Inhibitory Effect of Synthetic Flavone Derivatives on Pan-Aurora Kinases: Induction of G2/M Cell-Cycle Arrest and Apoptosis in HCT116 Human Colon Cancer Cells
Abstract
1. Introduction
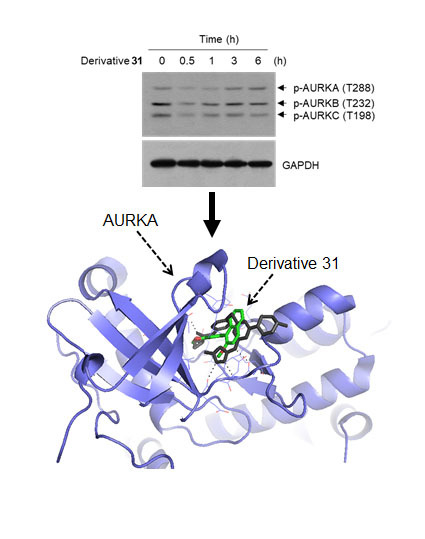
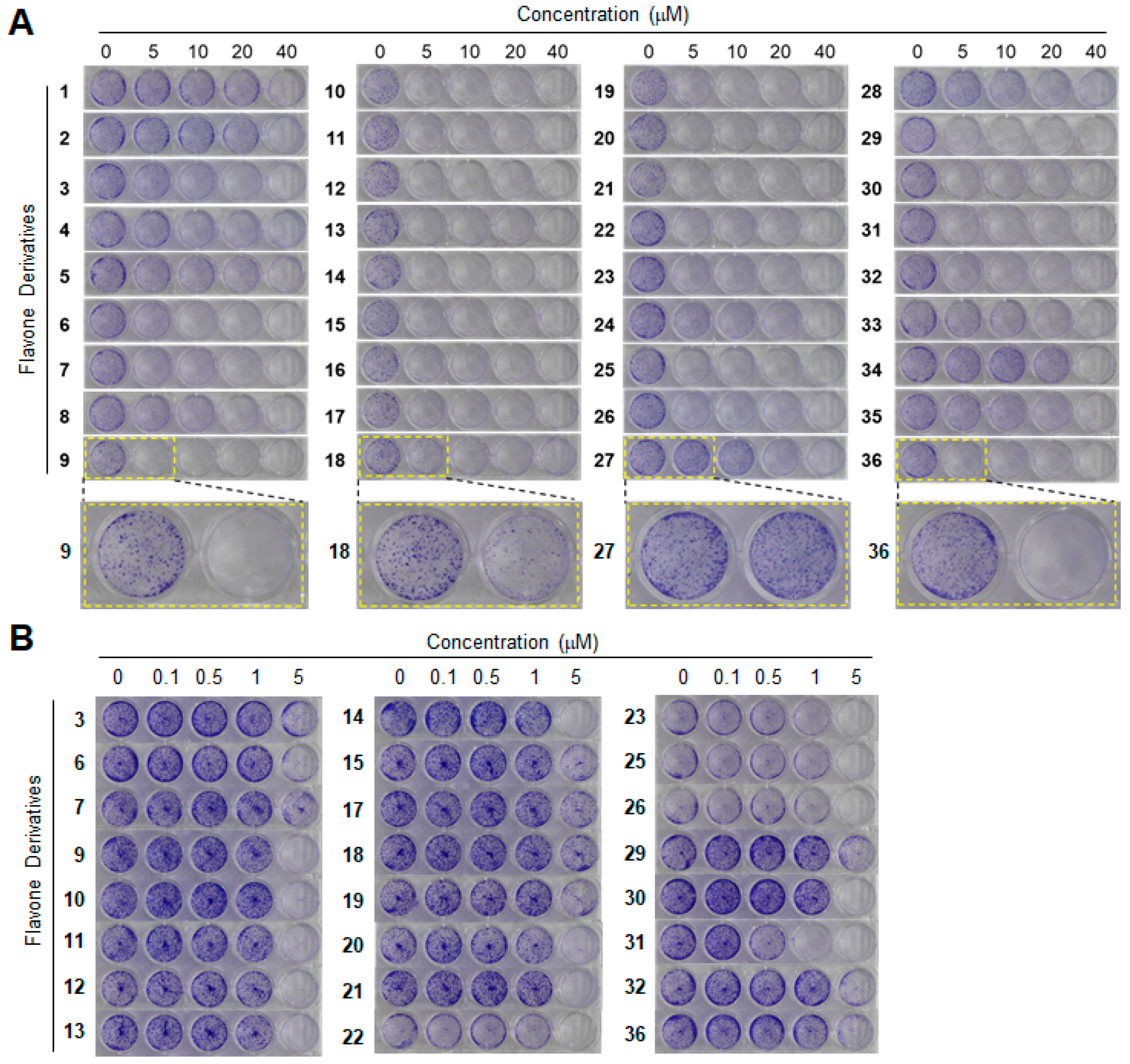
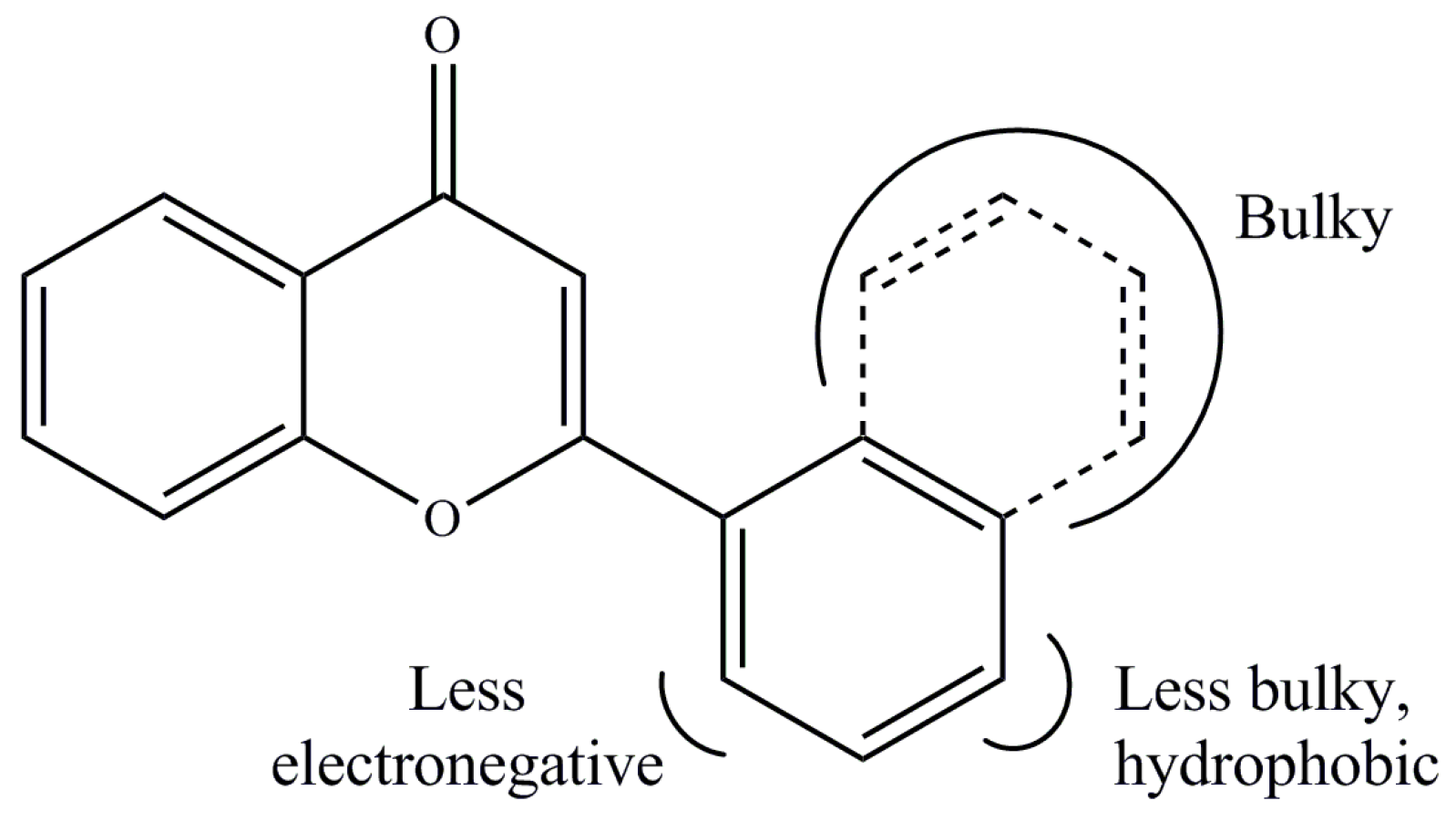
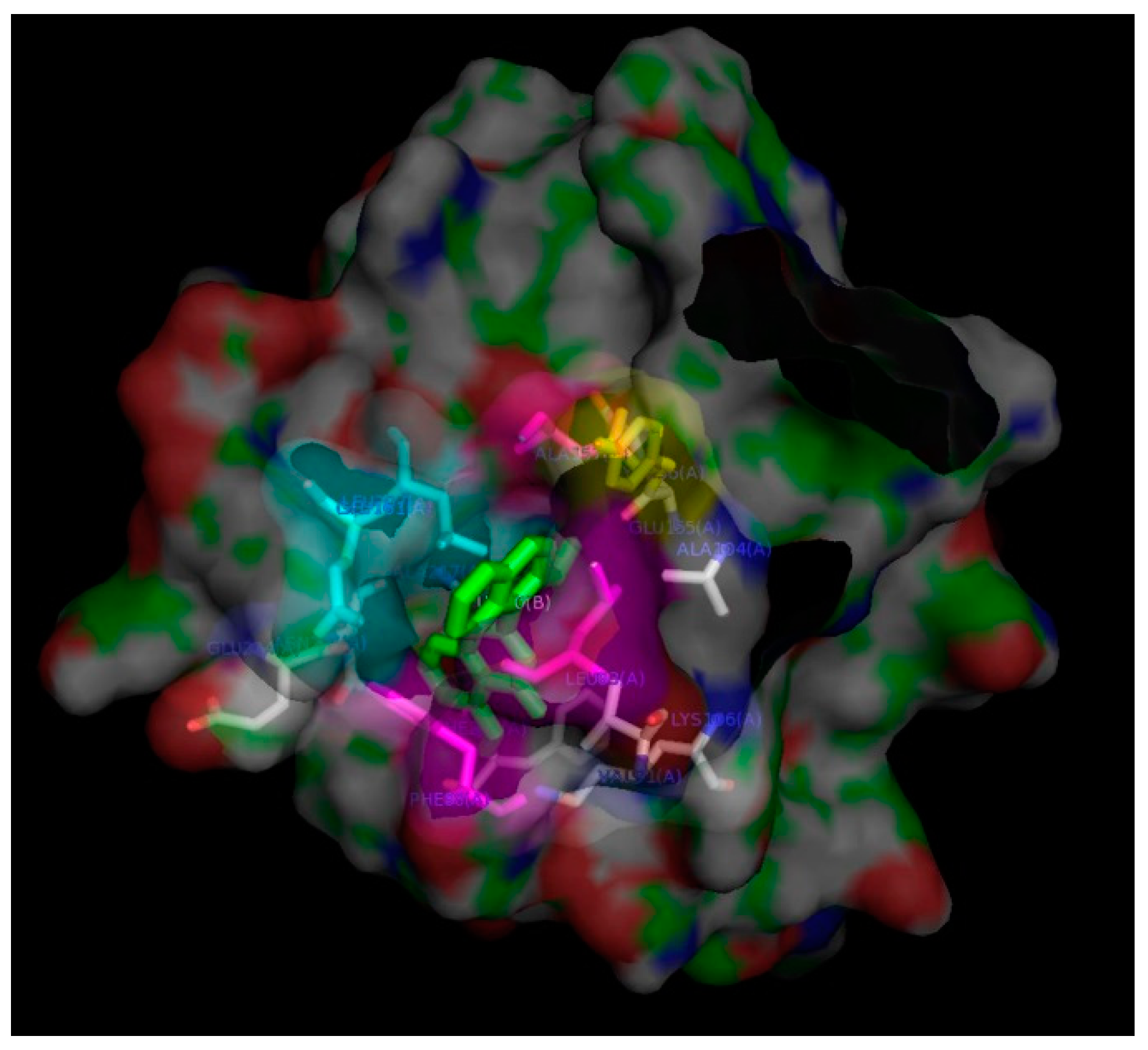
2. Results and Discussion
3. Materials and Methods
3.1. Preparation of 36 Synthetic Flavone Derivatives
3.2. Cell Culture
3.3. Clonogenic Long-Term Survival Assay
3.4. Quantitative Structure–Activity Relationship (QSAR)
3.5. Cell-Cycle Analysis by Flow Cytometry
3.6. Apoptosis Assay by Annexin V Staining
3.7. Western Blotting Analysis
3.8. In-Silico Docking
3.9. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| GI50 | cell growth inhibitory concentration |
| AURKA | aurora kinase A |
| AURKB | aurora kinase B |
| AURKC | aurora kinase C |
| CoMFA | comparative molecular field analysis |
| CoMSIA | comparative molecular similarity indices analysis |
| 3D-QSAR | Three-dimensional quantitative structure–activity relationship |
| PARP | poly(ADP-ribose) polymerase |
| TPB | 4-[(4-[2-(trifluoromethyl)phenyl]amino]pyrimidin-2-yl)amino]benzoic acid |
| FITC | fluorescein isothiocyanate |
References
- Lin, C.H.; Chang, C.Y.; Lee, K.R.; Lin, H.J.; Chen, T.H.; Wan, L. Flavones inhibit breast cancer proliferation through the Akt/FOXO3a signaling pathway. BMC Cancer 2015, 15, 958. [Google Scholar] [CrossRef] [PubMed]
- Gade, S.; Gandhi, N.M. Baicalein Inhibits MCF-7 Cell Proliferation In Vitro, Induces Radiosensitivity, and Inhibits Hypoxia Inducible Factor. J. Environ. Pathol. Toxicol. Oncol. 2015, 34, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Surichan, S.; Arroo, R.R.; Ruparelia, K.; Tsatsakis, A.M.; Androutsopoulos, V.P. Nobiletin bioactivation in MDA-MB-468 breast cancer cells by cytochrome P450 CYP1 enzymes. Food Chem. Toxicol. 2018, 113, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Dang, Q.; Song, W.; Xu, D.; Ma, Y.; Li, F.; Zeng, J.; Zhu, G.; Wang, X.; Chang, L.S.; He, D.; et al. Kaempferol suppresses bladder cancer tumor growth by inhibiting cell proliferation and inducing apoptosis. Mol. Carcinog. 2015, 54, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Samarghandian, S.; Afshari, J.T.; Davoodi, S. Chrysin reduces proliferation and induces apoptosis in the human prostate cancer cell line pc-3. Clinics 2011, 66, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Shin, S.Y.; Yong, Y.; Jung, H.; Ahn, S.; Lee, Y.H.; Lim, Y. Plant-derived flavones as inhibitors of aurora B kinase and their quantitative structure-activity relationships. Chem. Biol. Drug Des. 2015, 85, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Koh, E.J.; Jung, Y.; Shin, S.Y.; Lim, Y. 1H and 13C NMR spectral assignments of flavone derivatives. Magn. Reson. Chem. 2017, 55, 359–366. [Google Scholar] [CrossRef]
- Meyers, B.M.; Cosby, R.; Quereshy, F.; Jonker, D. Adjuvant Chemotherapy for Stage II and III Colon Cancer Following Complete Resection: A Cancer Care Ontario Systematic Review. Clin. Oncol. 2017, 29, 459–465. [Google Scholar] [CrossRef]
- Kannarkatt, J.; Joseph, J.; Kurniali, P.C.; Al-Janadi, A.; Hrinczenko, B. Adjuvant Chemotherapy for Stage II Colon Cancer: A Clinical Dilemma. J. Oncol. Pract. 2017, 13, 233–241. [Google Scholar] [CrossRef]
- Tsuji, Y.; Sugihara, K. Adjuvant chemotherapy for colon cancer: The difference between Japanese and western strategies. Expert. Opin. Pharmacother. 2016, 17, 783–790. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Shin, S.Y.; Ahn, S.; Koh, D.; Lee, Y.H.; Lim, Y. Biological evaluation of 2-pyrazolinyl-1-carbothioamide derivatives against HCT116 human colorectal cancer cell lines and elucidation on QSAR and molecular binding modes. Bioorg. Med. Chem. 2017, 25, 5423–5432. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.Y.; Park, J.; Han, Y.S.; Lee, Y.H.; Shin, S.Y.; Lim, Y. Synthesis and biological evaluation of hesperetin derivatives as agents inducing apoptosis. Bioorg. Med. Chem. 2017, 25, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.S.; Lim, Y.; Koh, D. Crystal structure of 2-(3,4-di-meth-oxy-phen-yl)-3-hy-droxy-4H-chromen-4-one. Acta Crystallogr. Sect. E Struct. Rep. Online 2014, 70. [Google Scholar] [CrossRef]
- Ahn, S.; Lim, Y.; Koh, D. Crystal structure of 2-(2,3-di-meth-oxy-naphthalen-1-yl)-3-hy-droxy-6-meth-oxy-4H-chromen-4-one. Acta Crystallogr. E Crystallogr. Commun. 2015, 71. [Google Scholar] [CrossRef]
- He, W.; Zhang, M.G.; Wang, X.J.; Zhong, S.; Shao, Y.; Zhu, Y.; Shen, Z.J. AURKA suppression induces DU145 apoptosis and sensitizes DU145 to docetaxel treatment. Am. J. Transl. Res. 2013, 5, 359–367. [Google Scholar]
- Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001, 2, 21–32. [Google Scholar] [CrossRef]
- Walter, A.O.; Seghezzi, W.; Korver, W.; Sheung, J.; Lees, E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 2000, 19, 4906–4916. [Google Scholar] [CrossRef]
- Bischoff, J.R.; Anderson, L.; Zhu, Y.; Mossie, K.; Ng, L.; Souza, B.; Schryver, B.; Flanagan, P.; Clairvoyant, F.; Ginther, C.; et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998, 17, 3052–3065. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef]
- Andree, H.A.; Reutelingsperger, C.P.; Hauptmann, R.; Hemker, H.C.; Hermens, W.T.; Willems, G.M. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 1990, 265, 4923–4928. [Google Scholar] [PubMed]
- McIlwain, D.R.; Berger, T.; Mak, T.W. Caspase functions in cell death and disease. Cold Spring Harb Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.J.; Shi, Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004, 5, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.P.; Zhu, J.Y.; Lawrence, H.R.; Pireddu, R.; Luo, Y.; Alam, R.; Ozcan, S.; Sebti, S.M.; Lawrence, N.J.; Schonbrunn, E. A novel mechanism by which small molecule inhibitors induce the DFG flip in Aurora A. ACS Chem. Biol. 2012, 7, 698–706. [Google Scholar] [CrossRef]
- Kramer, B.; Rarey, M.; Lengauer, T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins 1999, 37, 228–241. [Google Scholar] [CrossRef]
- Elkins, J.M.; Santaguida, S.; Musacchio, A.; Knapp, S. Crystal structure of human aurora B in complex with INCENP and VX-680. J. Med. Chem. 2012, 55, 7841–7848. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, B.S.; Ahn, S.; Koh, D.; Lee, Y.H.; Shin, S.Y.; Lim, Y. Anticancer and structure-activity relationship evaluation of 3-(naphthalen-2-yl)-N,5-diphenyl-pyrazoline-1-carbothioamide analogs of chalcone. Bioorg. Chem. 2016, 68, 166–176. [Google Scholar] [CrossRef]
- Shin, S.Y.; Jung, H.; Ahn, S.; Hwang, D.; Yoon, H.; Hyun, J.; Yong, Y.; Cho, H.J.; Koh, D.; Lee, Y.H.; et al. Polyphenols bearing cinnamaldehyde scaffold showing cell growth inhibitory effects on the cisplatin-resistant A2780/Cis ovarian cancer cells. Bioorg. Med. Chem. 2014, 22, 1809–1820. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, C.G.; Lim, Y.; Shin, S.Y. Aurora kinase A inhibitor TCS7010 demonstrates pro-apoptotic effect through the unfolded protein response pathway in HCT116 colon cancer cells. Oncol. Lett. 2017, 14, 6571–6577. [Google Scholar] [CrossRef]
- Shin, S.Y.; Ahn, S.; Yoon, H.; Jung, H.; Jung, Y.; Koh, D.; Lee, Y.H.; Lim, Y. Colorectal anticancer activities of polymethoxylated 3-naphthyl-5-phenylpyrazoline-carbothioamides. Bioorg. Med. Chem. Lett. 2016, 26, 4301–4309. [Google Scholar] [CrossRef]
- Gil, H.N.; Koh, D.; Lim, Y.; Lee, Y.H.; Shin, S.Y. The synthetic chalcone derivative 2-hydroxy-3′,5,5′-trimethoxychalcone induces unfolded protein response-mediated apoptosis in A549 lung cancer cells. Bioorg. Med. Chem. Lett. 2018, 28, 2969–2975. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
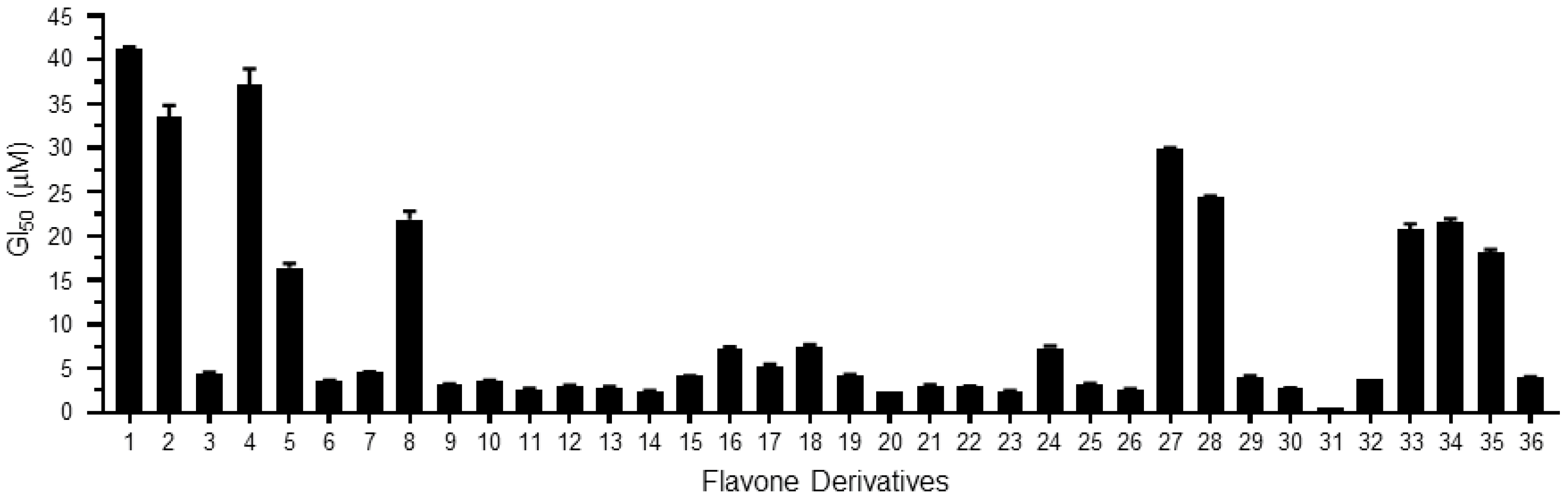
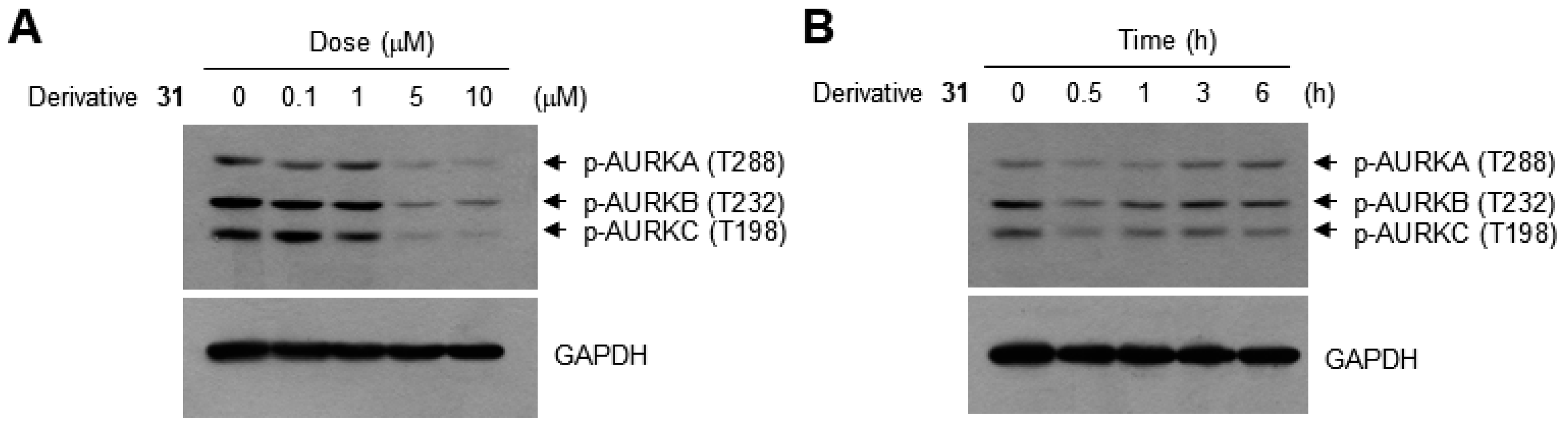
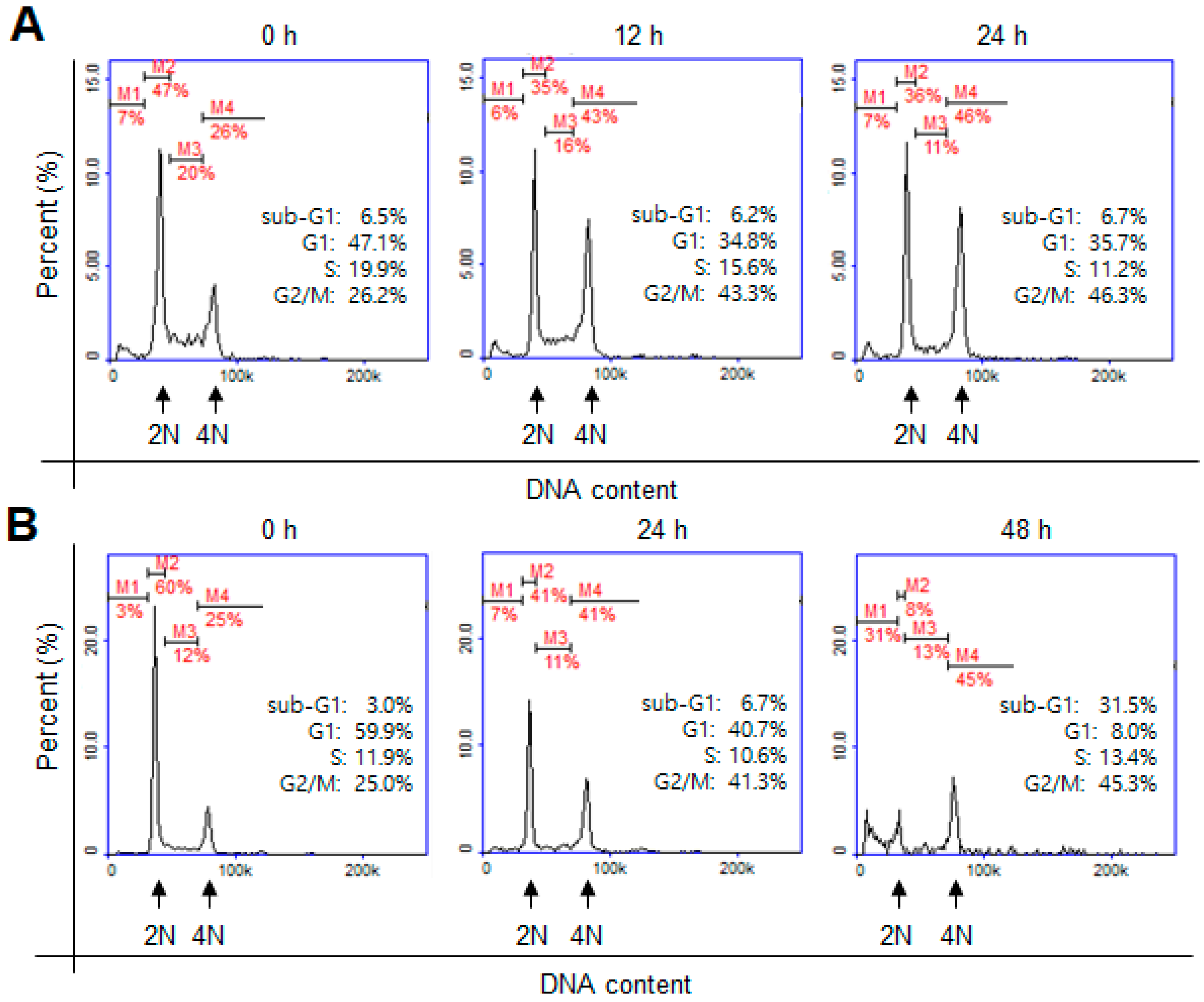
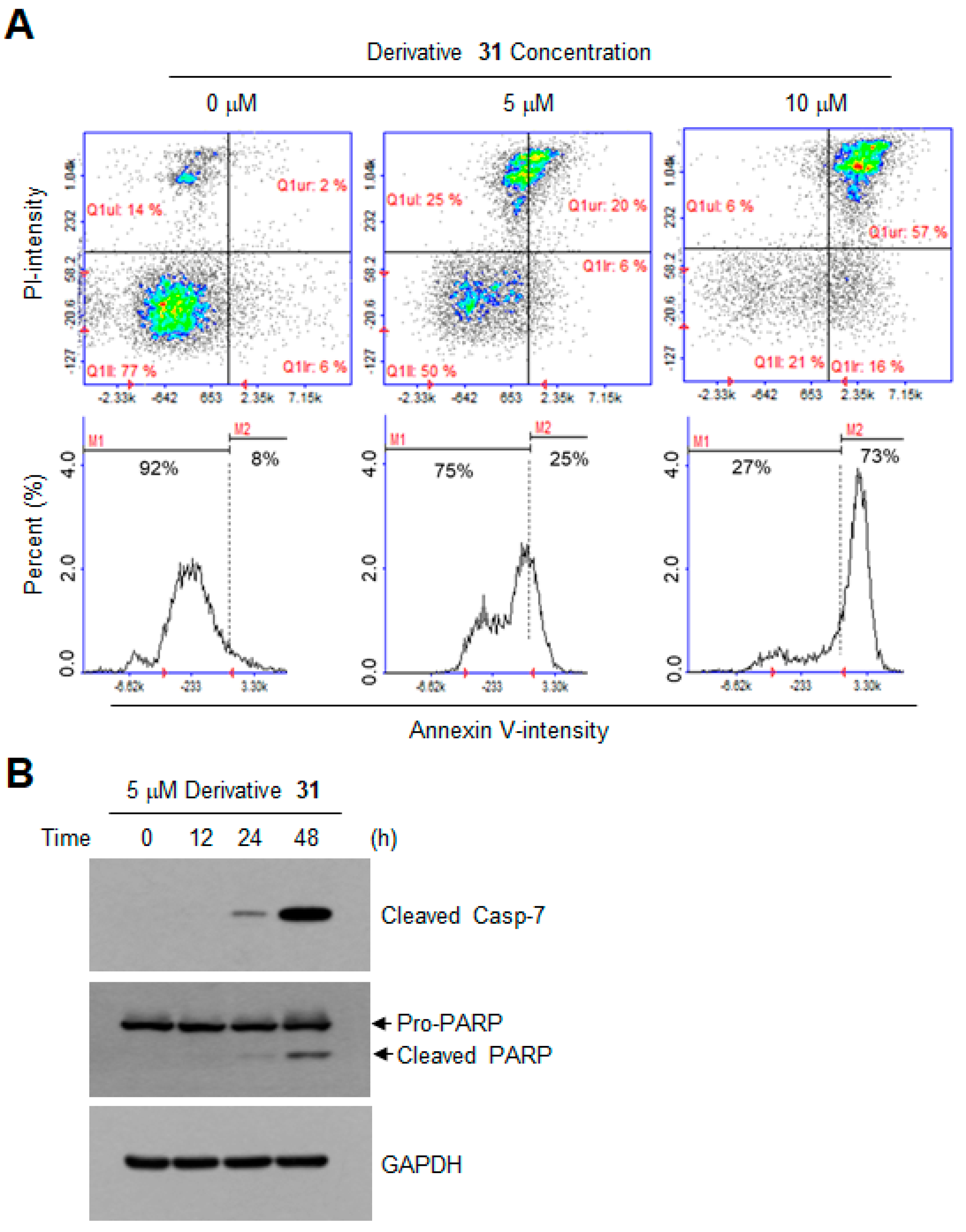
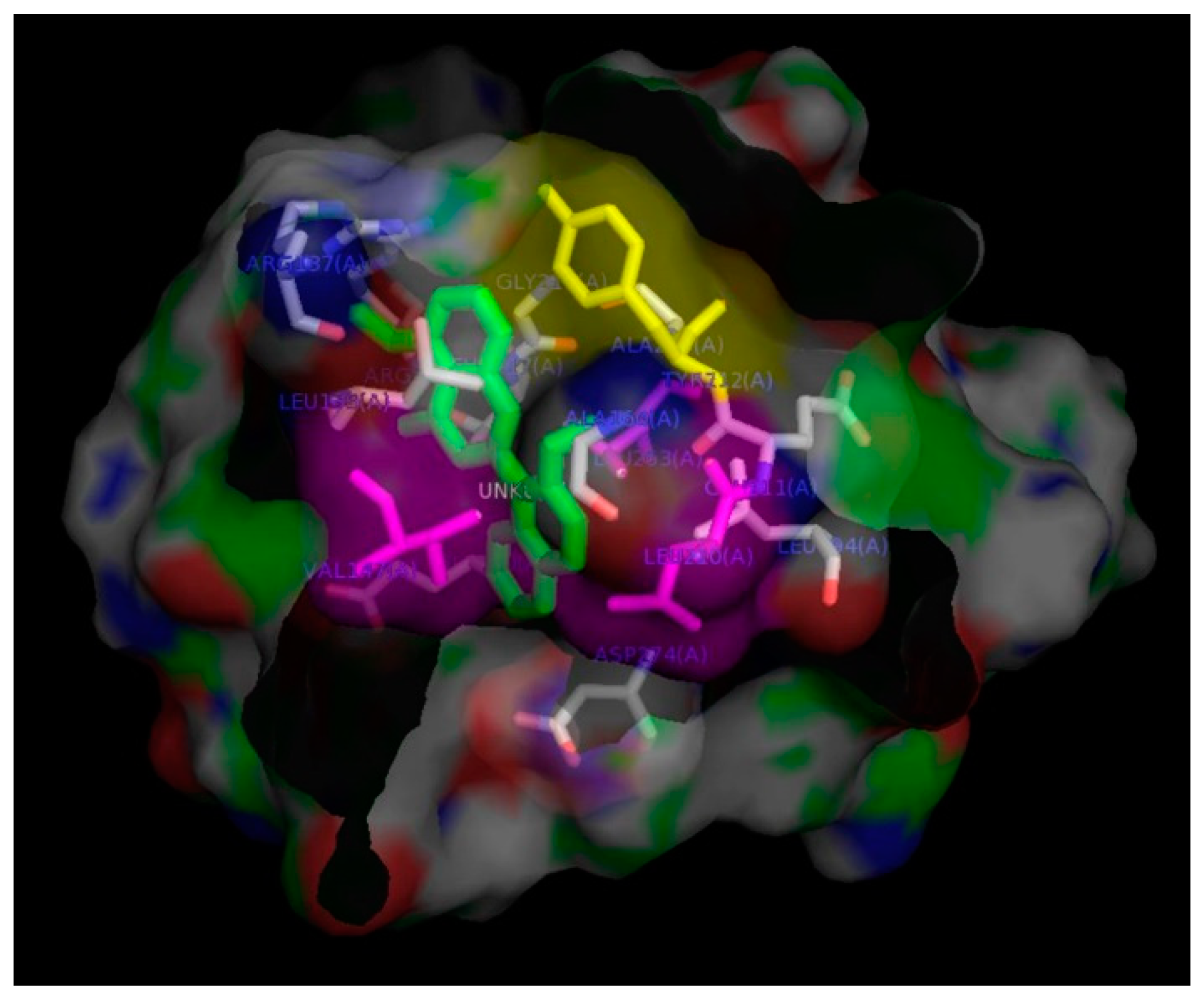
| Derivatives | Chemical Names | GI50 (μM) | pGI50 |
|---|---|---|---|
| 1 | 2-(2-fluorophenyl)-3-hydroxy-4H-chromen-4-one/2′-fluoroflavone | 41.19 | 1.39 |
| 2 | 2-(2-fluorophenyl)-3-hydroxy-6-nitro-4H-chromen-4-one/2′-fluoro-6-nitroflavone | 33.52 | 1.47 |
| 3 | 2-(4-fluorophenyl)-3-hydroxy-6-nitro-4H-chromen-4-one/4′-fluoro-6-nitroflavone | 4.49 | 2.35 |
| 4 | 3-hydroxy-2-(4-methoxyphenyl)-4H-chromen-4-one/4′-methoxyflavone | 37.18 | 1.43 |
| 5 | 3-hydroxy-2-(2-methoxyphenyl)-4H-chromen-4-one/2′-methoxyflavone | 16.44 | 1.78 |
| 6 | 2-(3,4-dimethoxyphenyl)-3-hydroxy-4H-chromen-4-one/3′,4′-dimethoxyflavone | 3.59 | 2.44 |
| 7 | 3-hydroxy-2-(2,4,6-trimethoxyphenyl)-4H-chromen-4-one/2′,4′,6′-trimethoxyflavone | 4.53 | 2.34 |
| 8 | 2-(2,4-dimethoxyphenyl)-3-hydroxy-4H-chromen-4-one/2′,4′-dimethoxyflavone | 21.92 | 1.66 |
| 9 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-3-hydroxy-4H-chromen-4-one/3-hydroxy-2′-(4-methoxystyryl)-flavone | 3.18 | 2.50 |
| 10 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-3-hydroxy-6-nitro-4H-chromen-4-one/3-hydroxy-6-nitro-2′-(4-methoxystyryl)-flavone | 3.63 | 2.44 |
| 11 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-6-bromo-3-hydroxy-4H-chromen-4-one/3-hydroxy-6-bromo-2′-(4-methoxystyryl)-flavone | 2.66 | 2.58 |
| 12 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-7-fluoro-3-hydroxy-4H-chromen-4-one/7-fluoro-3-hydroxy-2′-(4-methoxystyryl)-flavone | 3.07 | 2.51 |
| 13 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-6-chloro-3-hydroxy-4H-chromen-4-one/3-hydroxy-6-chloro-2′-(4-methoxystyryl)-flavone | 2.87 | 2.54 |
| 14 | 2-(6-(4-methoxystyryl)-2,4-dimethoxyphenyl)-6-fluoro-3-hydroxy-4H-chromen-4-one/3-hydroxy-6-fluoro-2′-(4-methoxystyryl)-flavone | 2.41 | 2.62 |
| 15 | 3-hydroxy-2-(naphthalen-1-yl)-4H-chromen-4-one/3-hydroxy-2′,3′-naphthoflavone | 4.14 | 2.38 |
| 16 | 3-hydroxy-6-methoxy-2-(naphthalen-1-yl)-4H-chromen-4-one/3-hydroxy-6-methoxy-2′,3′-naphthoflavone | 7.21 | 2.14 |
| 17 | 3-hydroxy-2-(2-methoxynaphthalen-1-yl)-4H-chromen-4-one/3-hydroxy-6′-methoxy-2′,3′-naphthoflavone | 5.28 | 2.28 |
| 18 | 2-(2,3-dimethoxynaphthalen-1-yl)-3-hydroxy-6-methoxy-4H-chromen-4-one/3-hydroxy-5′,6,6′-trimethoxy-2′,3′-naphthoflavone | 7.51 | 2.12 |
| 19 | 3-hydroxy-2-(4-methoxynaphthalen-1-yl)-4H-chromen-4-one/3-hydroxy-4′-methoxy-2′,3′-naphthoflavone | 4.28 | 2.37 |
| 20 | 3-hydroxy-2-(naphthalen-2-yl)-4H-chromen-4-one/3-hydroxy-3′,4′-naphthoflavone | 2.41 | 2.62 |
| 21 | 3-hydroxy-6-methoxy-2-(naphthalen-2-yl)-4H-chromen-4-one/3-hydroxy-6-methoxy-3′,4′-naphthoflavone | 3.07 | 2.51 |
| 22 | 2-(naphthalen-1-yl)-4H-chromen-4-one/2′,3′-naphthoflavone | 2.91 | 2.54 |
| 23 | 6-methoxy-2-(naphthalen-1-yl)-4H-chromen-4-one/6-methoxy-2′,3′-naphthoflavone | 2.41 | 2.62 |
| 24 | 5-methoxy-2-(naphthalen-1-yl)-4H-chromen-4-one/5-methoxy-2′,3′-naphthoflavone | 7.31 | 2.14 |
| 25 | 6,7-dimethoxy-2-(naphthalen-1-yl)-4H-chromen-4-one/6,7-dimethoxy-2′,3′-naphthoflavone | 3.26 | 2.49 |
| 26 | 7-methoxy-2-(naphthalen-1-yl)-4H-chromen-4-one/7-methoxy-2′,3′-naphthoflavone | 2.56 | 2.59 |
| 27 | 2-(naphthalen-2-yl)-4H-chromen-4-one/3′,4′-naphthoflavone | 29.86 | 1.52 |
| 28 | 6-methoxy-2-(naphthalen-2-yl)-4H-chromen-4-one/6-methoxy-3′,4′-naphthoflavone | 24.39 | 1.61 |
| 29 | 2-(2-methoxynaphthalen-1-yl)-4H-chromen-4-one/2′-methoxy-2′,3′-naphthoflavone | 4.06 | 2.39 |
| 30 | 6-methoxy-2-(2-methoxynaphthalen-1-yl)-4H-chromen-4-one/2′,6-dimethoxy-2′,3′-naphthoflavone | 2.78 | 2.56 |
| 31 | 5-methoxy-2-(2-methoxynaphthalen-1-yl)-4H-chromen-4-one/2′,5-dimethoxy-2′,3′-naphthoflavone | 0.49 | 3.31 |
| 32 | 6,7-dimethoxy-2-(2-methoxynaphthalen-1-yl)-4H-chromen-4-one/2′,6,7-trimethoxy-2′,3′-naphthoflavone | 3.80 | 2.42 |
| 33 | 2-(4-methoxynaphthalen-1-yl)-4H-chromen-4-one/4′-methoxy-2′,3′-naphthoflavone | 20.80 | 1.68 |
| 34 | 5,7-dimethoxy-2-(4-methoxynaphthalen-1-yl)-4H-chromen-4-one/4′,5,7-trimethoxy-2′,3′-naphthoflavone | 21.56 | 1.67 |
| 35 | 7-methoxy-2-(4-methoxynaphthalen-1-yl)-4H-chromen-4-one/4′,7-dimethoxy-2′,3′-naphthoflavone | 18.18 | 1.74 |
| 36 | 2-(2,3-dimethoxynaphthalen-1-yl)-7-methoxy-4H-chromen-4-one/2′,3′,7-trimethoxy-2′,3′-naphthoflavone | 3.95 | 2.40 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.Y.; Lee, Y.; Kim, B.S.; Lee, J.; Ahn, S.; Koh, D.; Lim, Y.; Lee, Y.H. Inhibitory Effect of Synthetic Flavone Derivatives on Pan-Aurora Kinases: Induction of G2/M Cell-Cycle Arrest and Apoptosis in HCT116 Human Colon Cancer Cells. Int. J. Mol. Sci. 2018, 19, 4086. https://doi.org/10.3390/ijms19124086
Shin SY, Lee Y, Kim BS, Lee J, Ahn S, Koh D, Lim Y, Lee YH. Inhibitory Effect of Synthetic Flavone Derivatives on Pan-Aurora Kinases: Induction of G2/M Cell-Cycle Arrest and Apoptosis in HCT116 Human Colon Cancer Cells. International Journal of Molecular Sciences. 2018; 19(12):4086. https://doi.org/10.3390/ijms19124086
Chicago/Turabian StyleShin, Soon Young, Youngshim Lee, Beom Soo Kim, Junho Lee, Seunghyun Ahn, Dongsoo Koh, Yoongho Lim, and Young Han Lee. 2018. "Inhibitory Effect of Synthetic Flavone Derivatives on Pan-Aurora Kinases: Induction of G2/M Cell-Cycle Arrest and Apoptosis in HCT116 Human Colon Cancer Cells" International Journal of Molecular Sciences 19, no. 12: 4086. https://doi.org/10.3390/ijms19124086
APA StyleShin, S. Y., Lee, Y., Kim, B. S., Lee, J., Ahn, S., Koh, D., Lim, Y., & Lee, Y. H. (2018). Inhibitory Effect of Synthetic Flavone Derivatives on Pan-Aurora Kinases: Induction of G2/M Cell-Cycle Arrest and Apoptosis in HCT116 Human Colon Cancer Cells. International Journal of Molecular Sciences, 19(12), 4086. https://doi.org/10.3390/ijms19124086