A Conserved Glycine Is Identified to be Essential for Desaturase Activity of IpFAD2s by Analyzing Natural Variants from Idesia polycarpa
Abstract
1. Introduction
2. Results
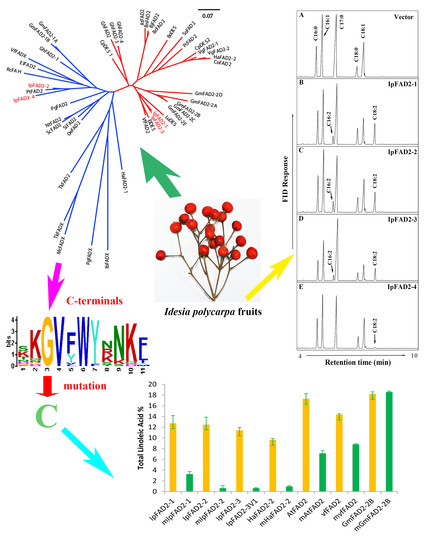
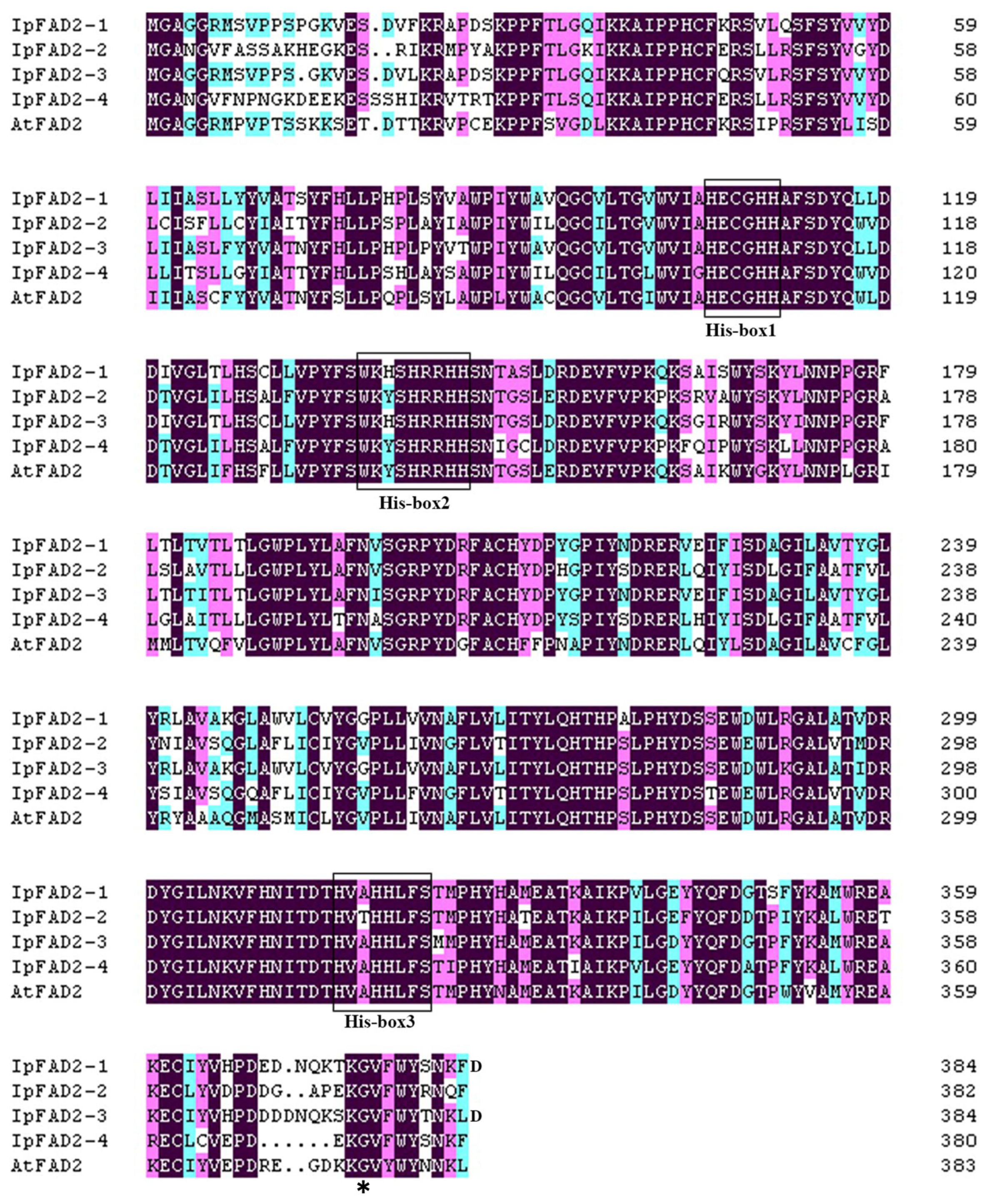
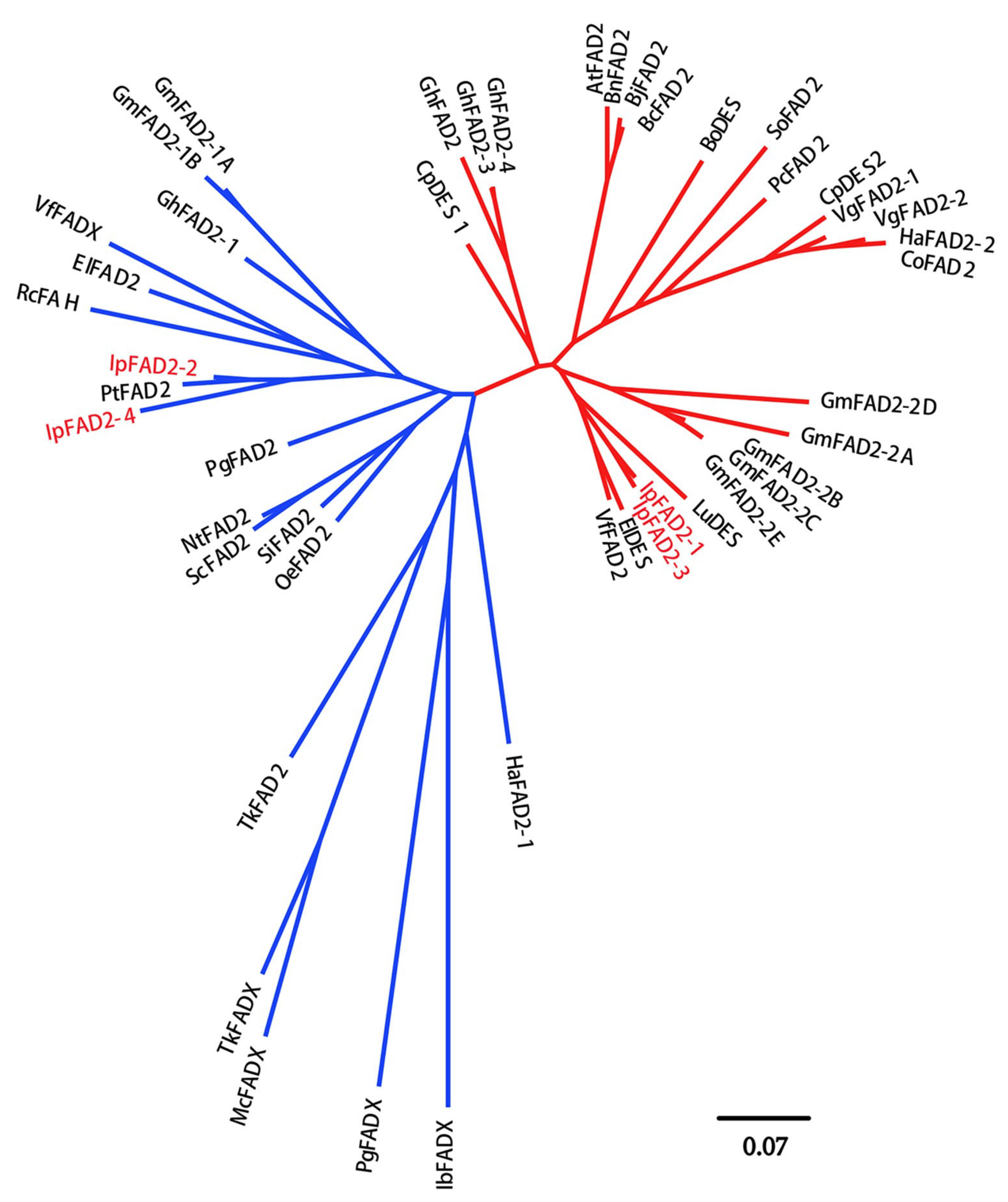
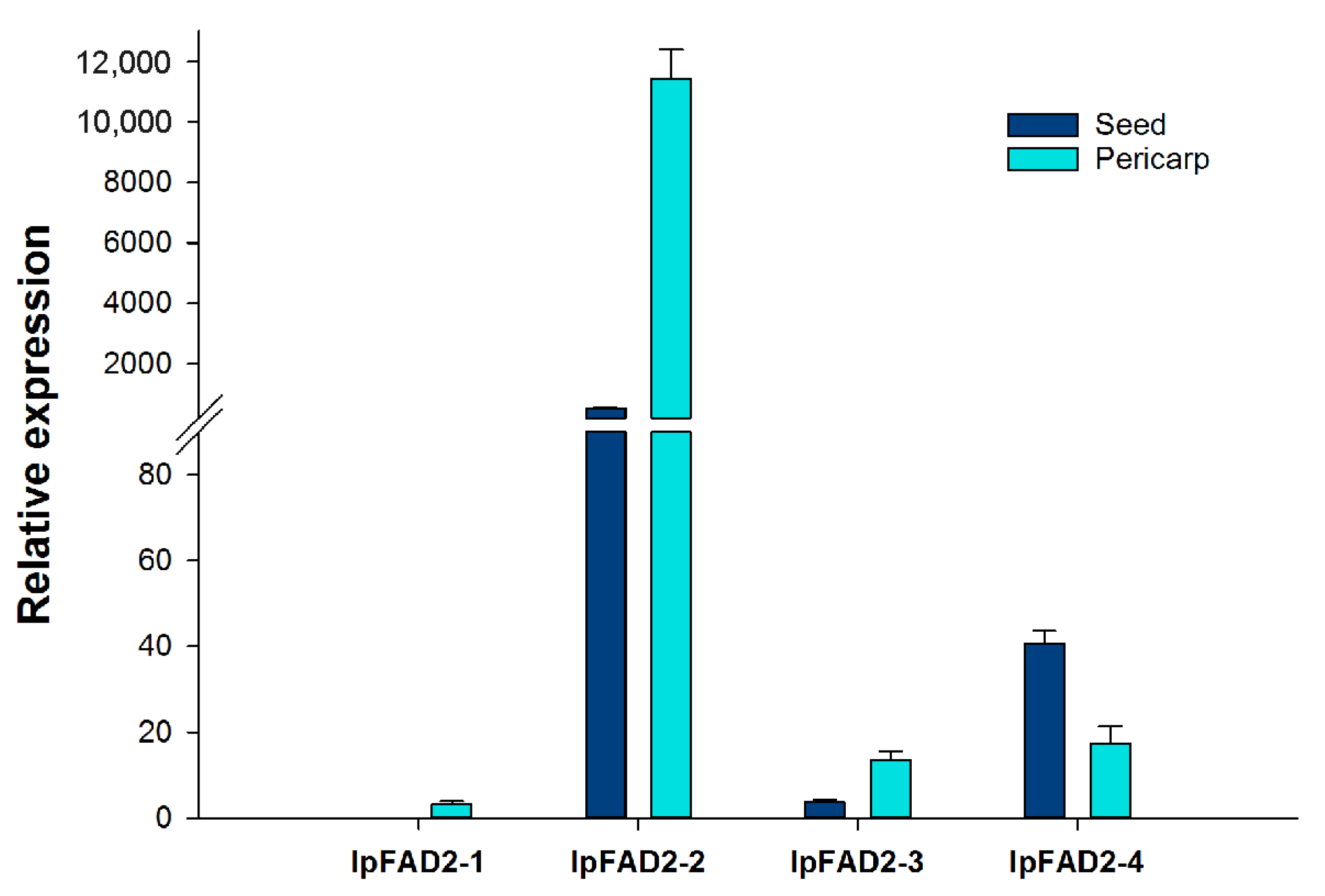
2.1. Isolation of FAD2 Orthologs from Idesia ploycarpa
2.2. Three IpFAD2s Possess Desaturase Activity
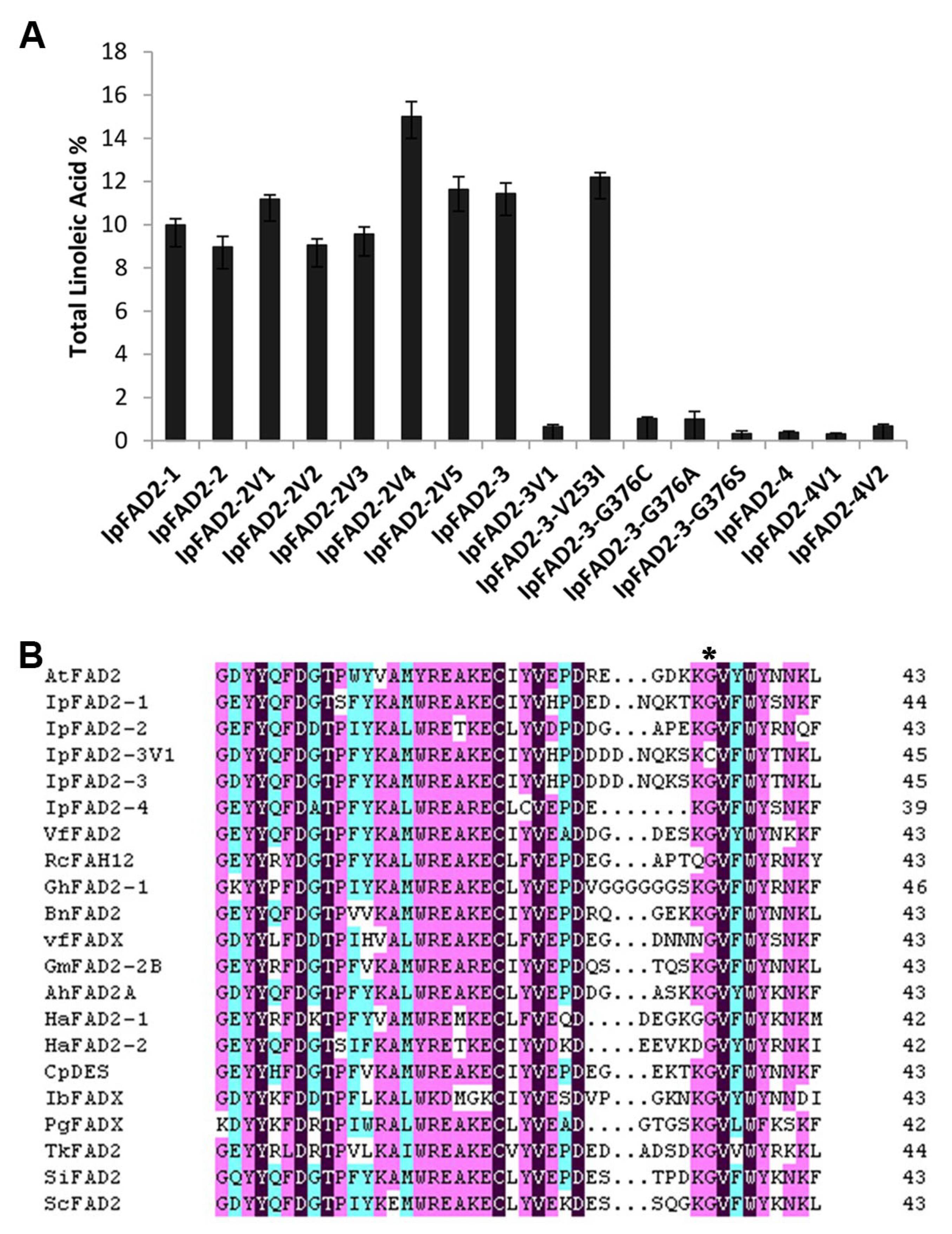
2.3. A Highly Conserved Glycine Residue Identified from the FAD2 Natural Variation is Required for FAD2 Desaturase Activity
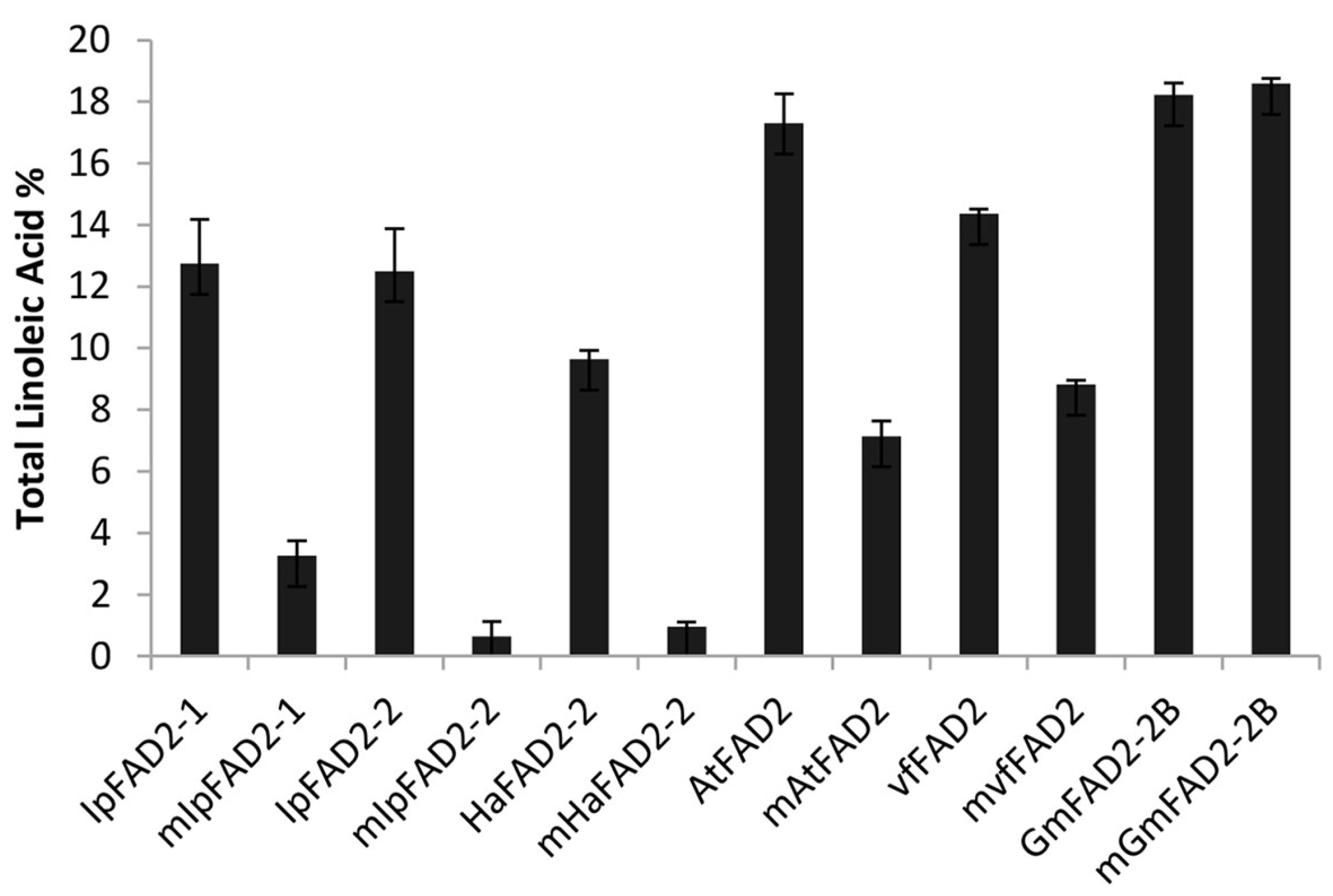
2.4. Gly376 Has Different Effects on the Activity of FAD2 Proteins among Different Species
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Total RNA Extraction and Complementary DNA Synthesis
4.3. Gene Cloning and Sequence Analysis
4.4. Real-Time Quantitative PCR
4.5. Site-Directed Mutagenesis
4.6. Yeast Transformation and Heterologous Expression of IpFAD2 Variants
4.7. Analysis of Fatty Acid Composition in Yeast
4.8. Subcellular Localization Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dyer, J.M.; Stymne, S.; Green, A.G.; Carlsson, A.S. High-value oils from plants. Plant J. 2008, 54, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Lummiss, J.A.M.; Oliveira, K.C.; Pranckevicius, A.M.T.; Santos, A.G.; dos Santos, E.N.; Fogg, D.E. Chemical Plants: High-Value Molecules from Essential Oils. J. Am. Chem. Soc. 2012, 134, 18889–18891. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, D.; Merker, A. Cloning and functional characterization of genes involved in fatty acid biosynthesis in the novel oilseed crop Lepidium campestre L. Plant Breed. 2011, 130, 407–409. [Google Scholar] [CrossRef]
- Yang, F.X.; Su, Y.Q.; Li, X.H.; Zhang, Q.; Sun, R.C. Preparation of biodiesel from Idesia polycarpa var. vestita fruit oil. Ind. Crop Prod. 2009, 29, 622–628. [Google Scholar] [CrossRef]
- Li, R.J.; Gao, X.; Li, L.M.; Liu, X.L.; Wang, Z.Y.; Lu, S.Y. De novo assembly and characterization of the fruit transcriptome of idesia polycarpa reveals candidate genes for lipid biosynthesis. Front. Plant Sci. 2016, 7, 801. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Wang, M.L.; Qiao, L.X.; Feng, S.P.; Khera, P.; Wang, H.; Tonnis, B.; Barkley, N.A.; Wang, J.P.; Holbrook, C.C.; et al. Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 2014, 15, 133. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Chang, Y.; Xu, W.L.; Cui, C.S.; Qu, S.P. Sequence variations in the FAD2 gene in seeded pumpkins. Genet. Mol. Res. 2015, 14, 17482–17488. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Mao, H.Z.; Chen, W.; Gao, S.Q.; Bai, Y.N.; Sun, Y.W.; Geng, Y.F.; Ye, J. Development of marker-free transgenic Jatropha plants with increased levels of seed oleic acid. Biotechnol. Biofuels 2012, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Miquel, M.; Browse, J. Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem. 1992, 267, 1502–1509. [Google Scholar] [PubMed]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 2010, 10, 195. [Google Scholar] [CrossRef]
- Janila, P.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular breeding for introgression of fatty acid desaturase mutant alleles (ahFAD2A and ahFAD2B) enhances oil quality in high and low oil containing peanut genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhou, X.R.; Wood, C.C.; Green, A.G.; Singh, S.P.; Liu, L.; Liu, Q. A large and functionally diverse family of Fad2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 2013, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.; Dowd, M.; Mack, B.; Gilbert, M.; Scheffler, B.; Ballard, L.; Frelichowski, J.; Mason, C. Naturally occurring high oleic acid cottonseed oil: Identification and functional analysis of a mutant allele of Gossypium barbadense fatty acid desaturase-2. Planta 2017, 245, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Sturtevant, D.; Horn, P.; Kennedy, C.; Hinze, L.; Percy, R.; Chapman, K. Lipid metabolites in seeds of diverse Gossypium accessions: Molecular identification of a high oleic mutant allele. Planta 2017, 245, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Unver, T.; Wu, Z.; Sterck, L.; Turktas, M.; Lohaus, R.; Li, Z.; Yang, M.; He, L.; Deng, T.; Escalante, F.J.; et al. Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. USA. 2017, 114, E9413–E9422. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 gene in plants: Occurrence, regulation, and role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Avelange-Macherel, M.H.; Macherel, D.; Wada, H.; Murata, N. Site-directed mutagenesis of histidine residues in the delta 12 acyl-lipid desaturase of Synechocystis. FEBS Lett. 1995, 361, 111–114. [Google Scholar] [CrossRef]
- Broun, P.; Shanklin, J.; Whittle, E.; Somerville, C. Catalytic plasticity of fatty acid modification enzymes underlying chemical diversity of plant lipids. Science 1998, 282, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.W.; Dyer, J.M.; Dhanoa, P.K.; Kim, P.K.; Andrews, D.W.; McNew, J.A.; Mullen, R.T. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 2004, 37, 156–173. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hornung, E.; Busch, S.; Kassner, N.; Ternes, P.; Braus, G.H.; Feussner, I. A small membrane-peripheral region close to the active center determines regioselectivity of membrane-bound fatty acid desaturases from Aspergillus nidulans. J. Biol. Chem. 2007, 282, 26666–26674. [Google Scholar] [CrossRef]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. BBA-Lipids Lipid Met. 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Phys. 1998, 49, 611–641. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Swift, D.; Sengoku, E.; Patel, M.; Teule, F.; Powell, G.; Moore, K.; Abbott, A. The high oleate trait in the cultivated peanut [Arachis hypogaea L.]. I. Isolation and characterization of two genes encoding microsomal oleoyl-PC desaturases. Mol. Gen. Genet. 2000, 263, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.; Carrero-Colon, M.; Crowe, M.; Gaskin, E.; Hudson, K. Novel FAD2-1A alleles confer an elevated oleic acid phenotype in soybean seeds. Crop Sci. 2016, 56, 226–231. [Google Scholar] [CrossRef]
- Ustun, R.; Uzun, B. Breeding for introgression of FAD2-1A and FAD2-1B genes to local soybean cultivars of Turkey. J. Biotechnol. 2017, 256, S103. [Google Scholar] [CrossRef]
- Dyer, J.M.; Chapital, D.C.; Kuan, J.C.W.; Mullen, R.T.; Turner, C.; McKeon, T.A.; Pepperman, A.B. Molecular analysis of a bifunctional fatty acid conjugase/desaturase from tung. Implications for the evolution of plant fatty acid diversity. Plant Physiol. 2002, 130, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E.; Browse, J. Arabidopsis Fad2 gene encodes the enzyme that is essential for polyunsaturated lipid-synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Kishore, K.; Sinha, S.K.; Kumar, R.; Gupta, N.C.; Dubey, N.; Sachdev, A. Isolation and characterization of microsomal omega-6-desaturase gene (fad2-1) from soybean. Indian J. Exp. Biol. 2007, 45, 390–397. [Google Scholar]
- Li, L.Y.; Wang, X.L.; Gai, J.Y.; Yu, D.Y. Molecular cloning and characterization of a novel microsomal oleate desaturase gene from soybean. J. Plant Physiol. 2007, 164, 1516–1526. [Google Scholar] [CrossRef]
- Pirtle, I.L.; Kongcharoensuntorn, W.; Nampaisansuk, M.; Knesek, J.E.; Chapman, K.D.; Pirtle, R.M. Molecular cloning and functional expression of the gene for a cotton Delta-12 fatty acid desaturase (FAD-2). BBA-Gene Struct. Expr. 2001, 1522, 122–129. [Google Scholar] [CrossRef]
- Sakai, H.; Kajiwara, S. Cloning and functional characterization of a Delta 12 fatty acid desaturase gene from the basidiomycete Lentinula edodes. Mol. Genet. Genomics 2005, 273, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Schierholt, A.; Becker, H.C.; Ecke, W. Mapping a high oleic acid mutation in winter oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2000, 101, 897–901. [Google Scholar] [CrossRef]
- Tang, G.Q.; Novitzky, W.P.; Griffin, H.C.; Huber, S.C.; Dewey, R.E. Oleate desaturase enzymes of soybean: Evidence of regulation through differential stability and phosphorylation. Plant J. 2005, 44, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Bruner, A.C.; Jung, S.; Abbott, A.G.; Powell, G.L. The naturally occurring high oleate oil character in some peanut varieties results from reduced oleoyl-PC desaturase activity from mutation of aspartate 150 to asparagine. Crop Sci. 2001, 41, 522–526. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]

| FAD2 Gene | SNPSite | Amino Acid Position | SNP Mutation | Amino Acid Mutation | Mutation Type |
|---|---|---|---|---|---|
| IpFAD2-1 | 201 | 67 | TAT→TAC | Tyr | S |
| (1158 bp) | 729 | 243 | GCA→GCG | Ala | S |
| 765 | 255 | TAT→TAC | Tyr | S | |
| IpFAD2-2 | 161 | 54 | TAT→TCT | Tyr→Ser | N |
| (1149 bp) | 206 | 69 | GCC→GTC | Ala→Val | N |
| 344 | 115 | CAG→CGG | Gln→Arg | N | |
| 372 | 124 | ATC→ATT | Ile | S | |
| 399 | 133 | TAC→TAT | Tyr | S | |
| 490 | 164 | AGT→AAT | Ser→Asn | N | |
| 612 | 204 | CGA→CGC | Arg | S | |
| 624 | 208 | CAC→CAT | His | S | |
| 727 | 243 | GTG→ATG | Val→Met | N | |
| 1041 | 347 | GAC→GAT | Asp | S | |
| 1092 | 364 | GTT→GTG | Val | S | |
| 1113 | 371 | CCA→CCC | Pro | S | |
| IpFAD2-3 | 690 | 230 | GGC→GGT | Gly | S |
| (1158 bp) | 696 | 232 | CTC→CTT | Leu | S |
| 729 | 243 | GTC→GTA | Val | S | |
| 757 | 253 | GTT→ATT | Val→Ile | N | |
| 1126 | 376 | GGC→TGC | Gly→Cys | N | |
| IpFAD2-4 | 92 | 31 | CCC→CTC | Pro→Leu | N |
| (1143 bp) | 212 | 71 | GCC→GTC | Ala→Val | N |
| 279 | 93 | CTA→CTC | Leu | S | |
| 451 | 151 | TGC→AGC | Cys→Ser | N | |
| 480 | 160 | CCA→CCG | Pro | S | |
| 491 | 164 | TTC→TCC | Phe→Ser | N | |
| 514 | 172 | CTC→TTC | Leu→Phe | N | |
| 531 | 177 | CCT→CCA | Pro | S | |
| 867 | 289 | GAA→GAT | Glu→Asp | N | |
| 999 | 333 | GCA→GCT | Ala | S | |
| 1002 | 334 | ACT→ACA | Thr | S | |
| 1094 | 365 | TGT→TAT | Cys→Tyr | N |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, P.; Zhang, L.; Feng, T.; Lu, W.; Zhao, H.; Li, J.; Lü, S. A Conserved Glycine Is Identified to be Essential for Desaturase Activity of IpFAD2s by Analyzing Natural Variants from Idesia polycarpa. Int. J. Mol. Sci. 2018, 19, 3932. https://doi.org/10.3390/ijms19123932
Wu P, Zhang L, Feng T, Lu W, Zhao H, Li J, Lü S. A Conserved Glycine Is Identified to be Essential for Desaturase Activity of IpFAD2s by Analyzing Natural Variants from Idesia polycarpa. International Journal of Molecular Sciences. 2018; 19(12):3932. https://doi.org/10.3390/ijms19123932
Chicago/Turabian StyleWu, Pan, Lingling Zhang, Tao Feng, Wenying Lu, Huayan Zhao, Jianzhong Li, and Shiyou Lü. 2018. "A Conserved Glycine Is Identified to be Essential for Desaturase Activity of IpFAD2s by Analyzing Natural Variants from Idesia polycarpa" International Journal of Molecular Sciences 19, no. 12: 3932. https://doi.org/10.3390/ijms19123932
APA StyleWu, P., Zhang, L., Feng, T., Lu, W., Zhao, H., Li, J., & Lü, S. (2018). A Conserved Glycine Is Identified to be Essential for Desaturase Activity of IpFAD2s by Analyzing Natural Variants from Idesia polycarpa. International Journal of Molecular Sciences, 19(12), 3932. https://doi.org/10.3390/ijms19123932