Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells
Abstract
1. Introduction
2. Results
2.1. The Kinetics of Focus Formation and Decay
2.2. Focus Area
2.3. Focus Intensity Dynamics
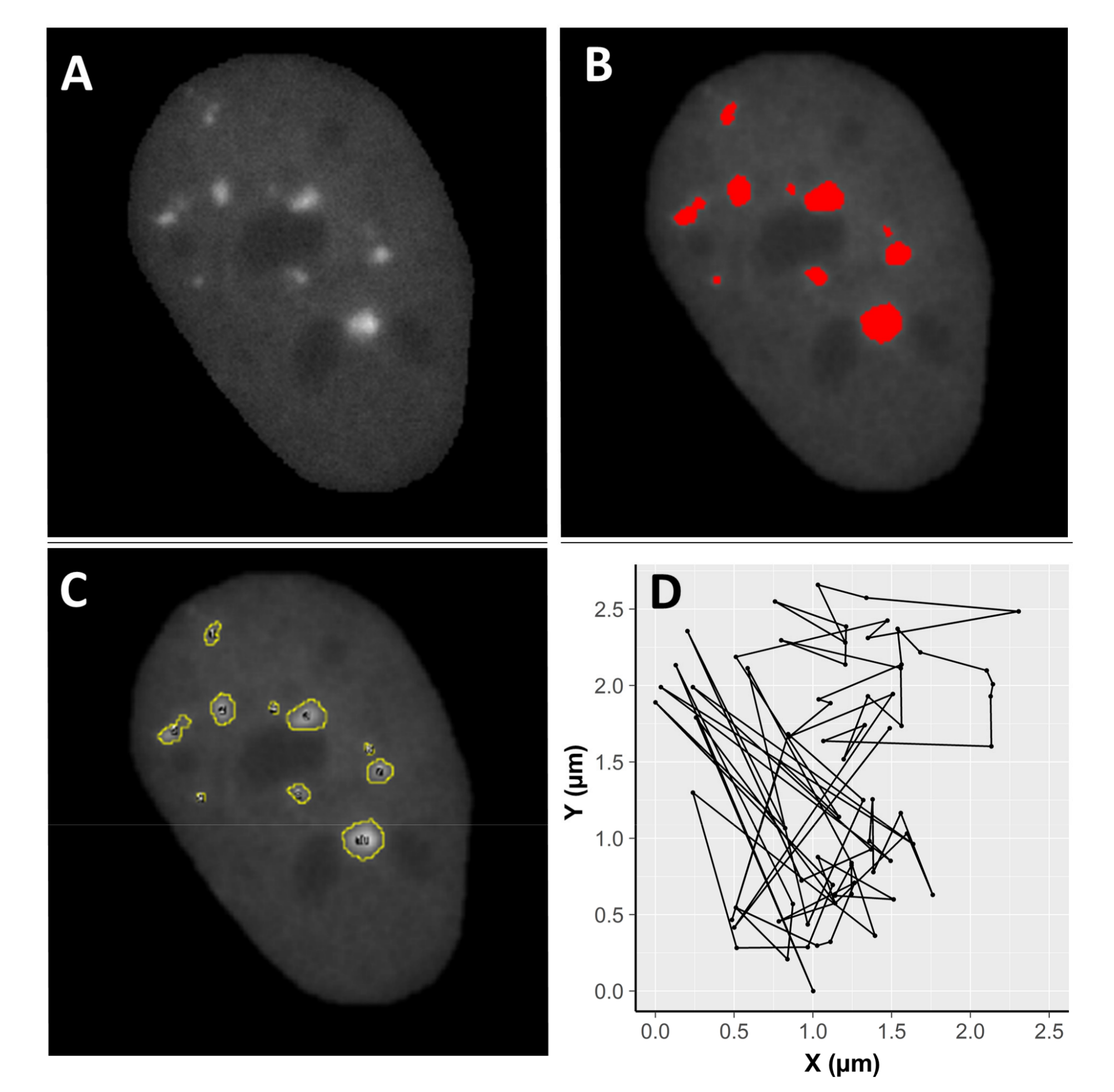
2.4. Focus Mobility
2.5. Focus Merging and Splitting
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell Line Authentication
4.2. Irradiation
4.3. Live Cell Imaging
4.4. Image Analysis
4.5. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Intrest
Abbreviations
| DSB | DNA double strand breaks |
| FPC | frequency per cell |
| GFP | green fluorescence protein |
| HRR | homologous recombination repair |
| LET | linear energy transfer |
| NHEJ | non-homologous end joining |
| p.r. | post radiationem |
| SD | standard deviation |
References
- Jeggo, P.A.; Lobrich, M. Radiation-induced DNA damage responses. Radiat. Prot. Dosim. 2006, 122, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., III; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Dantuma, N.P.; van Attikum, H. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016, 35, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Lukas, J.; Lukas, C.; Bartek, J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011, 13, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Pilch, D.R.; Redon, C.; Bonner, W.M. Histone H2AX in DNA damage and repair. Cancer Biol. Ther. 2003, 2, 233–235. [Google Scholar] [PubMed]
- Schultz, L.B.; Chehab, N.H.; Malikzay, A.; Halazonetis, T.D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000, 151, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Bekker-Jensen, S.; Lukas, C.; Melander, F.; Bartek, J.; Lukas, J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 2005, 170, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Lottersberger, F.; Karssemeijer, R.A.; Dimitrova, N.; de Lange, T. 53BP1 and the LINC Complex Promote Microtubule-Dependent DSB Mobility and DNA Repair. Cell 2015, 163, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Markova, E.; Schultz, N.; Belyaev, I.Y. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: Co-localization, relationship with DSB repair and clonogenic survival. Int. J. Radiat. Biol. 2007, 83, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Martin, O.A.; Ivashkevich, A.; Choo, S.; Woodbine, L.; Jeggo, P.A.; Martin, R.F.; Lobachevsky, P. Statistical analysis of kinetics, distribution and co-localisation of DNA repair foci in irradiated cells: Cell cycle effect and implications for prediction of radiosensitivity. DNA Repair 2013, 12, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Erixon, K.; Cedervall, B. Linear induction of DNA double-strand breakage with X-ray dose, as determined from DNA fragment size distribution. Radiat. Res. 1995, 142, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, D.T. Energy deposition stochastics and track structure: What about the target? Radiat. Prot. Dosim. 2006, 122, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; O’Neill, P.; Goodhead, D.T.; Terrissol, M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997, 71, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Mavragani, I.V.; Nikitaki, Z.; Souli, M.P.; Aziz, A.; Nowsheen, S.; Aziz, K.; Rogakou, E.; Georgakilas, A.G. Complex DNA Damage: A Route to Radiation-Induced Genomic Instability and Carcinogenesis. Cancers 2017, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Leatherbarrow, E.L.; Harper, J.V.; Cucinotta, F.A.; O’Neill, P. Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int. J. Radiat. Biol. 2006, 82, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Czub, J.; Wojcik, A. Gamma-H2AX foci in cells exposed to a mixed beam of X-rays and alpha particles. Genome Integr. 2012, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, F.; Campa, A.; Esposito, G.; Giardullo, P.; Belli, M.; Dini, V.; Meschini, S.; Simone, G.; Sorrentino, E.; Gerardi, S.; et al. Induction and Repair of DNA DSB as Revealed by H2AX Phosphorylation Foci in Human Fibroblasts Exposed to Low- and High-LET Radiation: Relationship with Early and Delayed Reproductive Cell Death. Radiat. Res. 2015, 183, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Costes, S.V.; Boissiere, A.; Ravani, S.; Romano, R.; Parvin, B.; Barcellos-Hoff, M.H. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat. Res. 2006, 165, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Sollazzo, A.; Brzozowska, B.; Cheng, L.; Lundholm, L.; Haghdoost, S.; Scherthan, H.; Wojcik, A. Alpha particles and X-rays interact in inducing DNA damage in U2OS cells. Radiat. Res. 2017, 188, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Voss, K.O.; Tobias, F.; Jakob, B.; Durante, M.; Taucher-Scholz, G. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res. 2013, 41, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Lorat, Y.; Timm, S.; Jakob, B.; Taucher-Scholz, G.; Rube, C.E. Clustered double-strand breaks in heterochromatin perturb DNA repair after high linear energy transfer irradiation. Radiother. Oncol. 2016, 121, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Plante, I.; Emfietzoglou, D.; Iliakis, G.; Georgakilas, A.G. Non-DSB clustered DNA lesions. Does theory colocalize with the experiment? Radiat. Phys. Chem. 2016, 128, 26–35. [Google Scholar] [CrossRef]
- Hendry, J.H.; Simon, S.L.; Wojcik, A.; Sohrabi, M.; Burkart, W.; Cardis, E.; Laurier, D.; Tirmarche, M.; Hayata, I. Human exposure to high natural background radiation: What can it teach us about radiation risks? J. Radiol. Prot. 2009, 29, A29–A42. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, D.T. Radiation protection aspects of the cosmic radiation exposure of aircraft crew. Radiat. Prot. Dosim. 2004, 109, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Takam, R.; Bezak, E.; Marcu, L.G.; Yeoh, E. Out-of-field neutron and leakage photon exposures and the associated risk of second cancers in high-energy photon radiotherapy: Current status. Radiat. Res. 2011, 176, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Yonai, S.; Furukawa, T.; Inaniwa, T. Measurement of neutron ambient dose equivalent in carbon-ion radiotherapy with an active scanned delivery system. Radiat. Prot. Dosim. 2014, 161, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Pachnerova-Brabcova, K.; Czub, J.; Braziewicz, J.; Nievaart, S.; Wojcik, A. Characterization of a setup for mixed beams exposure of cells to 241Am alpha particles and X-rays. Radiat. Prot. Dosim. 2012, 151, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Brehwens, K.; Haghdoost, S.; Nievaart, S.; Pachnerova-Brabcova, K.; Czub, J.; Braziewicz, J.; Wojcik, A. Micronuclei in human peripheral blood lymphocytes exposed to mixed beams of X-rays and alpha particles. Radiat. Environ. Biophys. 2012, 51, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Staaf, E.; Deperas-Kaminska, M.; Brehwens, K.; Haghdoost, S.; Czub, J.; Wojcik, A. Complex aberrations in lymphocytes exposed to mixed beams of 241Am alpha particles and X-rays. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013, 756, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Sollazzo, A.; Shakeri-Manesh, S.; Fotouhi, A.; Czub, J.; Haghdoost, S.; Wojcik, A. Interaction of low and high LET radiation in TK6 cells-mechanistic aspects and significance for radiation protection. J. Radiol. Prot. 2016, 36, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Coffman, V.C.; Wu, J.Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 2012, 37, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Dion, V.; Gasser, S.M. Chromatin movement in the maintenance of genome stability. Cell 2013, 152, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Friedland, W.; Dingfelder, M.; Kundrat, P.; Jacob, P. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2011, 711, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H.; Hieber, L.; Braselmann, H.; Meineke, V.; Zitzelsberger, H. Accumulation of DSBs in gamma-H2AX domains fuel chromosomal aberrations. Biochem. Biophys. Res. Commun. 2008, 371, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Neumaier, T.; Swenson, J.; Pham, C.; Polyzos, A.; Lo, A.T.; Yang, P.; Dyball, J.; Asaithamby, A.; Chen, D.J.; Bissell, M.J.; et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc. Natl. Acad. Sci. USA 2012, 109, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Schmid, T.E.; Dollinger, G.; Beisker, W.; Hable, V.; Greubel, C.; Auer, S.; Mittag, A.; Tarnok, A.; Friedl, A.A.; Molls, M.; et al. Differences in the kinetics of gamma-H2AX fluorescence decay after exposure to low and high LET radiation. Int. J. Radiat. Biol. 2010, 86, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Dion, V.; Kalck, V.; Horigome, C.; Towbin, B.D.; Gasser, S.M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 2012, 14, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Hubner, B.; Strickfaden, H.; Smeets, D.; Popken, J.; Sterr, M.; Markaki, Y.; Rippe, K.; Cremer, C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015, 589, 2931–2943. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Lukasova, E.; Stefancikova, L.; Baranova, E.; Falkova, I.; Jezkova, L.; Davidkova, M.; Bacikova, A.; Vachelova, J.; Michaelidesova, A.; et al. Heterochromatinization associated with cell differentiation as a model to study DNA double strand break induction and repair in the context of higher-order chromatin structure. Appl. Radiat. Isot. 2014, 83 Pt B, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Reindl, J.; Girst, S.; Walsh, D.W.; Greubel, C.; Schwarz, B.; Siebenwirth, C.; Drexler, G.A.; Friedl, A.A.; Dollinger, G. Chromatin organization revealed by nanostructure of irradiation induced gammaH2AX, 53BP1 and Rad51 foci. Sci. Rep. 2017, 7, 40616. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.R.K.; Simpson, P.J. FISH “painting” patterns resulting from complex exchanges. Mutat. Res. 1994, 312, 51–60. [Google Scholar] [CrossRef]
- Adhikary, A.; Becker, D.; Sevilla, M.D. Electron Spin Resonance of Radicals in Irradiated DNA. In Applications of EPR in Radiation Research; Lund, A., Shiotani, M., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 299–352. [Google Scholar]
- Sevilla, M.D.; Becker, D.; Kumar, A.; Adhikary, A. Gamma and Ion-Beam Irradiation of DNA: Free Radical Mechanisms, Electron Effects, and Radiation Chemical Track Structure. Radiat. Phys. Chem. 2016, 128, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Girst, S.; Hable, V.; Drexler, G.A.; Greubel, C.; Siebenwirth, C.; Haum, M.; Friedl, A.A.; Dollinger, G. Subdiffusion supports joining of correct ends during repair of DNA double-strand breaks. Sci. Rep. 2013, 3, 2511. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Durante, M.; Taucher-Scholz, G.; Jakob, B. ATM alters the otherwise robust chromatin mobility at sites of DNA double-strand breaks (DSBs) in human cells. PLoS ONE 2014, 9, e92640. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.A.; Stap, J.; Krawczyk, P.M.; van Oven, C.H.; Hoebe, R.A.; Essers, J.; Kanaar, R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 2004, 303, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.A.; Kanaar, R. Chromosomal organization: Mingling with the neighbors. PLoS Biol. 2006, 4, e155. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Elsner, M.; Kartberg, F.; Nilsson, T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004, 87, 3518–3524. [Google Scholar] [CrossRef] [PubMed]
- Brehwens, K.; Bajinskis, A.; Staaf, E.; Haghdoost, S.; Cederwall, B.; Wojcik, A. A new device to expose cells to changing dose rates of ionising radiation. Radiat. Prot. Dosim. 2012, 148, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Scherthan, H.; Adelfalk, C. Live cell imaging of meiotic chromosome dynamics in yeast. Methods Mol. Biol. 2011, 745, 537–548. [Google Scholar] [PubMed]
- Illner, D.; Scherthan, H. Ionizing irradiation-induced radical stress stalls live meiotic chromosome movements by altering the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 2013, 110, 16027–16032. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2016. Available online: https://www.R-project.org (accessed on 2 May 2017).
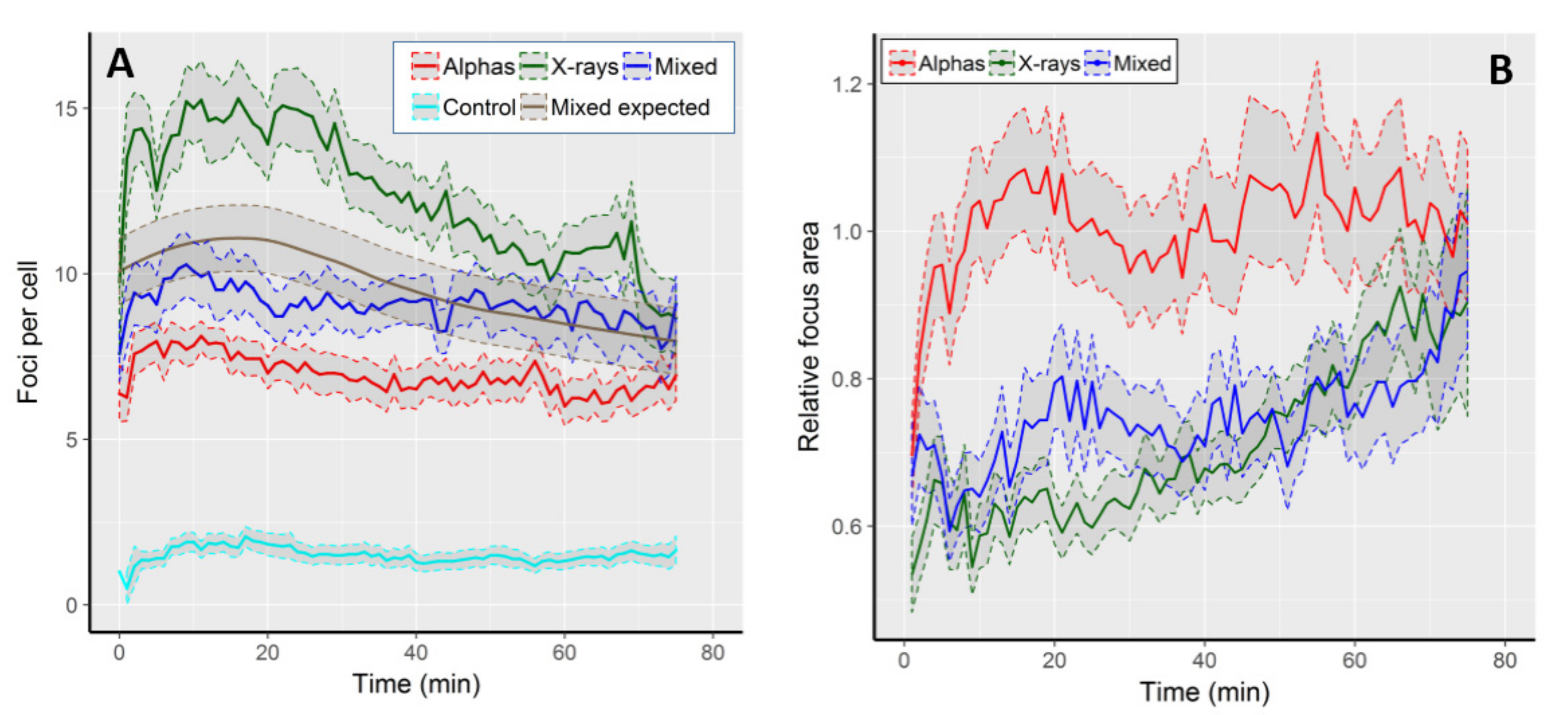
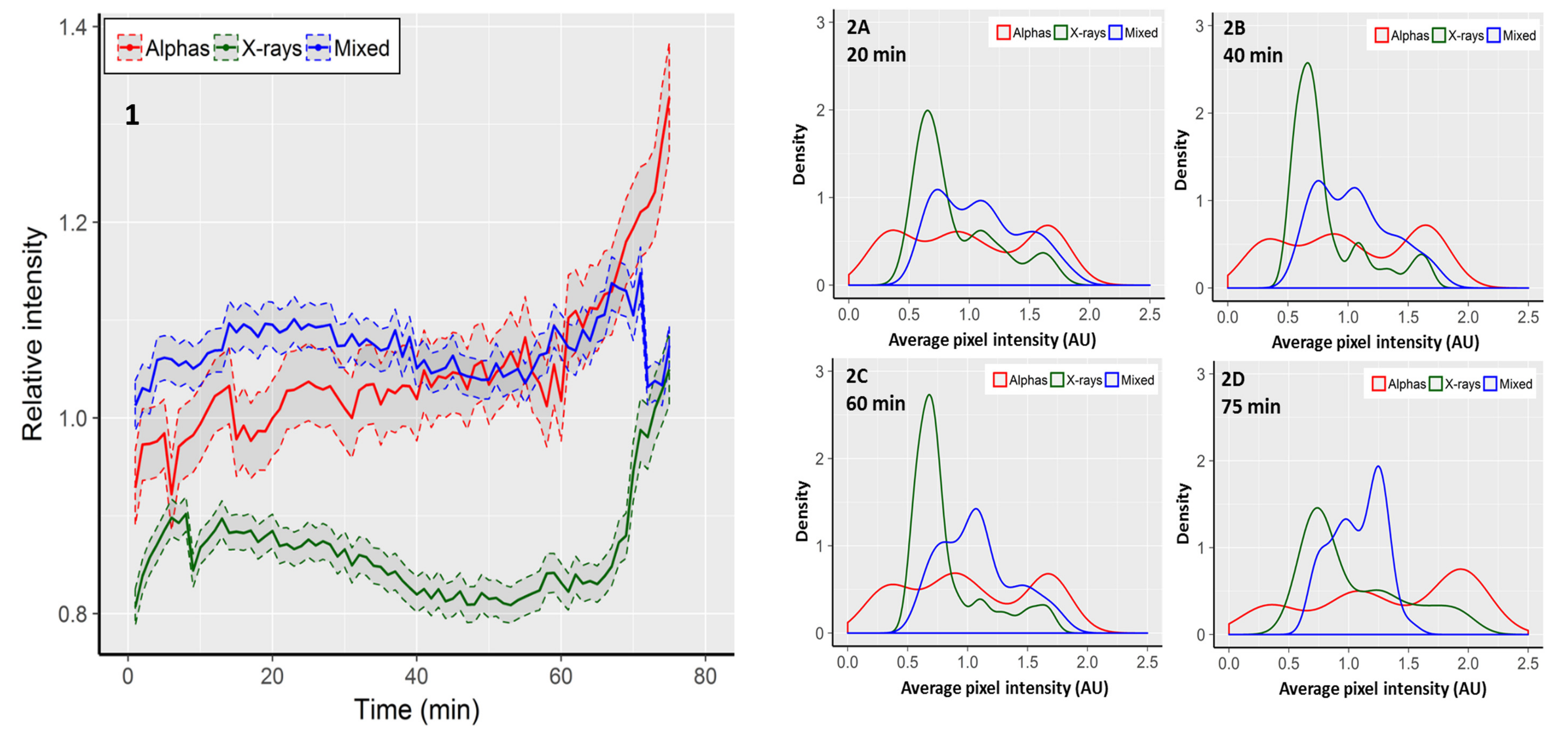
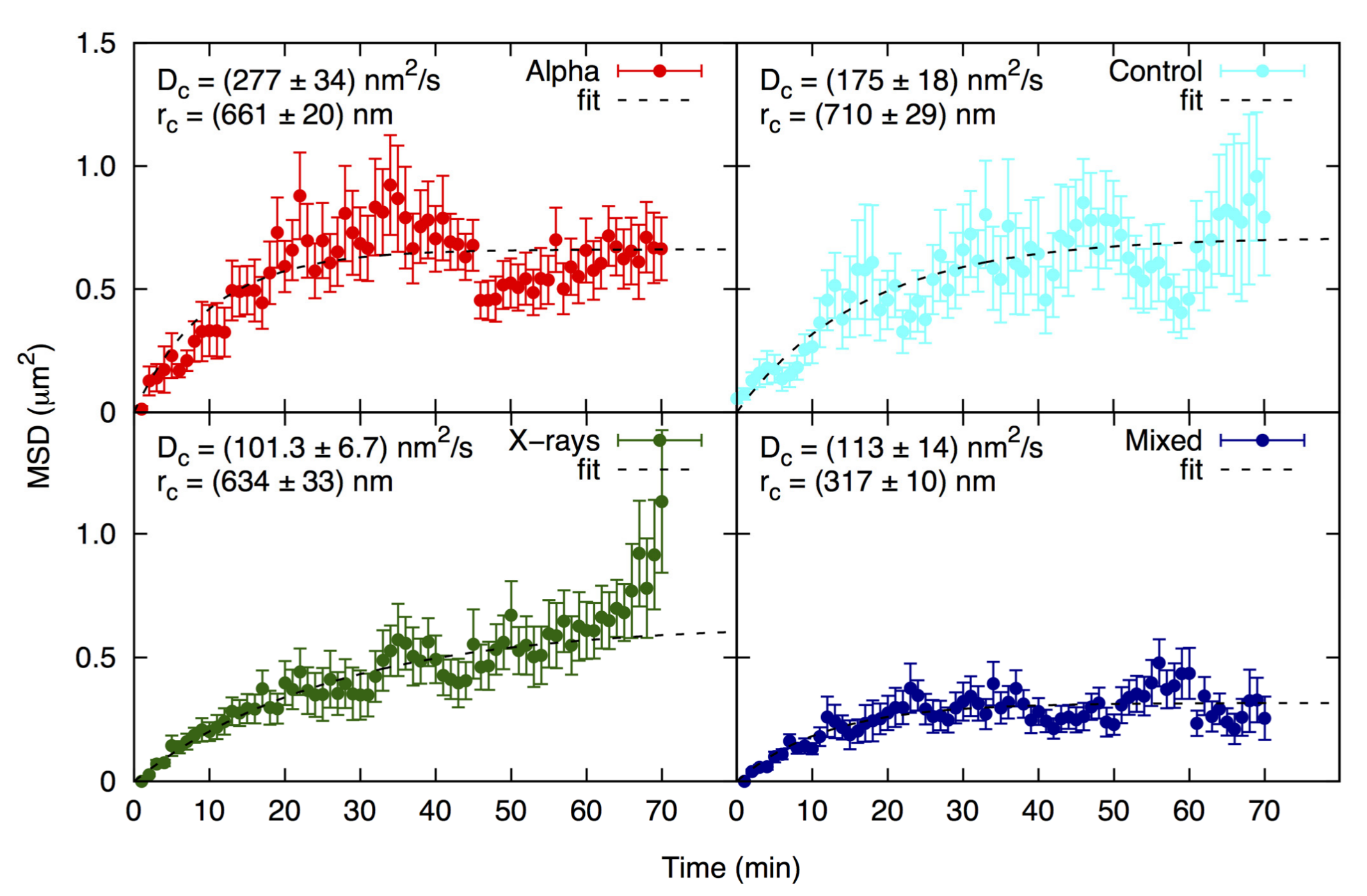
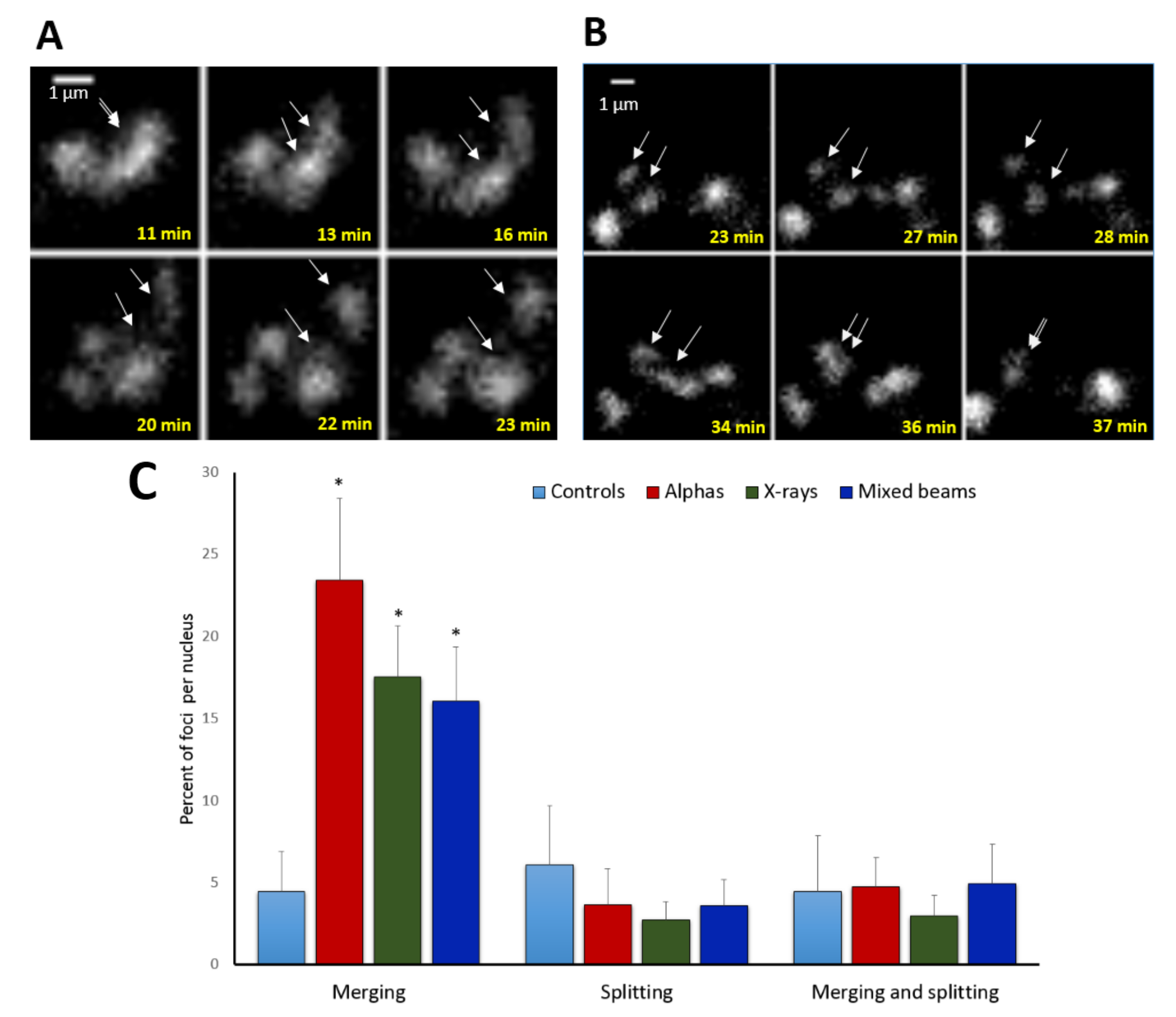
| Radiation Type | Initial FPC | Highest FPC (@ Time Point) | Final FPC | Max. Focus Increase 1 | Max. Focus Reduction 2 |
|---|---|---|---|---|---|
| Control | 1.5 ± 1.5 | 1.5 ± 1.5 | 1.5 ± 1.5 | - | - |
| Alpha particles | 6.4 ± 3.1 | 8.1 ± 2.2 (11 min) | 6.9 ± 3.1 | 5.4 | 1.2 |
| X-rays | 9.7 ± 4.3 | 15.3 ± 5.8 * (16 min) | 8.7 ± 4.2 | 10.2 | 1.8 |
| Mixed beams | 7.6 ± 3.4 | 10.3 ± 4.8 (9 min) | 9.1 ± 2.8 | 6.8 | 1.1 |
| Expected | 8.05 | 11.7 (15 min) | 7.8 | 7.8 | 1.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sollazzo, A.; Brzozowska, B.; Cheng, L.; Lundholm, L.; Scherthan, H.; Wojcik, A. Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells. Int. J. Mol. Sci. 2018, 19, 519. https://doi.org/10.3390/ijms19020519
Sollazzo A, Brzozowska B, Cheng L, Lundholm L, Scherthan H, Wojcik A. Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells. International Journal of Molecular Sciences. 2018; 19(2):519. https://doi.org/10.3390/ijms19020519
Chicago/Turabian StyleSollazzo, Alice, Beata Brzozowska, Lei Cheng, Lovisa Lundholm, Harry Scherthan, and Andrzej Wojcik. 2018. "Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells" International Journal of Molecular Sciences 19, no. 2: 519. https://doi.org/10.3390/ijms19020519
APA StyleSollazzo, A., Brzozowska, B., Cheng, L., Lundholm, L., Scherthan, H., & Wojcik, A. (2018). Live Dynamics of 53BP1 Foci Following Simultaneous Induction of Clustered and Dispersed DNA Damage in U2OS Cells. International Journal of Molecular Sciences, 19(2), 519. https://doi.org/10.3390/ijms19020519





