Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes
Abstract
1. Introduction
2. Results
2.1. Differential Clinical Characteristics and Leukocyte SIRT1 Gene Expression in the Studied Groups
2.1.1. GDM Patients Versus Control Subjects
2.1.2. GDM/SIRT1(↑) Patients Versus Control Subjects
2.2. Differential mRNA Expression of Diabetes-Related Genes in Leukocytes from GDM/SIRT1(↑) Patients Versus NGT Controls
2.3. Real-time PCR Verification of the Selected Genes Among Study Groups
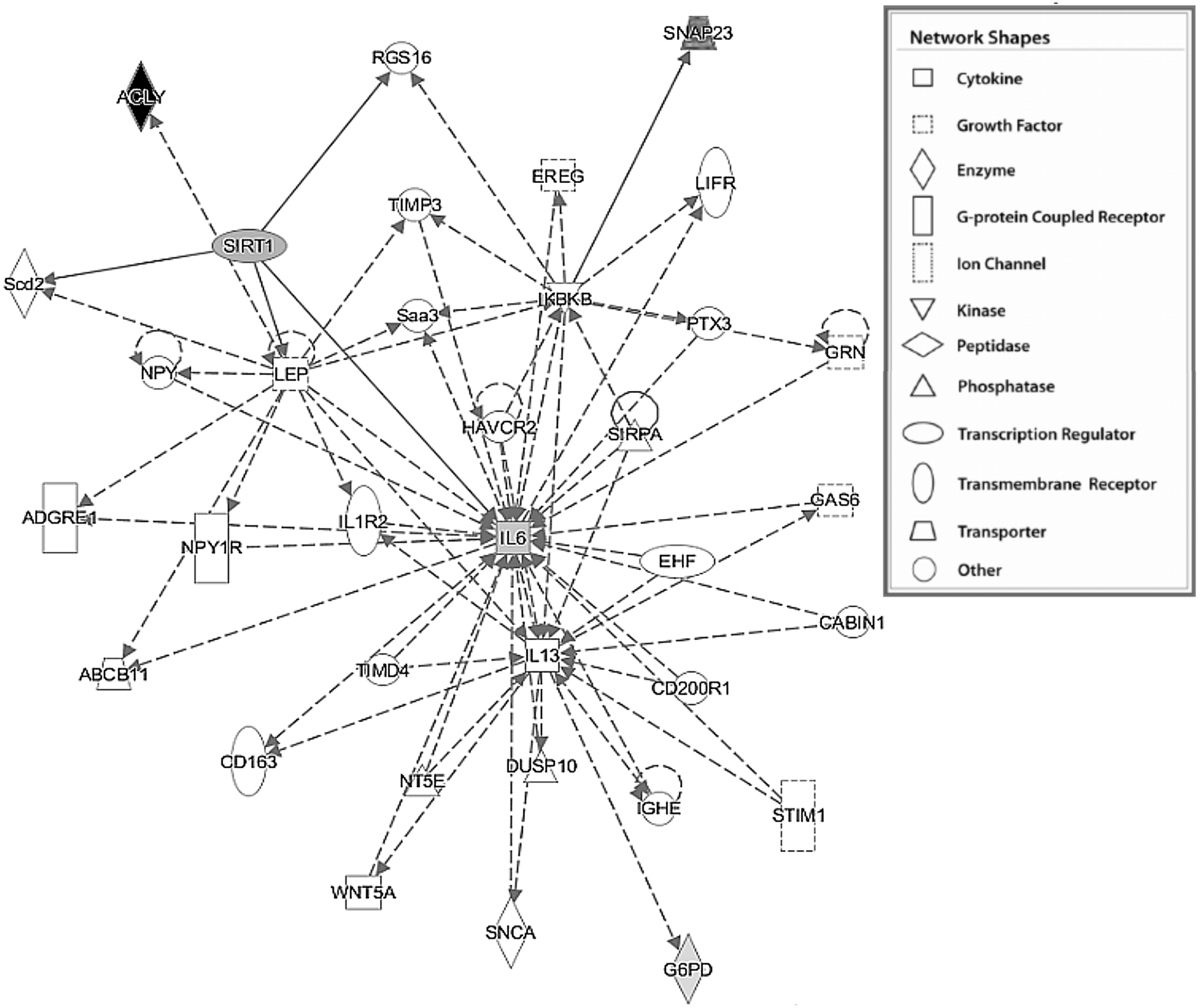
2.4. Functional analysis using Ingenuity Pathway Analysis (IPA)
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Anthropometric and Biochemical Data
4.3. Leukocytes Separation
4.4. RNA extraction and SIRT1 Gene Expression Assay
4.5. Human Diabetes RT² Profiler™ PCR Array
4.6. Quantitative RT-PCR Verification
4.7. Bioinformatics Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACL | ATP citrate lyase |
| COX2 | Cyclooxygenase 2 |
| ECFC | Endothelial colony-forming cell |
| GDM | Gestational diabetes mellitus |
| G6PD | Glucose-6-phosphate dehydrogenase |
| HFD | High fat diet |
| HUVEC | Human umbilical vein endothelial cells |
| IL6 | Interleukin 6 |
| IPA | Ingenuity Pathway Analysis |
| MAPK | Mitogen-activated protein kinases |
| NGT | Normal glucose tolerance |
| SIRT1 | Sirtuin 1 |
| SNAP23 | Synaptosomal-associated protein 23 |
| STACs | Sirtuin-activating compounds |
| T2DM | Type 2 diabetes mellitus |
References
- Coustan, D.R.; Lowe, L.P.; Metzger, B.E.; Dyer, A.R. The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study: Paving the way for new diagnostic criteria for gestational diabetes mellitus. Am. J. Obstet. Gynecol. 2010, 202, 654.e1-6. [Google Scholar] [CrossRef] [PubMed]
- Forsbach-Sánchez, G.; Tamez-Peréz, H.E.; Vazquez-Lara, J. Diabetes and pregnancy. Arch. Med. Res. 2005, 36, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.R.; Retnakaran, R.; Booth, G.L. Increased risk of cardiovascular disease in young women following gestational diabetes mellitus. Diabetes Care 2008, 31, 1668–1669. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, L.; Casas, J.P.; Hingorani, A.D.; Williams, D. Type 2 diabetes mellitus after gestational diabetes: A systematic review and meta-analysis. Lancet 2009, 373, 1773–1779. [Google Scholar] [CrossRef]
- Kautzky-Willer, A.; Pacini, G.; Tura, A.; Bieglmayer, C.; Schneider, B.; Ludvik, B.; Prager, R.; Waldhäusl, W. Increased plasma leptin in gestational diabetes. Diabetologia 2001, 44, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.; Cseh, K.; Baranyi, E.; Melczer, Z.; Speer, G.; Hajós, P.; Salamon, F.; Turi, Z.; Kovács, M.; Vargha, P.; et al. Tumor necrosis factor system in insulin resistance in gestational diabetes. Diabetes Res. Clin. Pract. 2002, 56, 93–99. [Google Scholar] [CrossRef]
- Retnakaran, R.; Hanley, A.J.; Raif, N.; Connelly, P.W.; Sermer, M.; Zinman, B. Reduced adiponectin concentration in women with gestational diabetes: A potential factor in progression to type 2 diabetes. Diabetes Care 2004, 27, 799–800. [Google Scholar] [CrossRef] [PubMed]
- Lappas, M.; Hiden, U.; Froehlich, J.; Desoye, G.; Haugel-de Mouzon, S.; Jawerbaum, A. The role of oxidative stress in the pathophysiology of gestational diabetes mellitus. Antioxid. Redox Signal. 2011, 15, 3061–4100. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, M.; Mac-Marcjanek, K.; Wozniak, L.A. Physiological and pathophysiological functions of SIRT1. Mini Rev. Med. Chem. 2009, 9, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiang, X.; Ma, H.; Wang, Y.; Xue, P.; Liu, Y. SIRT1 and insulin resistance. J. Diabetes Complicat. 2016, 30, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Banks, A.S.; Kon, N.; Knight, C.; Matsumoto, M.; Gutierrez-Juarez, R.; Rossetti, L.; Gu, W.; Accili, D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Pfluger, P.T.; Herranz, D.; Velasco-Miguel, S.; Serrano, M.; Tschöp, M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. USA 2008, 105, 9793–9798. [Google Scholar] [CrossRef] [PubMed]
- De Kreutzenberg, S.V.; Ceolotto, G.; Papparella, I.; Bortoluzzi, A.; Semplicini, A.; Dalla Man, C.; Cobelli, C.; Fadini, G.P.; Avogaro, A. Down-regulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: Potential biochemical mechanisms. Diabetes 2010, 59, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Balestrieri, M.L.; Servillo, L.; Esposito, A.; D’Onofrio, N.; Giovane, A.; Casale, R.; Barbieri, M.; Paolisso, P.; Rizzo, M.R.; Paolisso, G.; et al. Poor glycaemic control in type 2 diabetes patients reduces endothelial progenitor cell number by influencing SIRT1 signalling via platelet-activating factor receptor activation. Diabetologia 2013, 56, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006, 444, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Milne, J.C.; Lambert, P.D.; Schenk, S.; Carney, D.P.; Smith, J.J.; Gagne, D.J.; Jin, L.; Boss, O.; Perni, R.B.; Vu, C.B.; et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 2007, 450, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Davis, M.L.; Lainez, E.O.; Cote, A.M.; Johnson, M.O.; Gagne, D.J.; Vlasuk, G.P.; Ellis, J.L.; Suri, V. SRT2104, a novel small molecule SIRT1 activator ameliorates insulin resistance and promotes glucose utilization measured under a hyperinsulinemic-euglycemic clamp by enhancing both glycolysis and carbohydrate oxidation in mice fed a high fat diet. Diabetes 2011, 60 (Suppl. 1), 1007-P. [Google Scholar]
- Liu, K.; Zhou, R.; Wang, B.; Mi, M.T. Effect of resveratrol on glucose control and insulin sensitivity: A meta-analysis of 11 randomized controlled trials. Am. J. Clin. Nutr. 2014, 99, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Pacholec, M.; Bleasdale, J.E.; Chrunyk, B.; Cunningham, D.; Flynn, D.; Garofalo, R.S.; Griffith, D.; Griffor, M.; Loulakis, P.; Pabst, B.; et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010, 285, 8340–8351. [Google Scholar] [CrossRef] [PubMed]
- Timmers, S.; de Ligt, M.; Phielix, E.; van de Weijer, T.; Hansen, J.; Moonen-Kornips, E.; Schaart, G.; Kunz, I.; Hesselink, M.K.; Schrauwen-Hinderling, V.B.; et al. Resveratrol as add-on therapy in subjects with well-controlled type 2 diabetes: A randomized controlled trial. Diabetes Care 2016, 39, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Bordone, L.; Motta, M.C.; Picard, F.; Robinson, A.; Jhala, U.S.; Apfeld, J.; McDonagh, T.; Lemieux, M.; McBurney, M.; Szilvasi, A.; et al. 2006 Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006, 4, e31. [Google Scholar] [CrossRef]
- Frescas, D.; Valenti, L.; Accili, D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005, 280, 20589–20595. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Potthast, A.; Rohrbach, A.; Borns, K.; Das, A.M.; von Versen-Höynck, F. Gestational diabetes induces alterations of sirtuins in fetal endothelial cells. Pediatr. Res. 2016, 79, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Wan, J.; Li, H.; Ding, J.; Wang, Y.; Wang, X.; Li, M. Resveratrol relieves gestational diabetes mellitus in mice through activating AMPK. Reprod. Biol. Endocrinol. 2015, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.; Alzahrani, N.; Al-Sakkaf, K. The postpartum effect of maternal diabetes on the circulating levels of sirtuins and superoxide dismutase. FEBS Open Bio 2018, 8, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Moradipoor, S.; Ismail, P.; Etemad, A.; Wan Sulaiman, W.A.; Ahmadloo, S. Expression Profiling of Genes Related to Endothelial Cells Biology in Patients with Type 2 Diabetes and Patients with Prediabetes. BioMed Res. Int. 2016, 2016, 1845638. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.C.; Ma, J.; Tang, H.C.; Zheng, R.; Dempsey, A.A. The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Xu, W.; Chen, Y.; Li, Z.; Zeng, Y.; Fu, Y. The expression of Sirtuins 1 and 4 in peripheral blood leukocytes from patients with type 2 diabetes. Eur. J. Histochem. 2011, 2011, E10. [Google Scholar] [CrossRef] [PubMed]
- Friebe, D.; Neef, M.; Kratzsch, J.; Erbs, S.; Dittrich, K.; Garten, A.; Petzold-Quinque, S.; Blüher, S.; Reinehr, T.; Stumvoll, M.; et al. Leucocytes are a major source of circulating nicotinamide phosphoribosyltransferase (NAMPT)/pre-B cell colony (PBEF)/visfatin linking obesity and inflammation in humans. Diabetologia 2011, 54, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Alcendor, R.R.; Gao, S.; Zhai, P.; Zablocki, D.; Holle, E.; Yu, X.; Tian, B.; Wagner, T.; Vatner, S.F.; Sadoshima, J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007, 100, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, W.; Sikora, E.; Bielak-Zmijewska, A. Sirtuins, a promising target in slowing down the ageing process. Biogerontology 2017, 18, 447–476. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Seok, S.; Yau, P.; Li, X.; Kemper, B.; Kemper, J.K. 2017 Obesity and aging diminish sirtuin 1 (SIRT1)-mediated deacetylation of SIRT3, leading to hyperacetylation and decreased activity and stability of SIRT3. J. Biol. Chem. 2017, 292, 17312–17323. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.R.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Nisoli, E.; Tonello, C.; Cardile, A.; Cozzi, V.; Bracale, R.; Tedesco, L.; Falcone, S.; Valerio, A.; Cantoni, O.; Clementi, E.; et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 2005, 310, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Crujeiras, A.B.; Parra, D.; Goyenechea, E.; Martinez, J.A. Sirtuin gene expression in human mononuclear cells is modulated by caloric restriction. Eur. J. Clin. Investig. 2008, 38, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Engin-Ustun, Y.; Caglayan, E.K.; Kara, M.; Gocmen, A.Y.; Polat, M.F.; Aktulay, A. The effect of Ramadan fasting on sirtuin and visfatin levels. Interv. Med. Appl. Sci. 2016, 8, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Bori, Z.; Zhao, Z.; Koltai, E.; Fatouros, I.G.; Jamurtas, A.Z.; Douroudos, I.I.; Terzis, G.; Chatzinikolaou, A.; Sovatzidis, A.; Draganidis, D.; et al. The effects of aging, physical training, and a single bout of exercise on mitochondrial protein expression in human skeletal muscle. Exp. Gerontol. 2012, 47, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Wang, T.; Tung, Y.T.; Lin, W.T. Effect of Exercise Training on Skeletal Muscle SIRT1 and PGC-1α Expression Levels in Rats of Different Age. Int. J. Med. Sci. 2016, 13, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, S.; Arimoto, A.; Kuramoto, Y.; Kozako, T.; Honda, S.; Shimeno, H.; Soeda, S. Fasting promotes the expression of SIRT1, an NAD+-dependent protein deacetylase, via activation of PPARa in mice. Mol. Cell. Biochem. 2010, 339, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Kaviarasan, S.; Muniandy, S.; Qvist, R.; Ismail, I.S. Gene expression profile in leukocytes of type 2 diabetic subjects. Biomed. Res. 2009, 20, 158–165. [Google Scholar] [CrossRef]
- Manoel-Caetano, F.S.; Xavier, D.J.; Evangelista, A.F.; Takahashi, P.; Collares, C.V.; Puthier, D.; Foss-Freitas, M.C.; Foss, M.C.; Donadi, E.A.; Passos, G.A.; et al. Gene expression profiles displayed by peripheral blood mononuclear cells from patients with type 2 diabetes mellitus focusing on biological processes implicated on the pathogenesis of the disease. Gene 2012, 511, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Raatz, Y.; Ibrahim, S.; Feldmann, M.; Paleolog, E.M. Gene expression profiling and functional analysis of angiogenic markers in murine collagen induced arthritis. Arthritis Res. Ther. 2012, 14, R169. [Google Scholar] [CrossRef] [PubMed]
- Colomiere, M.; Permezel, M.; Riley, C.; Desoye, G.; Lappas, M. Defective insulin signaling in placenta from pregnancies complicated by gestational diabetes mellitus. Eur. J. Endocrinol. 2009, 160, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Kuzmicki, M.; Telejko, B.; Szamatowicz, J.; Zonenberg, A.; Nikolajuk, A.; Kretowski, A.; Gorska, M. High resistin and interleukin-6 levels are associated with gestational diabetes mellitus. Gynecol. Endocrinol. 2009, 25, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Morisset, A.S.; Dubé, M.C.; Côté, J.A.; Robitaille, J.; Weisnagel, S.J.; Tchernof, A. Circulating interleukin-6 concentrations during and after gestational diabetes mellitus. Acta Obstet. Gynecol. Scand. 2011, 90, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, D.G. Interleukin 6 stimulates hepatic glucose release from prelabeled glycogen pools. Am. J. Physiol. 1990, 258, E57–E64. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, C.; Papanicolaou, D.A.; Kyrou, I.; Defensor, R.; Mitsiadis, C.S.; Chrousos, G.P. Dose-dependent effects of recombinant human interleukin-6 on glucose regulation. J. Clin. Endocrinol. Metab. 1997, 82, 4167–4170. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Venugopal, S.K.; Singh, U.; Jialal, I. Hyperglycemia induces monocytic release of interleukin-6 via induction of protein kinase c-α and -β. Diabetes 2005, 54, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, M.; Fujisawa, K.; Uchimura, I.; Numano, F. The effect of glucose and advanced glycosylation end products on IL-6 production by human monocytes. Ann. N. Y. Acad. Sci. 1995, 748, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Turek, I.A.; Wozniak, L.A.; Cypryk, K.; Wojcik, M. Hyperglycaemia-induced oxidative stress in gestational diabetes mellitus (GDM). Diabetol. Klin. 2015, 4, 189–198. [Google Scholar] [CrossRef]
- Aslamy, A.; Thurmond, D.C. Exocytosis proteins as novel targets for diabetes prevention and/or remediation? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R739–R752. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Farimani, A.; Saidijam, M.; Goodarzi, M.T.; Yadegar Azari, R.; Asadi, S.; Zarei, S.; Shabab, N. Effect of Resveratrol Supplementation on the SNARE Proteins Expression in Adipose Tissue of Stroptozotocin-Nicotinamide Induced Type 2 Diabetic Rats. Iran. J. Med. Sci. 2015, 40, 248–255. [Google Scholar] [PubMed]
- Zhang, Z.; Apse, K.J.P.; Stanton, R.C. High glucose inhibits glucose 6-phosphate dehydrogenase via cAMP in aortic endo thelial cells. J. Biol. Chem. 2000, 275, 40042–40047. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Osborne, B.W.; Stanton, R.C. Diabetes causes inhibition of glucose-6-phosphate dehydrogenase via activation of protein kinase A which contributes to oxidative stress in rat kidney cortex. Am. J. Physiol. Ren. Physiol. 2005, 289, F1040–F1047. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Flores, M.; Ibáñez-Hernández, M.A.; Galván, R.E.; Gutiérrez, M.; Durán-Reyes, G.; Medina-Navarro, R.; Pascoe-Lira, D.; Ortega-Camarillo, C.; Vilar-Rojas, C.; Cruz, M.; et al. Glucose-6-phosphate dehydrogenase activity and NADPH/NADP1 ratio in liver and pancreas are dependent on the severity of hyperglycemia in rat. Life Sci. 2006, 78, 2601–2607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, Z.; Zhu, B.; Hu, J.; Liew, C.W.; Zhang, Y.; Leopold, J.A.; Handy, D.E.; Loscalzo, J.; Stanton, R.C. Increasing glucose 6-phosphate dehydrogenase activity restores redox balance in vascular endothelial cells exposed to high glucose. PLoS ONE 2012, 7, e49128. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, L.A.; Chistiakova, O.V. The regulation of glucose-6-phosphate dehydrogenase and glycogen synthase activities by insulin superfamily peptides in myometrium of pregnant women and its impairments under different types of diabetes mellitus. Biomed. Khim. 2009, 55, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Heymann, A.D.; Cohen, Y.; Chodick, G. 2012 Glucose-6-phosphate dehydrogenase deficiency and type 2 diabetes. Diabetes Care 2012, 35, e58. [Google Scholar] [CrossRef] [PubMed]
- Yazgan, Ü.C.; Taşdemir, E.; Bilgin, H.M.; Deniz Obay, B.; Şermet, A.; Elbey, B. Comparison of the anti-diabetic effects of resveratrol, gliclazide and losartan in streptozotocin-induced experimental diabetes. Arch. Physiol. Biochem. 2015, 121, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Gribble, A.D.; Ife, R.J.; Shaw, A.; McNair, D.; Novelli, C.E.; Bakewell, S.; Shah, V.P.; Dolle, R.E.; Groot, P.H.; Pearce, N.; et al. ATP-Citrate lyase as a target for hypolipidemic intervention. 2. Synthesis and evaluation of (3R,5S)-omega-substituted-3-carboxy-3,5-dihydroxyalkanoic acids and their gamma-lactone prodrugs as inhibitors of the enzyme in vitro and in vivo. J. Med. Chem. 1998, 41, 3582–3595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Jiang, L.; Wang, J.; Li, S.; Yu, Y.; You, J.; Zeng, R.; Gao, X.; Rui, L.; Li, W.; et al. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology 2009, 49, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Chypre, M.; Zaidi, N.; Smans, K. ATP-citrate lyase: A mini-review. Biochem. Biophys. Res. Commun. 2012, 422, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Shin, H.T.; Seo, Y.H.; Byun, H.O.; Yoon, S.H.; Lee, I.K.; Hyun, D.H.; Chung, H.Y.; Yoon, G. Sterol regulatory element-binding protein (SREBP)-1-mediated lipogenesis is involved in cell senescence. J. Biol. Chem. 2010, 285, 29069–29077. [Google Scholar] [CrossRef] [PubMed]
- Hata, K.; Andoh, A.; Shimada, M.; Fujino, S.; Bamba, S.; Araki, Y.; Okuno, T.; Fujiyama, Y.; Bamba, T. IL-17 stimulates inflammatory responses via NF-kappaB and MAP kinase pathways in human colonic myofibroblasts. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 282, G1035–G1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Andoh, A.; Inatomi, O.; Bamba, S.; Takayanagi, A.; Shimizu, N.; Fujiyama, Y. Interleukin-17 and lipopolysaccharides synergistically induce cyclooxygenase-2 expression in human intestinal myofibroblasts. J. Gastroenterol. Hepatol. 2005, 20, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Grover, M.; Gibbons, S.J.; Nair, A.A.; Bernard, C.E.; Zubair, A.S.; Eisenman, S.T.; Wilson, L.A.; Miriel, L.; Pasricha, P.J.; Parkman, H.P.; et al. Transcriptomic signatures reveal immune dysregulation in human diabetic and idiopathic gastroparesis. BMC Med. Genom. 2018, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, T.; Varastehpour, A.; Catalano, P.; Hauguel-de Mouzon, S. Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 2003, 52, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Wang, D.P.; Zhang, L.L.; Zhang, F.; Wang, D.M.; Zhang, W.Y. Genomic expression profiles of blood and placenta reveal significant immune-related pathways and categories in Chinese women with gestational diabetes mellitus. Diabet. Med. 2011, 28, 237–246. [Google Scholar] [CrossRef] [PubMed]
- The Polish Diabetes Association 2011. Clinical recommendations for the management of patients with diabetes. Diabetol. Prakt. 2011, 12 (Suppl. A), A1–A46. [Google Scholar]
- Wojcik, M.; Zieleniak, A.; Mac-Marcjanek, K.; Wozniak, L.A.; Cypryk, K. The elevated gene expression level of the A(2B) adenosine receptor is associated with hyperglycemia in women with gestational diabetes mellitus. Diabetes Metab. Res. Rev. 2014, 30, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, M.; Zieleniak, A.; Zurawska-Klis, M.; Cypryk, K.; Wozniak, L.A. Increased expression of immune-related genes in leukocytes of patients with diagnosed gestational diabetes mellitus (GDM). Exp. Biol. Med. 2016, 241, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. 1985 Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Siegel, S.; Castellan, N.J. Nonparametric Statistics for the Behavioral Sciences, 2nd ed.; International ed.; McGraw-Hill: New York, NY, USA, 1988; p. 399. [Google Scholar]


| Variable | NGT (n = 41) | GDM (n = 122) | p # | GDM/SIRT1(↔) (n = 92) | GDM/SIRT1(↑) (n = 30) | p ## |
|---|---|---|---|---|---|---|
| Age [years] | 29.0 (26.0–33.0) | 30.5 (27.0–33.0) | 0.156 | 31.0 (27.5–33.0) | 29.5 (26.0–33.0) | 0.265 |
| Pre-pregnancy BMI [kg/m2] | 24.2 (21.4–26.6) | 24.3 (21.1–27.7) | 0.648 | 24.0 (21.1–27.7) | 24.6 (21.9–27.7) | 0.845 |
| Pregnancy BMI [kg/m2] | 28.0 (25.0–30.5) | 27.5 (24.5–32.1) | 0.905 | 27.5 (24.2–32.1) | 28.1 (25.0–32.7) | 0.932 |
| Gestational weight gain [kg] | 10.0 (6.0–12.0) | 8.2 (6.0–12.0) | 0.244 | 8.0 (6.0–11.8) | 8.9 (6.0–13.0) | 0.500 |
| TGs [mg/dL] | 214.5 (179.0–277.1) | 217.9 (175.5–259.0) | 0.477 | 217.9 (174.0–259.0) | 217.8 (175.5–264.0) | 0.772 |
| TC [mg/dL] | 261.3 (238.5–282.5) | 251.1 (226.0–282.3) | 0.303 | 254.0 (226.0–283.0) | 249.7 (219.0–275.0) | 0.570 |
| HDL-C [mg/dL] | 76.3 (60.6–85.5) | 70.5 (59.3–82.5) | 0.810 | 74.0 (61.0–84.0) | 65.0 (53.0–73.0) | 0.279 |
| LDL-C [mg/dL] | 141.5 (123.5–168.5) | 136.0 (114.0–158.0) | 0.547 | 134.5 (114.0–158.0) | 145.0 (133.0–154.0) | 0.571 |
| HbA1c [%] | 5.3 (5.0–5.6) | 5.4 (5.2–5.7) | 0.090 | 5.4 (5.2–5.7) | 5.4 (5.2–5.7) | 0.223 |
| FPG [mg/dL] | 81.0 (74.5–84.0) | 88.0 (80.0–97.0) | 0.000 | 88.0 (79.5–97.5) | 92.0 (80.0–97.0) | 0.000 A,B |
| 1-h OGTT [mg/dL] | 164.5 (126.0–185.0) | 183.0 (168.0–202.0) | 0.000 | 182.0 (168.0–202.0) | 189.0 (160.0–200.0) | 0.000 A,B |
| 2-h OGTT [mg/dL] | 122.0 (102.0–132.0) | 157.0 (148.0–176.0) | 0.000 | 156.0 (147.0–173.5) | 158.0 (152.0–178.0) | 0.000 A,B |
| CRP [mg/L] | 3.9 (2.1–8.3) | 3.3 (2.0–5.9) | 0.308 | 3.4 (2.0–6.3) | 3.1 (2.3–5.8) | 0.591 |
| Insulin [µlU/mL] | 7.1 (1.7–10.1) | 5.2 (2.8–8.5) | 0.602 | 5.2 (2.9–8.8) | 5.2 (2.4–8.5) | 0.790 |
| HOMA-IR | 1.3 (0.5–2.2) | 1.2 (0.6–1.9) | 0.972 | 1.2 (0.6–1.9) | 1.3 (0.6–1.9) | 0.995 |
| SIRT1 | 0.59 (0.00–1.00) | 0.78 (0.27–1.41) | 0.041 | 0.56 (0.16–0.89) | 1.81 (1.54–2.28) | 0.000 B,C |
| Variable | NGT+GDM (n = 163) | NGT+GDM/SIRT1(↔) (n = 133) | NGT+GDM/SIRT1(↑) (n = 71) | |||
|---|---|---|---|---|---|---|
| R (95% CI) | p | R (95% CI) | p | R (95%CI) | p | |
| Age [years] | −0.03 (−0.18, 0.13) | 0.721 | 0.01 (−0.16, 0.18) | 0.938 | −0.02 (−0.25, 0.22) | 0.888 |
| Pre−pregnancy BMI [kg/m2] | 0.10 (−0.05, 0.25) | 0.197 | 0.12 (−0.06, 0.28) | 0.187 | 0.12 (−0.11, 0.35) | 0.325 |
| Pregnancy BMI [kg/m2] | 0.04 (−0.11, 0.20) | 0.581 | 0.04 (−0.14, 0.21) | 0.679 | 0.07 (−0.17, 0.30) | 0.553 |
| Gestational weight gain [kg] | −0.12 (−0.27, 0.03) | 0.127 | −0.16 (−0.32, 0.02) | 0.069 | −0.12 (−0.34, 0.12) | 0.318 |
| TC [mg/dL] | −0.01 (−0.20, 0.19) | 0.919 | 0.03 (−0.19, 0.24) | 0.815 | −0.02 (−0.29, 0.26) | 0.911 |
| TGs [mg/dL] | 0.06 (−0.14, 0.25) | 0.575 | 0.09 (−0.12, 0.30) | 0.401 | 0.06 (−0.22, 0.33) | 0.695 |
| HDL-C [mg/dL] | −0.17 (−0.36, 0.02) | 0.080 | −0.10 (−0.31, 0.11) | 0.345 | −0.25 (−0.49, 0.03) | 0.082 |
| LDL-C [mg/dL] | −0.00 (−0.20, 0.19) | 0.969 | −0.08 (−0.29, 0.14) | 0.474 | 0.01 (−0.27, 0.29) | 0.942 |
| HbA1c [%] | 0.04 (−0.12, 0.20) | 0.616 | 0.03 (−0.15, 0.20) | 0.774 | 0.31 (0.08, 0.51) | 0.010 * |
| FPG [mg/dL] | 0.07 (−0.09, 0.22) | 0.406 | −0.03 (−0.21, 0.15) | 0.733 | 0.41 (0.19, 0.59) | 0.000 * |
| 1-h OGTT [mg/dL] | −0.00 (−0.17, 0.17) | 0.993 | −0.05 (−0.24, 0.14) | 0.615 | 0.32 (0.06, 0.53) | 0.015 * |
| 2-h OGTT [mg/dL] | 0.23 (0.07, 0.37) | 0.005 * | 0.08 (−0.09, 0.25) | 0.368 | 0.76 (0.64, 0.85) | 0.000 * |
| CRP [mg/L] | −0.01 (−0.18, 0.16) | 0.922 | 0.04 (−0.15, 0.22) | 0.702 | −0.01 (−0.26, 0.24) | 0.924 |
| Insulin [µlU/mL] | −0.13 (−0.29, 0.04) | 0.138 | −0.12 (−0.30, 0.07) | 0.196 | −0.15 (−0.38, 0.10) | 0.233 |
| HOMA-IR | −0.08 (−0.25, 0.10) | 0.386 | −0.11 (−0.30, 0.09) | 0.285 | −0.06 (−0.31, 0.19) | 0.623 |
| Variable | DF | MS | F | p |
|---|---|---|---|---|
| Age | 1 | 0.34 | 0.74 | 0.392 |
| Pre-pregnancy BMI | 1 | 0.11 | 0.24 | 0.627 |
| Pregnancy BMI | 1 | 0.15 | 0.32 | 0.574 |
| Gestational weight gain | 1 | 0.29 | 0.62 | 0.431 |
| Status | 1 | 2.17 | 4.72 | 0.031 |
| Unigene | GenBank | Symbol | Description | FC | |
|---|---|---|---|---|---|
| Receptors, Transporters & Channels * | |||||
| Hs,431279 | NM_006178 | NSF | N-ethylmaleimide-sensitive factor | 0.48 | |
| Hs,511149 | NM_003825 | SNAP23 | Synaptosomal-associated protein, 23 kDa | 3.90 | |
| Hs,515104 | NM_006949 | STXBP2 | Syntaxin binding protein 2 | 0.50 | |
| Metabolic Enzymes | |||||
| Hs,387567 | NM_001096 | ACLY | ATP citrate lyase | 0.48 | |
| Hs,461047 | NM_000402 | G6PD | Glucose-6-phosphate dehydrogenase | 2.81 | |
| Hs,524418 | NM_005276 | GPD1 | Glycerol-3-phosphate dehydrogenase 1 (soluble) | 0.49 | |
| Cytokines & Growth Factors | |||||
| Hs,654458 | NM_000600 | IL6 | Interleukin 6 (interferon, beta 2) | 2.07 | |
| Signal Transduction | |||||
| Hs,471508 | NM_005544 | IRS1 | Insulin receptor substrate 1 | 0.49 | |
| Hs,442344 | NM_003749 | IRS2 | Insulin receptor substrate 2 | 2.31 | |
| Transcription Factors | |||||
| Hs,32938 | NM_000209 | PDX1 | Pancreatic and duodenal homeobox 1 | 0.33 | |
| Hs,592123 | NM_004176 | SREBF1 | Sterol regulatory element binding transcription factor 1 | 0.32 | |
| Symbol | Description | PCR Array | RT-qPCR Verification | |
|---|---|---|---|---|
| GDM/SIRT1(↑) vs. NGT | GDM/SIRT1(↑) vs. NGT | GDM/SIRT1(↔) vs. NGT | ||
| Receptors, Transporters & Channels § | ||||
| SNAP23 | Synaptosomal-associated protein, 23 kDa | 3.90 | 6.55 * | 3.93 |
| Metabolic enzyme | ||||
| ACLY | ATP citrate lyase | 0.48 | 0.63 * | 1.11 |
| G6PD | Glucose-6-phosphate dehydrogenase | 2.81 | 1.52 * | 1.06 |
| Cytokines & Growth Factors | ||||
| IL6 | Interleukin 6 (interferon, beta 2) | 2.07 | 2.28 * | 1.88 * |
| Signal Transduction | ||||
| IRS2 | Insulin receptor substrate 2 | 2.31 | 2.62 | 1.69 |
| Transcription Factors | ||||
| SREBF1 | Sterol regulatory element binding transcription factor 1 | 0.32 | 0.33 | 0.68 |
| Category | Top Function | p Range | Number of Targets |
|---|---|---|---|
| Disease and disorder | |||
| 1 | Inflammatory response | 6.60 × 10−6–3.24 × 10−2 | 3 |
| 2 | Cardiovascular disease | 2.54 × 10−5–4.55 × 10−2 | 3 |
| 3 | Organismal injury and abnormalities | 2.54 × 10−5–4.87 × 10−2 | 4 |
| 4 | Immunological disease | 9.57 × 10−5–2.33 × 10−2 | 2 |
| 5 | Inflammatory disease | 9.57 × 10−5−1.53 × 10−2 | 2 |
| Molecular and cellular function | |||
| 1 | Cell-to cell signaling and interaction | 6.60 × 10−6–3.15 × 10−2 | 2 |
| 2 | Cellular development | 2.10 × 10−5–4.70 × 10−2 | 4 |
| 3 | Drug metabolism | 3.82 × 10−5–1.41 × 10−2 | 2 |
| 4 | Molecular transport | 3.82 × 10−5–3.04 × 10−2 | 5 |
| 5 | Small molecule biochemistry | 3.82 × 10−5–3.04 × 10−2 | 4 |
| Physiological system development and function | |||
| 1 | Hematological system development and function | 6.60 × 10−6–4.82 × 10−2 | 4 |
| 2 | Immune cell trafficking | 6.60 × 10−6–3.24 × 10−2 | 2 |
| 3 | Cardiovascular system development and function | 2.54 × 10−5–4.44 × 10−2 | 3 |
| 4 | Organ morphology | 2.54 × 10−5–4.44 × 10−2 | 3 |
| 5 | Organismal development | 2.54 × 10−5–4.23 × 10−2 | 3 |
| Pathways | p |
|---|---|
| Sirtuin signaling pathway | 4.20 × 10−5 |
| Acetyl-CoA biosynthesis III (from citrate) | 3.01 × 10−4 |
| Pentose phosphate pathway (oxidative branch) | 1.20 × 10−3 |
| Pentose phosphate pathway | 3.01 × 10−3 |
| Differential regulation of cytokine production in macrophage and T helper cells by IL-17A and IL-17F | 5.41 × 10−3 |
| Gene Symbol | Primer Sequence 5’→3’ | Amplicon Size (bp) |
|---|---|---|
| Semi-quantitative PCR | ||
| SIRT1 | F: TCACCACCAGATTCTTCAGTG R: CCTCTTGATCATCTCCATCAGTC | 544 |
| GAPDH | F: CTGCACCACCAACTGCTTAG R: GTTGCTGTAGCCAAATTCGTTG | 514 |
| RT-qPCR | ||
| ACLY | F: TCTTTGTGCTGGGAAGGAGT R: GTAGGGTGCCTACTGCTATG | 90 |
| G6PD | F: ACGTCCGTGATGAGAAGGTC R: GTGGGGTCGTCCAGGTAC | 133 |
| IL6 | F: CCTGAGAAAGGAGACATGTAACAAG R: AAGGTTCAGGTTGGTTTTCTGCC | 79 |
| IRS2 | F: CGGTGAGTTCTACGGGTACAT R: TCAGGGTGTATTCATCCAGCG | 194 |
| SNAP23 | F: TAGCCATTGAGTCTCAGGATG R: GGTTTAGTTGTTCCTTTTGTTCA | 72 |
| SREBF1 | F: ACAGTGACTTCCCTGGCCTAT R: GCATGGACGGGTACATCTTCAA | 222 |
| GAPDH | F: GGTGGTCTCCTCTGACTTCAACA R: GTTGCTGTAGCCAAATTCGTTGT | 27 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mac-Marcjanek, K.; Zieleniak, A.; Zurawska-Klis, M.; Cypryk, K.; Wozniak, L.; Wojcik, M. Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes. Int. J. Mol. Sci. 2018, 19, 3826. https://doi.org/10.3390/ijms19123826
Mac-Marcjanek K, Zieleniak A, Zurawska-Klis M, Cypryk K, Wozniak L, Wojcik M. Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes. International Journal of Molecular Sciences. 2018; 19(12):3826. https://doi.org/10.3390/ijms19123826
Chicago/Turabian StyleMac-Marcjanek, Katarzyna, Andrzej Zieleniak, Monika Zurawska-Klis, Katarzyna Cypryk, Lucyna Wozniak, and Marzena Wojcik. 2018. "Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes" International Journal of Molecular Sciences 19, no. 12: 3826. https://doi.org/10.3390/ijms19123826
APA StyleMac-Marcjanek, K., Zieleniak, A., Zurawska-Klis, M., Cypryk, K., Wozniak, L., & Wojcik, M. (2018). Expression Profile of Diabetes-Related Genes Associated with Leukocyte Sirtuin 1 Overexpression in Gestational Diabetes. International Journal of Molecular Sciences, 19(12), 3826. https://doi.org/10.3390/ijms19123826







