas, H.; Aukrust, I.; Grimmer, S.; Strand, E.; Flatmark, T.; Vedeler, A. Annexin A2 recognises a specific region in the 3′-UTR of its cognate messenger RNA. Biochim. Biophys. Acta 2006, 1763, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Aukrust, I.; Hollas, H.; Strand, E.; Evensen, L.; Trave, G.; Flatmark, T.; Vedeler, A. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J. Mol. Biol. 2007, 368, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Anji, A.; Kumari, M. A cis-acting region in the N-methyl-d-aspartate R1 3′-untranslated region interacts with the novel RNA-binding proteins β subunit of α glucosidase II and annexin A2—Effect of chronic ethanol exposure in vivo. Eur. J. Neurosci. 2011, 34, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Vedeler, A.; Hollas, H.; Grindheim, A.K.; Raddum, A.M. Multiple roles of annexin A2 in post-transcriptional regulation of gene expression. Curr. Protein Pept. Sci. 2012, 13, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Kwak, H.; Park, M.W.; Jeong, S. Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE 2011, 6, e24067. [Google Scholar] [CrossRef] [PubMed]
- Sharathchandra, A.; Lal, R.; Khan, D.; Das, S. Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol. 2012, 9, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Aukrust, I.; Rosenberg, L.A.; Ankerud, M.M.; Bertelsen, V.; Hollas, H.; Saraste, J.; Grindheim, A.K.; Vedeler, A. Post-translational modifications of Annexin A2 are linked to its association with perinuclear nonpolysomal mRNP complexes. FEBS Open Bio 2017, 7, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.E.; Huntzinger, E.; Izaurralde, E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 2013, 768, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.N.; Antonangeli, F. LncRNAs: New players in apoptosis control. Int. J. Cell Biol. 2014, 2014, 473857. [Google Scholar] [CrossRef] [PubMed]
- Stachurska, A.; Zorro, M.M.; van der Sijde, M.R.; Withoff, S. Small and long regulatory RNAs in the immune system and immune diseases. Front. Immunol. 2014, 5, 513. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.K.; Wolfson, B.; Zhou, Q. Cellular, physiological and pathological aspects of the long non-coding RNA NEAT1. Front. Biol. 2016, 11, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, Z. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017, 50. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Quiskamp, N.; Poeter, M.; Raabe, C.A.; Hohenester, U.M.; Konig, S.; Gerke, V.; Rescher, U. The tumor suppressor annexin A10 is a novel component of nuclear paraspeckles. Cell. Mol. Life Sci. 2014, 71, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Kim, P.J.; Jung, K.H.; Noh, J.H.; Eun, J.W.; Bae, H.J.; Xie, H.J.; Shan, J.M.; Ping, W.Y.; Park, W.S.; et al. Decreased expression of annexin A10 in gastric cancer and its overexpression in tumor cell growth suppression. Oncol. Rep. 2010, 24, 607–612. [Google Scholar] [PubMed]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, D.; Wu, W.; Wu, S.; Qian, J.; Hao, Y.; Yan, F.; Zhu, P.; Wu, J.; Huang, G.; et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017, 77, 6704–6716. [Google Scholar] [CrossRef] [PubMed]
- Solbak, S.M.O.; Abdurakhmanov, E.; Vedeler, A.; Danielson, U.H. Characterization of interactions between hepatitis C virus NS5B polymerase, annexin A2 and RNA—Effects on NS5B catalysis and allosteric inhibition. Virol. J. 2017, 14, 236. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Hennessy, E.J. Extracellular communication via microRNA: Lipid particles have a new message. J. Lipid Res. 2013, 54, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.; Di Bella, S.; Nigita, G.; Macca, V.; Lagana, A.; Giugno, R.; Pulvirenti, A.; Ferro, A. miRandola: Extracellular circulating microRNAs database. PLoS ONE 2012, 7, e47786. [Google Scholar] [CrossRef] [PubMed]
- Ajit, S.K. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012, 12, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Welsh, J.W.; Calin, G.A. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012, 195, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Grasedieck, S.; Sorrentino, A.; Langer, C.; Buske, C.; Dohner, H.; Mertens, D.; Kuchenbauer, F. Circulating microRNAs in hematological diseases: Principles, challenges and perspectives. Blood 2013. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Sita-Lumsden, A.; Dart, D.A.; Waxman, J.; Bevan, C.L. Circulating microRNAs as potential new biomarkers for prostate cancer. Br. J. Cancer 2013. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Hergenreider, E.; Heydt, S.; Treguer, K.; Boettger, T.; Horrevoets, A.J.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Boriachek, K.; Islam, M.N.; Moller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J.A. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. small 2017. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, Y.; Ono, K.; Horie, T.; Nishi, H.; Nagao, K.; Kinoshita, M.; Watanabe, S.; Baba, O.; Kojima, Y.; Shizuta, S.; et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 2011, 4, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Babiychuk, E.B.; Monastyrskaya, K.; Draeger, A. Fluorescent annexin A1 reveals dynamics of ceramide platforms in living cells. Traffic 2008, 9, 1757–1775. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Fonsato, V.; Collino, F.; Herrera, M.B.; Cavallari, C.; Deregibus, M.C.; Cisterna, B.; Bruno, S.; Romagnoli, R.; Salizzoni, M.; Tetta, C.; et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells 2012, 30, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Heusermann, W.; Hean, J.; Trojer, D.; Steib, E.; von Bueren, S.; Graff-Meyer, A.; Genoud, C.; Martin, K.; Pizzato, N.; Voshol, J.; et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell. Biol. 2016, 213, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Morgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signalling and lipid raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013. [Google Scholar] [CrossRef] [PubMed]
- Desrochers, L.M.; Antonyak, M.A.; Cerione, R.A. Extracellular vesicles: Satellites of information transfer in cancer and stem cell biology. Dev. Cell. 2016, 37, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Vickers, K.C. Intercellular transport of microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metast. Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Demory Beckler, M.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-dependent sorting of miRNA to exosomes. eLife 2015, 4, e07197. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; Katsuda, T.; Gailhouste, L.; Kosaka, N.; Ochiya, T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015, 589 (24 Pt B), 4071–4078. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Beheshti, E.; Choi, W.; Weiswald, L.B.; Kharmate, G.; Ghaffari, M.; Roshan-Moniri, M.; Hassona, M.D.; Chan, L.; Chin, M.Y.; Tai, I.T.; et al. Exosomes confer pro-survival signals to alter the phenotype of prostate cells in their surrounding environment. Oncotarget 2016, 7, 14639–14658. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.J.; Rescher, U.; Gerke, V.; Moss, S.E. Annexin-actin interactions. Traffic 2004, 5, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Faure, A.V.; Migne, C.; Devilliers, G.; Ayala-Sanmartin, J. Annexin 2 “secretion” accompanying exocytosis of chromaffin cells: Possible mechanisms of annexin release. Exp. Cell Res. 2002, 276, 79–89. [Google Scholar] [CrossRef] [PubMed]
- White, I.J.; Bailey, L.M.; Aghakhani, M.R.; Moss, S.E.; Futter, C.E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Leoni, G.; Neumann, P.A.; Kamaly, N.; Quiros, M.; Nishio, H.; Jones, H.R.; Sumagin, R.; Hilgarth, R.S.; Alam, A.; Fredman, G.; et al. Annexin A1-containing extracellular vesicles and polymeric nanoparticles promote epithelial wound repair. J. Clin. Investig. 2015, 125, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.; van Adrichem, N.P.; van Wijnen, M.; Wauben, M.H.; Stout, T.A.; Stoorvogel, W. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 2012, 86, 82. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Yang, X.; Hoyer, F.F.; Paul, K.; Heiermann, N.; Becher, M.U.; Abu Hussein, N.; Kebschull, M.; Bedorf, J.; Franklin, B.S.; et al. Endothelial microparticle uptake in target cells is annexin I/phosphatidylserine receptor dependent and prevents apoptosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- D’Acquisto, F.; Perretti, M.; Flower, R.J. Annexin-A1: A pivotal regulator of the innate and adaptive immune systems. Br. J. Pharmacol. 2008, 155, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.A.; Vago, J.P.; Teixeira, M.M.; Sousa, L.P. Annexin A1 and the Resolution of inflammation: modulation of neutrophil recruitment, apoptosis, and clearance. J. Immunol. Res. 2016, 2016, 8239258. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.H.; Chien, H.Y.; Shih, C.H.; Lai, S.L.; Li, I.T.; Hsu, S.C.; Kou, Y.R.; Hsu, H.C. Annexin A1 mediates the anti-inflammatory effects during the granulocytic differentiation process in all-trans retinoic acid-treated acute promyelocytic leukemic cells. J. Cell. Physiol. 2012, 227, 3661–3669. [Google Scholar] [CrossRef] [PubMed]
- Morel, E.; Parton, R.G.; Gruenberg, J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev. Cell. 2009, 16, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.D.; Fang, Y.T.; Cheng, Y.L.; Lin, C.F.; Hsu, L.J.; Wang, S.Y.; Anderson, R.; Chang, C.P.; Lin, Y.S. Exophagy of annexin A2 via RAB11, RAB8A and RAB27A in IFN-gamma-stimulated lung epithelial cells. Sci. Rep. 2017, 7, 5676. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Mathias, R.A.; Greening, D.W.; Gopal, S.K.; Ji, H.; Kapp, E.A.; Coleman, B.M.; Hill, A.F.; Kusebauch, U.; Hallows, J.L.; et al. Oncogenic H-RAS reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol. Cell. Proteom. 2013, 12, 2148–2159. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J.K. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Sakwe, A.M.; Koumangoye, R.; Guillory, B.; Ochieng, J. Annexin A6 contributes to the invasiveness of breast carcinoma cells by influencing the organization and localization of functional focal adhesions. Exp. Cell Res. 2011, 317, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Leca, J.; Martinez, S.; Lac, S.; Nigri, J.; Secq, V.; Rubis, M.; Bressy, C.; Serge, A.; Lavaut, M.N.; Dusetti, N.; et al. Cancer-associated fibroblast-derived annexin A6+ extracellular vesicles support pancreatic cancer aggressiveness. J. Clin. Investig. 2016, 126, 4140–4156. [Google Scholar] [CrossRef] [PubMed]
- Shiromizu, T.; Kume, H.; Ishida, M.; Adachi, J.; Kano, M.; Matsubara, H.; Tomonaga, T. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci. Rep. 2017, 7, 12782. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Yan, X.; Yao, X.; Zhang, Y.; Shan, Y.; Mao, N.; Yang, Y.; Pan, L. Secretion of annexin A3 from ovarian cancer cells and its association with platinum resistance in ovarian cancer patients. J. Cell. Mol. Med. 2012, 16, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Krohn, J.B.; Hutcheson, J.D.; Martinez-Martinez, E.; Aikawa, E. Extracellular vesicles in cardiovascular calcification: Expanding current paradigms. J. Physiol. 2016, 594, 2895–2903. [Google Scholar] [CrossRef] [PubMed]
- New, S.E.; Goettsch, C.; Aikawa, M.; Marchini, J.F.; Shibasaki, M.; Yabusaki, K.; Libby, P.; Shanahan, C.M.; Croce, K.; Aikawa, E. Macrophage-derived matrix vesicles: An alternative novel mechanism for microcalcification in atherosclerotic plaques. Circ. Res. 2013, 113, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Kapustin, A.N.; Shanahan, C.M. Emerging roles for vascular smooth muscle cell exosomes in calcification and coagulation. J. Physiol. 2016, 594, 2905–2914. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Rashdan, N.A.; Zhu, D.; Milne, E.M.; Ajuh, P.; Milne, G.; Helfrich, M.H.; Lim, K.; Prasad, S.; Lerman, D.A.; et al. End stage renal disease-induced hypercalcemia may promote aortic valve calcification via Annexin VI enrichment of valve interstitial cell derived-matrix vesicles. J. Cell. Physiol. 2017, 232, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Thery, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D'Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell. Sci. 2010, 123 Pt 10, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell. Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, S.S.; Mengistab, A.T.; Tauscher, A.N.; LaVail, J.; Basbaum, C. The microvesicle as a vehicle for EMMPRIN in tumor-stromal interactions. Oncogene 2004, 23, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Bianco, F.; Pravettoni, E.; Colombo, A.; Schenk, U.; Moller, T.; Matteoli, M.; Verderio, C. Astrocyte-derived ATP induces vesicle shedding and IL-1 β release from microglia. J. Immunol. 2005, 174, 7268–7277. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.L.; Francis, S.E.; Dower, S.K.; Crossman, D.C. Secretion of intracellular IL-1 receptor antagonist (type 1) is dependent on P2X7 receptor activation. J. Immunol. 2004, 173, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.L.; Lin, Y.C.; Mochitate, K.; Grinnell, F. Stress-relaxation of fibroblasts in collagen matrices triggers ectocytosis of plasma membrane vesicles containing actin, annexins II and VI, andβ 1 integrin receptors. J. Cell. Sci. 1993, 105 Pt 1, 167–177. [Google Scholar] [PubMed]
- Babiychuk, E.B.; Monastyrskaya, K.; Potez, S.; Draeger, A. Intracellular Ca(2+) operates a switch between repair and lysis of streptolysin O-perforated cells. Cell Death Differ. 2009, 16, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Potez, S.; Luginbuhl, M.; Monastyrskaya, K.; Hostettler, A.; Draeger, A.; Babiychuk, E.B. Tailored protection against plasmalemmal injury by annexins with different Ca2+ sensitivities. J. Biol. Chem. 2011, 286, 17982–17991. [Google Scholar] [CrossRef] [PubMed]
- Frislev, H.S.; Boye, T.L.; Nylandsted, J.; Otzen, D. Liprotides kill cancer cells by disrupting the plasma membrane. Sci. Rep. 2017, 7, 15129. [Google Scholar] [CrossRef] [PubMed]
- Mirzapoiazova, T.; Lennon, F.E.; Mambetsariev, B.; Allen, M.; Riehm, J.; Poroyko, V.A.; Singleton, P.A. Extracellular vesicles from caveolin-enriched microdomains regulate hyaluronan-mediated sustained vascular integrity. Int. J. Cell Biol. 2015, 2015, 481493. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monastyrskaya, K. Functional Association between Regulatory RNAs and the Annexins. Int. J. Mol. Sci. 2018, 19, 591. https://doi.org/10.3390/ijms19020591
Monastyrskaya K. Functional Association between Regulatory RNAs and the Annexins. International Journal of Molecular Sciences. 2018; 19(2):591. https://doi.org/10.3390/ijms19020591
Chicago/Turabian StyleMonastyrskaya, Katia. 2018. "Functional Association between Regulatory RNAs and the Annexins" International Journal of Molecular Sciences 19, no. 2: 591. https://doi.org/10.3390/ijms19020591
APA StyleMonastyrskaya, K. (2018). Functional Association between Regulatory RNAs and the Annexins. International Journal of Molecular Sciences, 19(2), 591. https://doi.org/10.3390/ijms19020591



