RNA Sequencing-Based Bulked Segregant Analysis Facilitates Efficient D-genome Marker Development for a Specific Chromosomal Region of Synthetic Hexaploid Wheat
Abstract
1. Introduction
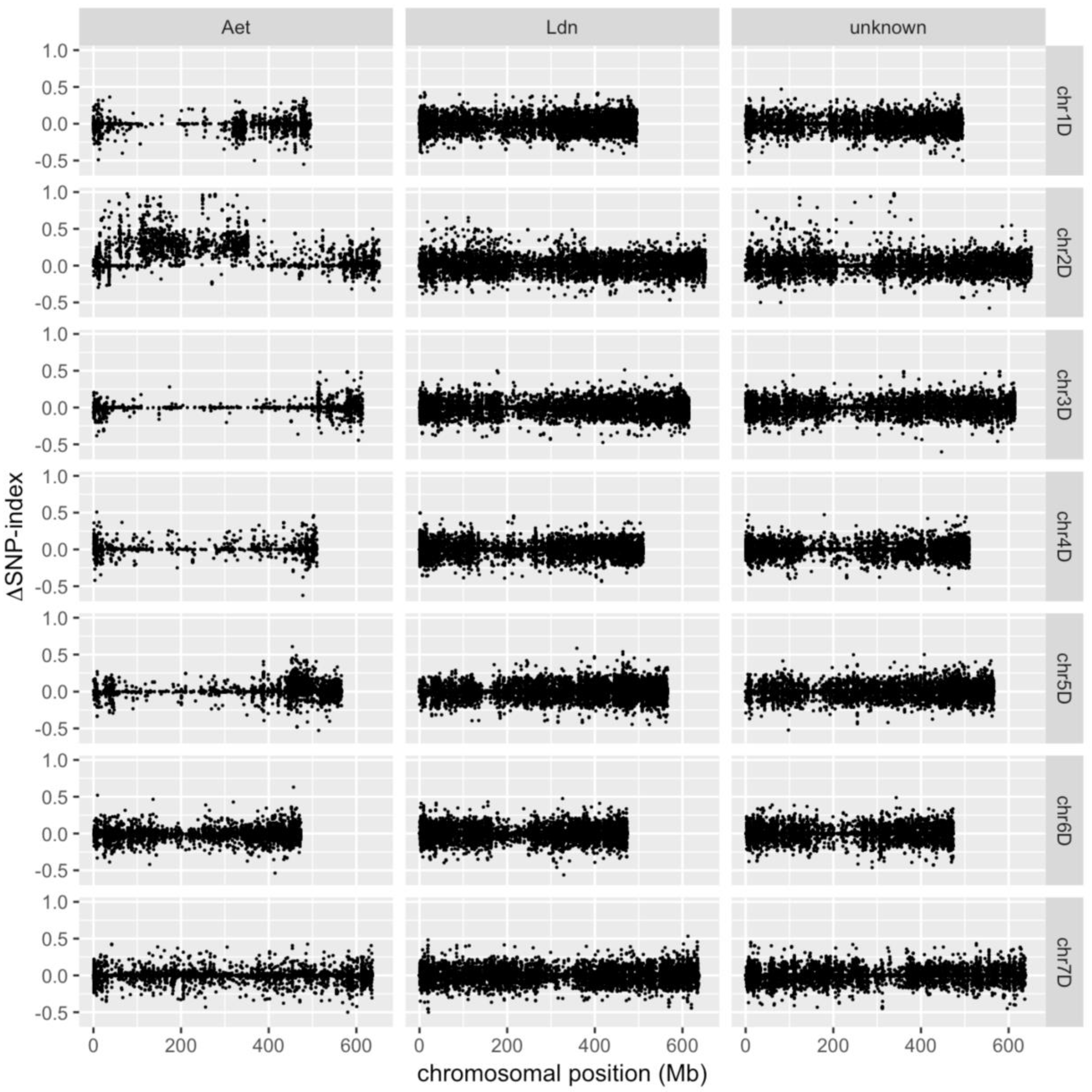
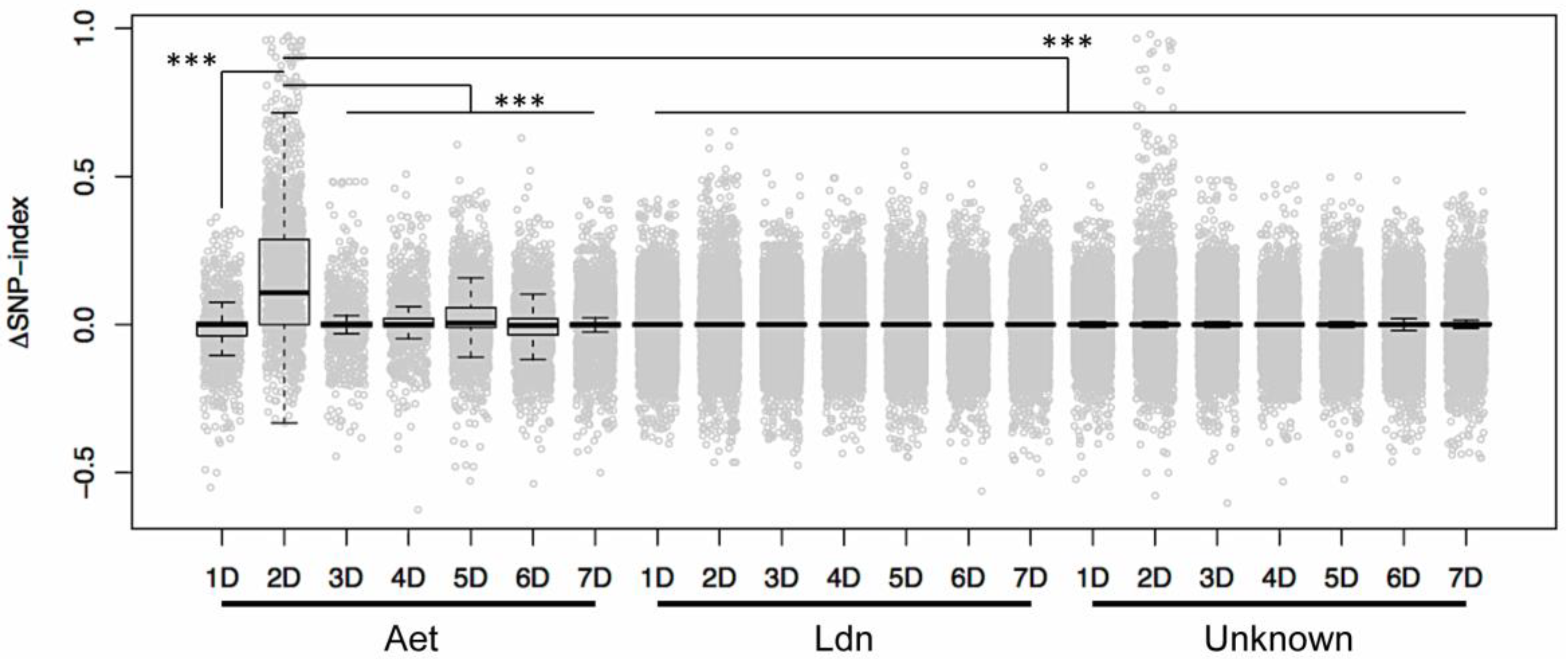
2. Results and Discussion
3. Materials and Methods
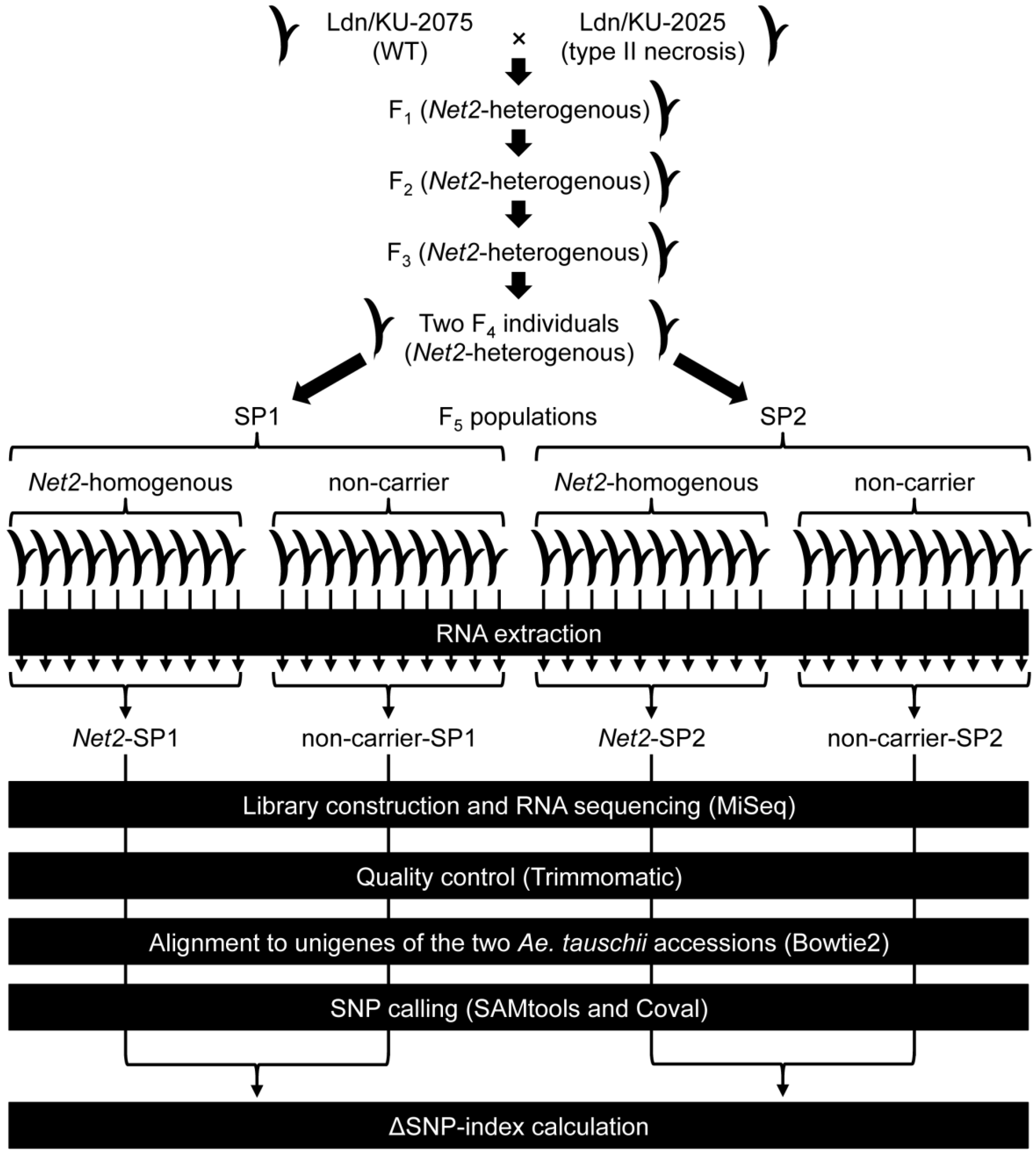
3.1. Plant Materials
3.2. Library Construction and RNA Sequencing
3.3. Alignment of RNA-seq Reads to de Novo Assembled Transcripts of the Parental Ae. tauschii Accessions
3.4. Identification of D-genome Specific SNPs and Calculation of ΔSNP-Index
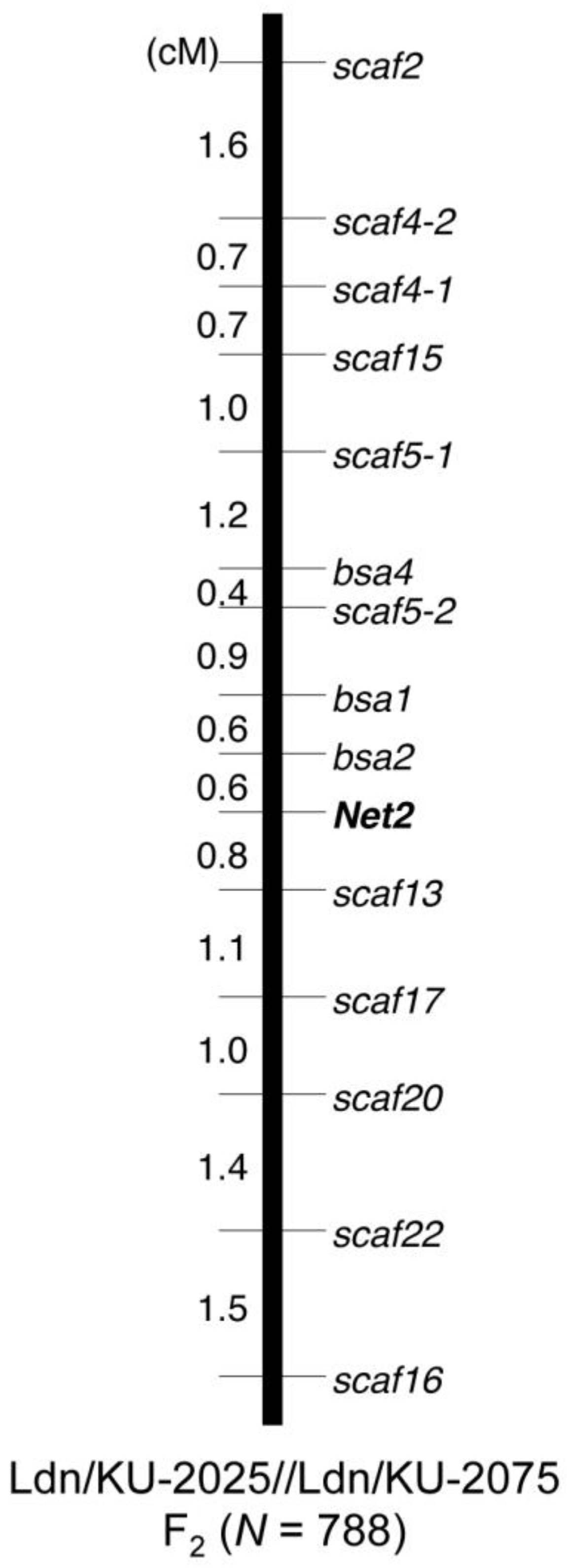
3.5. Molecular Marker Development and Genotyping
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| RNA-seq | RNA sequencing |
| NGS | next-generation sequencing |
| BSA | bulked segregant analysis |
| SNP | single nucleotide polymorphism |
| Indel | insertion/deletion |
| NR | non-redundant |
| SP | Segregating Population |
| dCAPS | derived cleaved amplified polymorphic sequence |
| Ldn | Langdon |
References
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, M.C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Nasuda, S. Durum wheat as a candidate for the unknown female progenitor of bread wheat: An empirical study with a highly fertile F1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 2004, 109, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Zohary, D.; Harlan, J.R.; Vardi, A. The wild diploid progenitors of wheat and their breeding value. Euphytica 1969, 18, 58–65. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Rosas, V.; Roldan, S. Conservation of the genetic variation of Triticum tauschii (Coss.) Schmalh. (Aegilops squarrosa auct. non L.) in synthetic hexaploid wheats (T. turgidum L. s.lat. x T. tauschii; 2n=6x=42, AABBDD) and its potential utilization for wheat improvement. Genet. Resour. Crop Evol. 1996, 43, 129–134. [Google Scholar] [CrossRef]
- Jones, H.; Gosman, N.; Horsnell, R.; Rose, G.A.; Everst, L.A.; Bentley, A.R.; Tha, S.; Uauy, C.; Kowalski, A.; Novoselovic, D.; et al. Strategy for exploiting exotic germplasm using genetic, morphological, and environmental diversity: The Aegilops tauschii Coss. example. Theor. Appl. Genet. 2013, 126, 1793–1808. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Yorgancilar, A.; Baenziger, P.S. Genome-wide association study reveals favorable alleles associated with common bunt resistance in synthetic hexaploid wheat. Euphytica 2018, 214, 200. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Baenziger, P.S. Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int. J. Mol. Sci. 2018, 19, 3011. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-wide association study reveals novel genomic regions associated with 10 grain minerals in synthetic hexaploid wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef] [PubMed]
- Gorafi, Y.S.A.; Kim, J.S.; Elbashir, A.A.E.; Tsujimoto, H. A population of wheat multiple synthetic derivatives: An effective platform to explore, harness and utilize genetic diversity of Aegilops tauschii for wheat improvement. Theor. Appl. Genet. 2018, 131, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Dreisigacker, S.; Melchinger, A.E.; Reif, J.C.; Mujeeb-Kazi, A.; Van Ginkel, M.; Hoisington, D.; Warburton, M.L. Quantifying novel sequence variation and selective advantage in synthetic hexaploid wheats and their backcross-derived lines using SSR markers. Mol. Breed. 2005, 15, 1–10. [Google Scholar] [CrossRef]
- Jafarzadeh, J.; Bonnett, D.; Jannink, J.L.; Akdemir, D.; Dreisigacker, S.; Sorrells, M.E. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS ONE 2016, 11, e0162860. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Poland, J.; Baenziger, P.S. Unlocking the novel genetic diversity and population structure of synthetic hexaploid wheat. BMC Genom. 2018, 19, 591. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.C.; He, Z.; Rajaram, S. Wheat genetic resources in the post-genomics era: Promise and challenges. Ann. Bot. 2018, 121, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Van Slageren, M.W. Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae); Wageningen Agricultural University: Wageningen, The Netherlands, 1994; pp. 326–344. ISBN 90-6754-377-2. [Google Scholar]
- Matsuoka, Y.; Takumi, S.; Kawahara, T. Flowering time diversification and dispersal in central Eurasian wild wheat Aegilops tauschii Coss.: Genealogical and ecological framework. PLoS ONE 2008, 3, e3138. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, N.; Yamasaki, M.; Matsuoka, Y.; Kawahara, T.; Takumi, S. Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: Implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 2010, 19, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Kawahara, T.; Takumi, S. Intraspecific lineage divergence and its association with reproductive trait change during species range expansion in central Eurasian wild wheat Aegilops tauschii Coss. (Poaceae). BMC Evol. Biol. 2015, 15, 213. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Takumi, S. The role of reproductive isolation in allohexaploid speciation pattern: Empirical insights from the progenitors of common wheat. Sci. Rep. 2017, 7, 16004. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, N.; Hosogi, N.; Park, P.; Takumi, S. Hypersensitive response-like reaction is associated with hybrid necrosis in interspecific crosses between tetraploid wheat and Aegilops tauschii Coss. PLoS ONE 2010, 5, e11326. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, N.; Shitsukawa, N.; Hosogi, N.; Park, P.; Takumi, S. Autoimmune response and repression of mitotic cell division occur in inter-specific crosses between tetraploid wheat and Aegilops tauschii Coss. that show low temperature-induced hybrid necrosis. Plant J. 2011, 68, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; Nishijima, R.; Iehisa, J.C.M.; Takumi, S. Fine mapping and genetic association analysis of Net2, the causative D-genome locus of low temperature-induced hybrid necrosis in interspecific crosses between tetraploid wheat and Aegilops tauschii. Genetica 2016, 144, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, G.; Saito, M.; Tanaka, T.; Katayose, Y.; Kanamori, H.; Kurita, K.; Nakamura, T. An efficient approach for the development of genome-specific markers in allohexaploid wheat (Triticum aestivum L.) and its application in the construction of high-density linkage maps of the D genome. DNA Res. 2018, 25, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Iehisa, J.C.M.; Shimizu, A.; Sato, K.; Nasuda, S.; Takumi, S. Discovery of high-confidence single nucleotide polymorphisms from large-scale de novo analysis of leaf transcripts of Aegilops tauschii, a wild wheat progenitor. DNA Res. 2012, 19, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Iehisa, J.C.M.; Shimizu, A.; Sato, K.; Nishijima, R.; Sakaguchi, K.; Matsuda, R.; Nasuda, S.; Takumi, S. Genome-wide marker development for the wheat D genome based on single nucleotide polymorphisms identified from transcripts in the wild wheat progenitor Aegilops tauschii. Theor. Appl. Genet. 2014, 127, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, R.; Yoshida, K.; Motoi, Y.; Sato, K.; Takumi, S. Genome-wide identification of novel genetic markers from RNA sequencing assembly of diverse Aegilops tauschii accessions. Mol. Genet. Genom. 2016, 291, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Schneeberge, K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014, 15, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef] [PubMed]
- Su, A.; Song, W.; Xing, J.; Zhao, Y.; Zhang, R.; Li, C.; Duan, M.; Luo, M.; Shi, Z.; Zhao, J. Identification of genes potentially associated with the fertility instability of S-type cytoplasmic male sterility in maize via bulked segregant RNA-Seq. PLoS ONE 2016, 11, e0163489. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhu, J.; Su, H.; Huang, M.; Wang, H.; Ding, S.; Zhang, B.; Luo, A.; Wei, S.; Tian, X.; et al. Bulked segregant RNA-seq revealed differential expression and SNPs of candidate genes associated with waterlogging tolerance in maize. Front. Plant Sci. 2017, 8, 1022. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Trick, M.; Adamski, N.M.; Mugford, S.G.; Jiang, C.C.; Febrer, M.; Uauy, C. Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 2012, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gonzalez, R.H.; Segovia, V.; Bird, N.; Fenwick, P.; Holdgate, S.; Bery, S.; Jack, P.; Caccamo, M.; Uauy, C. RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol. J. 2015, 13, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Xie, J.; Hu, J.; Qiu, D.; Liu, Z.; Li, J.; Li, M.; Zhang, H.; Yang, L.; Liu, H.; et al. Development of molecular markers linked to powdery mildew resistance gene Pm4b by combining SNP discovery from transcriptome sequencing data with bulked segregant analysis (BSR-Seq) in wheat. Front. Plant Sci. 2018, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.-C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Ywardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Yoshida, K.; Takumi, S. Hybrid incompatibilities in interspecific crosses between tetraploid wheat and its wild relative Aegilops umbellulata. Plant Mol. Biol. 2017, 95, 625–645. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Yoshida, K.; Mizuno, N.; Nasuda, S.; Sato, K.; Takumi, S. Origin of the wheat B-genome chromosomes conferred by RNA sequencing analysis of leaf transcripts in the section Sitopsis species of Aegilops. DNA Res. 2018. under review. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Subgroup 1000 Genome Project Data Processing. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Natsume, S.; Yoshida, K.; MacLean, D.; Cano, L.; Kamoun, S.; Terauchi, R. Coval: Improving alignment quality and variant calling accuracy for next-generation sequencing data. PLoS ONE 2013, 8, e75402. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, T.; Murai, K.; Takumi, S. Distinct genetic regulation of flowering time and grain-filling period based on empirical study of D genome diversity in synthetic hexaploid wheat lines. Breed. Sci. 2011, 61, 130–141. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com/ (accessed on 1 November 2016).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 1 November 2016).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
| Samples | Total Read Pairs | Filtered Read Pairs (%) a | Aligned to the Ae. tauschii Transcripts b (%) c | |
|---|---|---|---|---|
| KU-2075 | KU-2025 | |||
| Synthetic hexaploids | ||||
| non-carrier-SP1-1st | 4,202,114 | 2,799,202 (66.61%) | 2,020,405 (72.18%) | 1,930,400.5 (68.96%) |
| non-carrier-SP1-2nd | 4,059,840 | 2,858,956 (70.42%) | 2,062,291 (72.13%) | 1,971,110.5 (68.95%) |
| non-carrier-SP2-1st | 4,492,358 | 2,953,088 (65.74%) | 2,164,656 (73.3%) | 2,083,120 (70.54%) |
| non-carrier-SP2-2nd | 4,115,352 | 2,864,271 (69.60%) | 2,098,383 (73.26%) | 2,020,752.5 (70.55%) |
| Net2-SP1-1st | 4,710,499 | 3,148,652 (66.84%) | 2,208,392 (70.14%) | 2,110,452.5 (67.03%) |
| Net2-SP1-2nd | 4,403,630 | 3,108,568 (70.59%) | 2,178,403 (70.08%) | 2,082,004 (66.98%) |
| Net2-SP2-1st | 4,828,182 | 3,249,596 (67.30%) | 2,420,056 (74.47%) | 2,348,814 (72.28%) |
| Net2-SP2-2nd | 5,216,082 | 3,709,478 (71.12%) | 2,763,471 (74.5%) | 2,684,019 (72.36%) |
| Tetraploid wheat | ||||
| cv. Langdon | 6,316,174 | 4,372,660 (69.23%) | 2,974,277 (68.02%) | 2,661,487 (60.87%) |
| Transcripts a | KU-2075 | KU-2025 | ||
|---|---|---|---|---|
| The Number of SNP | Total | Anchored to the Genome b (%) | Total | Anchored to the Genome b (%) |
| Synthetic hexaploids | ||||
| non-carrier-SP1 | 277,605 | 275,799 (99.35%) | 262,966 | 261,128 (99.30%) |
| non-carrier-SP2 | 276,564 | 274,772 (99.35%) | 269,175 | 267,249 (99.28%) |
| Net2-SP1 | 318,046 | 315,859 (99.31%) | 296,819 | 294,739 (99.30%) |
| Net2-SP2 | 298,496 | 296,419 (99.30%) | 285,798 | 283,684 (99.26%) |
| Tetraploid wheat | ||||
| cv. Langdon | 429,346 | 421,957 (98.28%) | 350,871 | 345,657 (98.51%) |
| Chr. | D-genome-Specific | Homoeologous | Unclassified | Total |
|---|---|---|---|---|
| 1D | 1674 | 29,307 | 11,760 | 42,741 |
| 2D | 2295 | 33,822 | 13,975 | 50,092 |
| 3D | 1611 | 31,932 | 15,532 | 49,075 |
| 4D | 1698 | 28,781 | 11,776 | 42,255 |
| 5D | 2936 | 34,961 | 14,336 | 52,233 |
| 6D | 3730 | 24,534 | 10,966 | 39,230 |
| 7D | 3983 | 28,593 | 11,606 | 44,182 |
| Total | 17,927 | 211,930 | 89,951 | 319,808 |
| Marker Name | Primer Sequence (5′ to 3′) | Restriction Enzyme |
|---|---|---|
| bsa1 | TCATGACCTGCTGGTTTGTT | StyI |
| GATTCCAATGTTATTTCTGAACCCT | ||
| bsa2 | TCACAACATTCGCAGGTCAT | HpaII |
| TGGTTCTGTTGATCTCACTGCC | ||
| bsa4 | ACAAGTCGGATATCGCCAAA | HinfI |
| CAGCTAAAAACTGTTTGCTTGAGA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishijima, R.; Yoshida, K.; Sakaguchi, K.; Yoshimura, S.-i.; Sato, K.; Takumi, S. RNA Sequencing-Based Bulked Segregant Analysis Facilitates Efficient D-genome Marker Development for a Specific Chromosomal Region of Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3749. https://doi.org/10.3390/ijms19123749
Nishijima R, Yoshida K, Sakaguchi K, Yoshimura S-i, Sato K, Takumi S. RNA Sequencing-Based Bulked Segregant Analysis Facilitates Efficient D-genome Marker Development for a Specific Chromosomal Region of Synthetic Hexaploid Wheat. International Journal of Molecular Sciences. 2018; 19(12):3749. https://doi.org/10.3390/ijms19123749
Chicago/Turabian StyleNishijima, Ryo, Kentaro Yoshida, Kohei Sakaguchi, Shin-ichi Yoshimura, Kazuhiro Sato, and Shigeo Takumi. 2018. "RNA Sequencing-Based Bulked Segregant Analysis Facilitates Efficient D-genome Marker Development for a Specific Chromosomal Region of Synthetic Hexaploid Wheat" International Journal of Molecular Sciences 19, no. 12: 3749. https://doi.org/10.3390/ijms19123749
APA StyleNishijima, R., Yoshida, K., Sakaguchi, K., Yoshimura, S.-i., Sato, K., & Takumi, S. (2018). RNA Sequencing-Based Bulked Segregant Analysis Facilitates Efficient D-genome Marker Development for a Specific Chromosomal Region of Synthetic Hexaploid Wheat. International Journal of Molecular Sciences, 19(12), 3749. https://doi.org/10.3390/ijms19123749





