Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella)
Abstract
1. Introduction
2. Results
2.1. Enrichment and Morphological Identification of Head Kidney Primary Cells
2.2. Survey on the Proposed Macrophage Markers Expression and the Respiratory Burst Activity
2.3. Characteristics of the Plastid Genomes
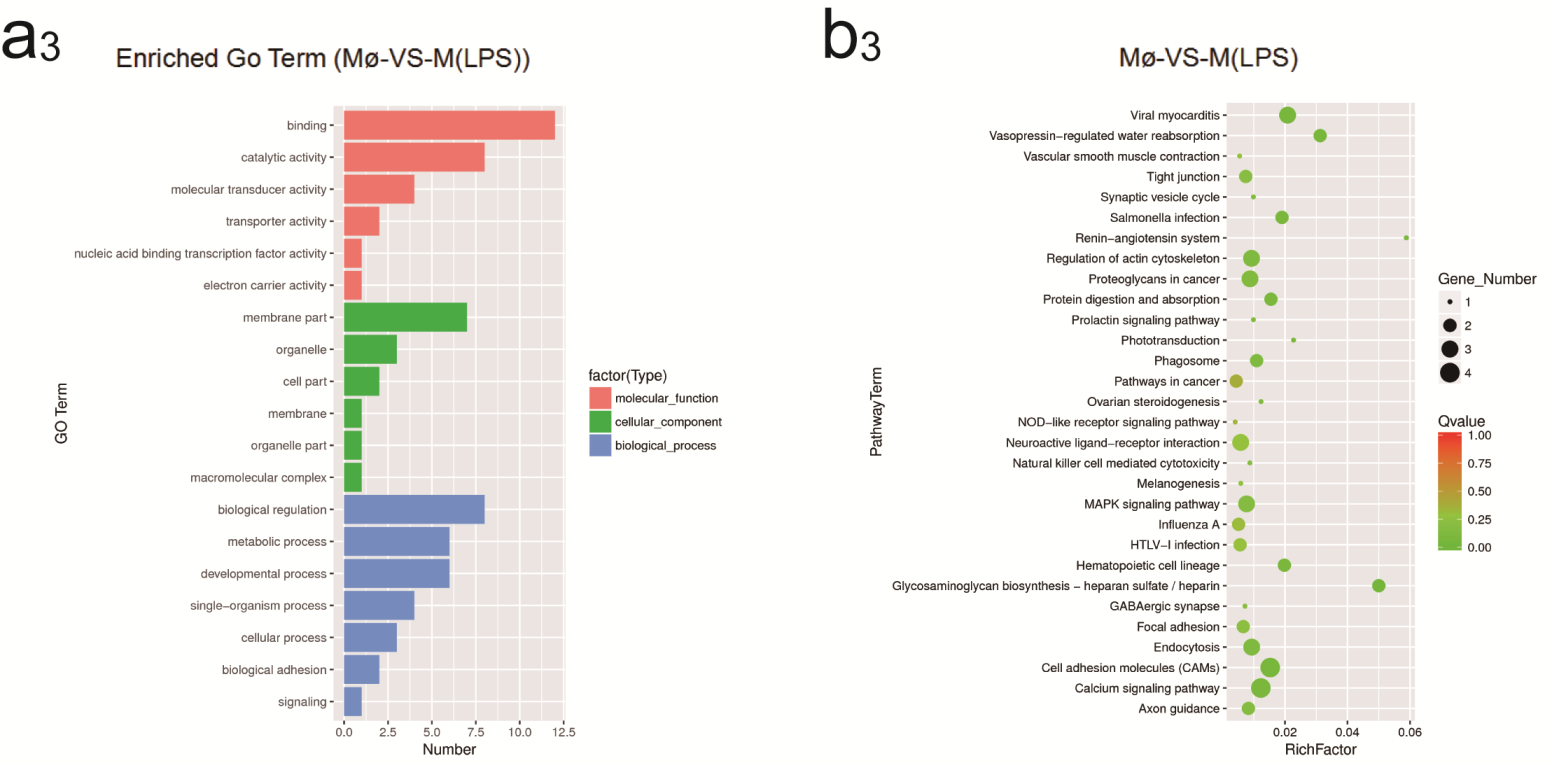
2.4. Identification of GO and KEGG Enrichment Analysis
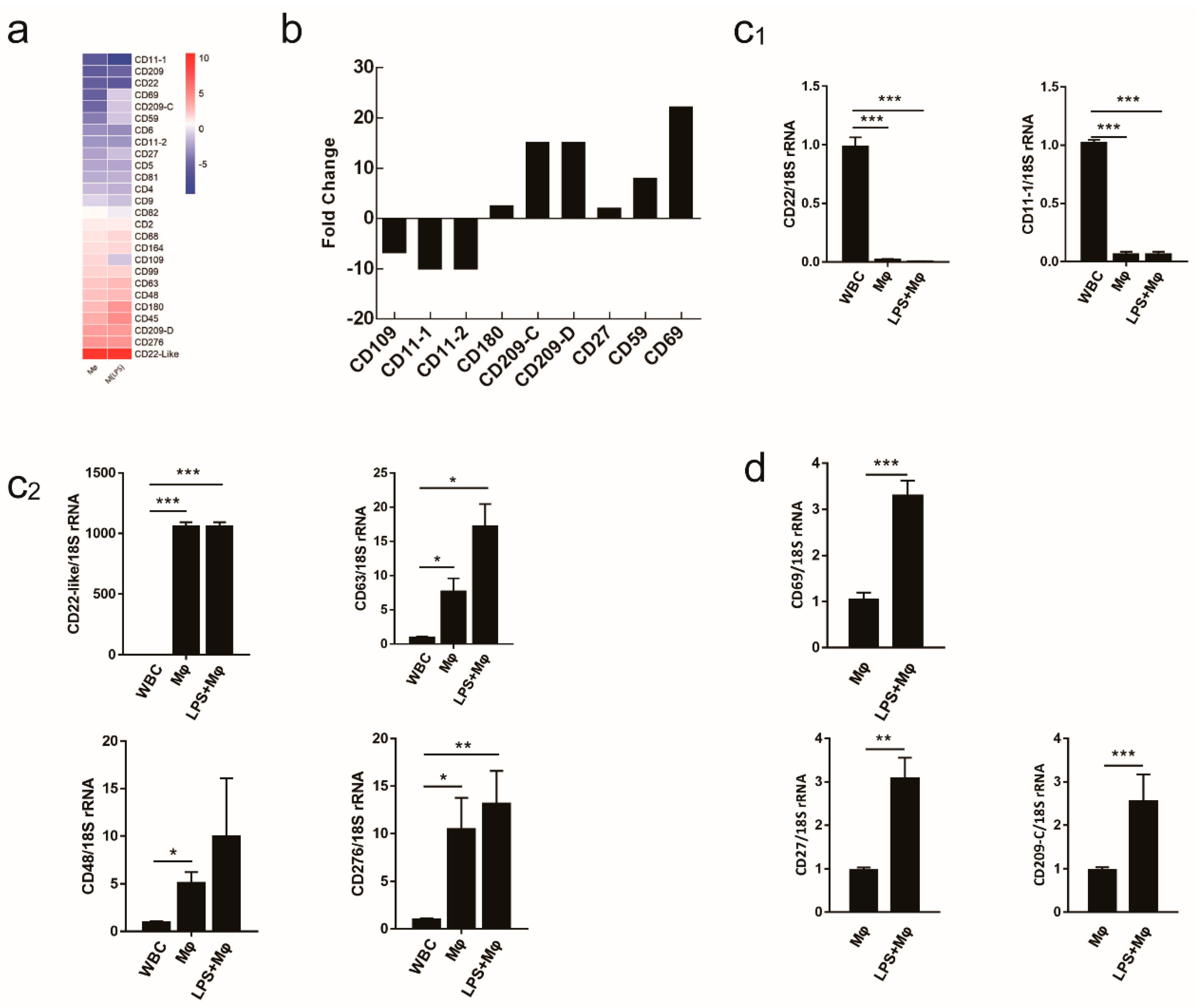
2.5. Evaluating the Use of CD Molecule Transcripts as Macrophages Markers
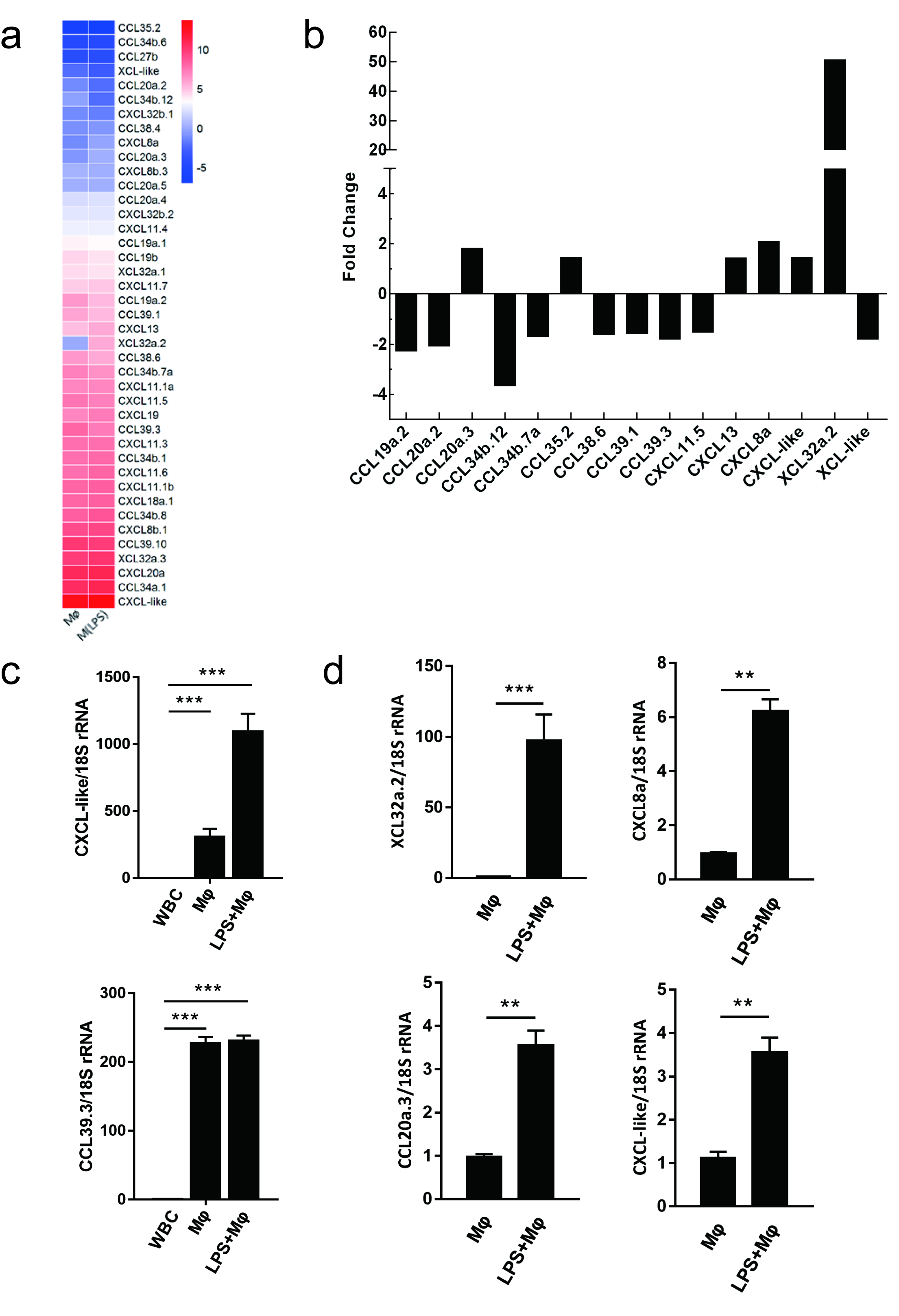
2.6. Evaluating the Use of Chemokine Transcripts as Macrophage Markers
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Fish Rearing
4.3. Isolation of Head-Kidney Leukocytes and Macrophages
4.4. Respiratory Burst Activity
4.5. RNA Extraction and RNA Sequencing by Illumina Hiseq
4.6. Differential Gene Expression Analysis
4.7. Confirmation of Gene Expression Profiles by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability
Acknowledgments
Conflicts of Interest
References
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Hume, D.A.; Allan, W.; Fabrus, B.; Weidemann, M.J.; Hapel, A.J.; Bartelmez, S. Regulation of proliferation of bone marrow-derived macrophages. Lymphokine Res. 1987, 6, 127–139. [Google Scholar] [PubMed]
- McDonald, L.; Mehrotra, M.; LaRue, A. Hematopoietic origin of murine lung fibroblasts. Stem Cells Int. 2015, 2015, 159713. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Mulero, V.; Meseguer, J.; Ayala, A.G. Professional phagocytic granulocytes of the bony fish gilthead seabream display functional adaptation to testicular microenvironment. J. Leukoc. Biol. 2005, 78, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Hsieh, M.S.; Liao, H.A.; Yang, Y.J.; Ho, Y.J.; Lin, L.I. Zebrafish as a Model for the Study of Human Myeloid Malignancies. Biomed Res. Int. 2015, 2015, 641475. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Chen, Q.; Rong, Y.J.; Chen, F.; Chen, J. CXCR3.1 and CXCR3.2 Differentially Contribute to Macrophage Polarization in Teleost Fish. J. Immunol. 2017, 198, 4692–4706. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.N.; Bi, C.S.; Gao, L.N.; An, Y.; Chen, F.; Chen, F.M. Macrophage polarization in human gingival tissue in response to periodontal disease. Oral Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.; Martinez-Pomares, L.; Stacey, M.; Lin, H.; Brown, G.; Gordon, S. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2017, 199, 2194–2201. [Google Scholar] [CrossRef] [PubMed]
- Hume, D.A. The many alternative faces of macrophage activation. Front. Immunol. 2015, 6, 370. [Google Scholar] [CrossRef] [PubMed]
- Kollipara, R.K.; Perumal, N.B. Motif prediction to distinguish LPS-stimulated pro-inflammatory vs. antibacterial macrophage genes. Immunome Res. 2010, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.; Marinov, G.; Chin, Y.; Lim, Y.; Ea, C. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci. Rep. 2017, 7, 12227. [Google Scholar] [CrossRef] [PubMed]
- Menchetti, L.; Barbato, O.; Filipescu, I.; Traina, G.; Leonardi, L.; Polisca, A.; Troisi, A.; Guelfi, G.; Piro, F.; Brecchia, G. Effects of local lipopolysaccharide administration on the expression of Toll-like receptor 4 and pro-inflammatory cytokines in uterus and oviduct of rabbit does. Theriogenology 2017, 107, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.H.Y.; Bebien, M.; Didierlaurent, A.; Nebauer, R.; Hussell, T.; Broide, D.; Karin, M.; Lawrence, T. An antiinflammatory role for IKK beta through the inhibition of “classical” macrophage activation. J. Exp. Med. 2008, 205, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.G.; Du, Y.; Perrigoue, J.G.; Zaph, C.; Taylor, J.J.; Goldschmidt, M.; Swain, G.P.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.; et al. Alternatively activated macrophage-derived RELM-alpha is a negative regulator of type 2 inflammation in the lung. J. Exp. Med. 2009, 206, 937–952. [Google Scholar] [CrossRef] [PubMed]
- El Kasmi, K.C.; Qualls, J.E.; Pesce, J.T.; Smith, A.M.; Thompson, R.W.; Henao-Tamayo, M.; Basaraba, R.J.; Koenig, T.; Schleicher, U.; Koo, M.-S.; et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008, 9, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, E.; Zordan, P.; Sciorati, C.; Rovere-Querini, P.; Brunelli, S. Macrophage plasticity in skeletal muscle repair. Biomed Res. Int. 2014, 2014, 560629. [Google Scholar] [CrossRef] [PubMed]
- Kreider, T.; Anthony, R.M.; Urban, J.F., Jr.; Gause, W.C. Alternatively activated macrophages in helminth infections. Curr. Opin. Immunol. 2007, 19, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.M.; Urban, J.F., Jr.; Alem, F.; Hamed, H.A.; Rozo, C.T.; Boucher, J.-L.; van Rooijen, N.; Gause, W.C. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat. Med. 2006, 12, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative Activation of Macrophages: An Immunologic Functional Perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.D.; Jiang, C.C.; Matta, B.; Tietzel, I.; Watkins, S.K.; Suttles, J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J. Immunol. 2005, 175, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.D.; Suttles, J. Functional plasticity of macrophages: Reversible adaptation to changing microenvironments. J. Leukoc. Biol. 2004, 76, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Wang, B.; Yang, F.; Chen, L. Blockade of Interleukin-7 Receptor Shapes Macrophage Alternative Activation and Promotes Functional Recovery After Spinal Cord Injury. Neuroscience 2018, 371, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Barreda, D.; Belosevic, M. Transcriptional regulation of hemopoiesis. Dev. Comp. Immunol. 2001, 25, 763–789. [Google Scholar] [CrossRef]
- Murray, P.; Allen, J.; Biswas, S.; Fisher, E.; Gilroy, D.; Goerdt, S.; Gordon, S.; Hamilton, J.; Ivashkiv, L.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Hu, Y.; Wei, X.; Xiao, X.; Jakovlic, I.; Liu, X.; Su, J.; Yuan, G. Chemotactic effect of beta-defensin 1 on macrophages in Megalobrama arnblycephala. Fish Shellfish Immunol. 2018, 74, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lu, X.; Li, M.; Chen, J. Molecular cloning, pathologically-correlated expression and functional characterization of the colonystimulating factor 1 receptor (CSF-1R) gene from a teleost, Plecoglossus altivelis. Zool. Res. 2016, 37, 96–102. [Google Scholar] [PubMed]
- Deng, F.; Chen, W.; Liu, L.; Wang, L.; Chen, X. The expression and molecular mechanism of M1 macrophages in rheumatic valvular disease. Zhonghua Wai Ke Za Zhi 2012, 50, 933–937. [Google Scholar] [PubMed]
- Wiegertjes, G.F.; Wentzel, A.S.; Spaink, H.P.; Elks, P.M.; Fink, I.R. Polarization of immune responses in fish: The ‘macrophages first’ point of view. Mol. Immunol. 2016, 69, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Sudan, B.; Wacker, M.A.; Wilson, M.E.; Graff, J.W. A systematic approach to identify markers of distinctly activated human macrophages. Front. Immunol. 2015, 6, 253. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.W.; Wan, Q.Y.; Xiao, X.; Ji, J.F.; Su, J.G. A systematic investigation on the composition, evolution and expression characteristics of chemokine superfamily in grass carp Ctenopharyngodon Idella. Dev. Comp. Immunol. 2018, 82, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Holden, J.A.; Attard, T.J.; Laughton, K.M.; Mansell, A.; O’Brien-Simpson, N.M.; Reynolds, E.C. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect. Immun. 2014, 82, 4190–4203. [Google Scholar] [CrossRef] [PubMed]
- Lachmandas, E.; Boutens, L.; Ratter, J.; Hijmans, A.; Hooiveld, G.; Joosten, L.; Rodenburg, R.; Fransen, J.; Houtkooper, R.; van Crevel, R.; et al. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat. Microbiol. 2016, 2, 16246. [Google Scholar] [CrossRef] [PubMed]
- Sepulcre, M.P.; Alcaraz-Perez, F.; Lopez-Munoz, A.; Roca, F.J.; Meseguer, J.; Cayuela, M.L.; Mulero, V. Evolution of lipopolysaccharide (LPS) recognition and signaling: Fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 2009, 182, 1836–1845. [Google Scholar] [CrossRef] [PubMed]
- Holen, E.; Lie, K.K.; Araujo, P.; Olsvik, P.A. Pathogen recognition and mechanisms in Atlantic cod (Gadus morhua) head kidney cells Bacteria (LPS) and virus (poly I:C) signals through different pathways and affect distinct genes. Fish Shellfish Immunol. 2012, 33, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Seppola, M.; Mikkelsen, H.; Johansen, A.; Steiro, K.; Myrnes, B.; Nilsen, I.W. Ultrapure LPS induces inflammatory and antibacterial responses attenuated in vitro by exogenous sera in Atlantic cod and Atlantic salmon. Fish Shellfish Immunol. 2015, 44, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Wang, Q.; Li, J.; Wang, X.; Wang, Y.; Du, H. Iron reduces M1 macrophage polarization in RAW264.7 macrophages associated with inhibition of STAT1. Mediat. Inflamm. 2017, 2017, 8570818. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, Y.; Zhao, X.; Wang, H.; Wang, L.; Yuan, G.; Asim, M.; Wang, W.; Zeng, L.; Liu, X.; et al. Oligochitosan stimulated phagocytic activity of macrophages from blunt snout bream (Megalobrama amblycephala) associated with respiratory burst coupled with nitric oxide production. Dev. Comp. Immunol. 2014, 47, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Peng, X.; Lv, B.; Wang, P.; Zhao, X.; Yu, B. Geraniin inhibits LPS-induced THP-1 macrophages switching to M1 phenotype via SOCS1/NF-κB pathway. Inflammation 2016, 39, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yin, L.; Feng, S.; Wang, X.; Yang, K.; Zhang, A.; Zhou, H. Dual-parallel inhibition of IL-10 and TGF-β1 controls LPS-induced inflammatory response via NF-κB signaling in grass carp monocytes/macrophages. Fish Shellfish Immunol. 2015, 44, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Kalish, S.; Lyamina, S.; Usanova, E.; Manukhina, E.; Larionov, N.; Malyshev, I. Macrophages reprogrammed in vitro towards the M1 phenotype and activated with LPS extend lifespan of mice with ehrlich ascites carcinoma. Med. Sci. Monit. Basic Res. 2015, 21, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Guo, Y.; So, K.; Vijayan, M.; Guo, Y.; Wong, V.; Yao, Y.; Lee, K.; Chiu, P.; Yeung, W. Soluble human leukocyte antigen G5 polarizes differentiation of macrophages toward a decidual macrophage-like phenotype. Hum. Reprod. 2015, 30, 2263–2274. [Google Scholar] [CrossRef] [PubMed]
- Soldano, S.; Pizzorni, C.; Paolino, S.; Trombetta, A.; Montagna, P.; Brizzolara, R.; Ruaro, B.; Sulli, A.; Cutolo, M. Alternatively activated (M2) macrophage phenotype is inducible by endothelin-1 in cultured human macrophages. PLoS ONE 2016, 11, e0166433. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Goldenberg, D.; Michel, R.; Rossi, D.; Wallace, D.; Chang, C. Trogocytosis of multiple B-cell surface markers by CD22 targeting with epratuzumab. Blood 2013, 122, 3020–3029. [Google Scholar] [CrossRef] [PubMed]
- Bachanova, V.; Frankel, A.; Cao, Q.; Lewis, D.; Grzywacz, B.; Verneris, M.; Ustun, C.; Lazaryan, A.; McClune, B.; Warlick, E.; et al. Phase I study of a bispecific ligand-directed toxin targeting CD22 and CD19 (DT2219) for refractory B-cell malignancies. Clin. Cancer Res. 2015, 21, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Hassuna, N.; Monk, P.; Ali, F.; Read, R.; Partridge, L. A role for the tetraspanin proteins in Salmonella infection of human macrophages. J. Infect. 2017, 75, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Abadía-Molina, A.; Ji, H.; Faubion, W.; Julien, A.; Latchman, Y.; Yagita, H.; Sharpe, A.; Bhan, A.; Terhorst, C. CD48 controls T-cell and antigen-presenting cell functions in experimental colitis. Gastroenterology 2006, 130, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Möller, J.; Lühmann, T.; Chabria, M.; Hall, H.; Vogel, V. Macrophages lift off surface-bound bacteria using a filopodium-lamellipodium hook-and-shovel mechanism. Sci. Rep. 2013, 3, 2884. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, S.; Kasuya, Y.; Kuroda, F.; Tanaka, K.; Tsuyusaki, J.; Yamauchi, K.; Matsunaga, H.; Iwamura, C.; Nakayama, T.; Tatsumi, K. Role of CD69 in acute lung injury. Life Sci. 2012, 90, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Tsertsvadze, T.; Mitskevich, N.; Bilanishvili, A.; Girdaladze, D.; Porakishvili, N. Phagocytosis and expression of FCg-receptors and CD180 on monocytes in chronic lymphocytic leukemia. Georgian Med. News 2017, 270, 88–93. [Google Scholar]
- Yu, C.; Micaroni, M.; Puyskens, A.; Schultz, T.; Yeo, J.; Stanley, A.; Lucas, M.; Kurihara, J.; Dobos, K.; Stow, J.; et al. RP105 engages phosphatidylinositol 3-kinase p110δ to facilitate the trafficking and secretion of cytokines in macrophages during mycobacterial infection. J. Immunol. 2015, 195, 3890–3900. [Google Scholar] [CrossRef] [PubMed]
- Lo, D.; Feng, L.; Li, L.; Carson, M.; Crowley, M.; Pauza, M.; Nguyen, A.; Reilly, C. Integrating innate and adaptive immunity in the whole animal. Immunol. Rev. 1999, 169, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Li, L.; Cai, J.; Huang, W.; Qin, B.; Zhang, S. ORF3 of hepatitis E Virus inhibits the expression of proinflammatory cytokines and chemotactic factors in LPS-stimulated human PMA-THP1 cells by inhibiting NF-κB pathway. Viral Immunol. 2016, 29, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Haghnegahdar, H.; Du, J.; Wang, D.; Strieter, R.M.; Burdick, M.D.; Nanney, L.B.; Cardwell, N.; Luan, J.; Shattuck-Brandt, R.; Richmond, A. The tumorigenic and angiogenic effects of MGSA/GRO proteins in melanoma. J. Leukoc. Biol. 2000, 67, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Davidson, G.; Lin, S.; Secombes, C.; Ellis, A. Detection of specific and ‘constitutive’ antibody secreting cells in the gills, head kidney and peripheral blood leucocytes of dab (Limanda limanda). Vet. Immunol. Immunopathol. 1997, 58, 363–374. [Google Scholar] [CrossRef]
- Bai, Z.; Yin, Y.; Hu, S.; Wang, G.; Zhang, X.; Li, J. Identification of genes involved in immune response, microsatellite, and SNP markers from expressed sequence tags generated from hemocytes of freshwater pearl mussel (Hyriopsis cumingii). Mar. Biotechnol. 2009, 11, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Su, J. Transcriptome analysis provides insights into the regulatory function of alternative splicing in antiviral immunity in grass carp (Ctenopharyngodon idella). Sci. Rep. 2015, 5, 12946. [Google Scholar] [CrossRef] [PubMed]
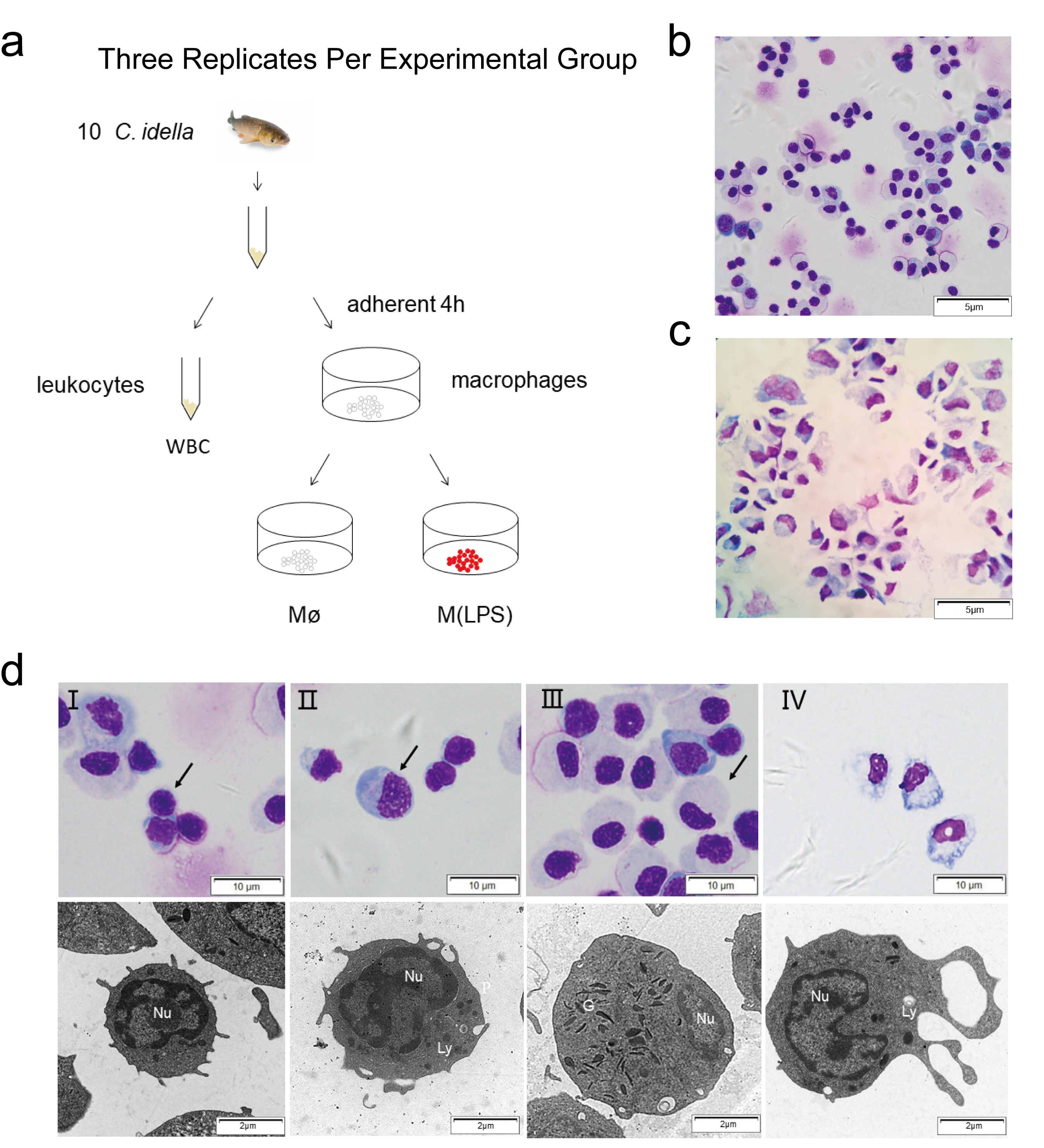
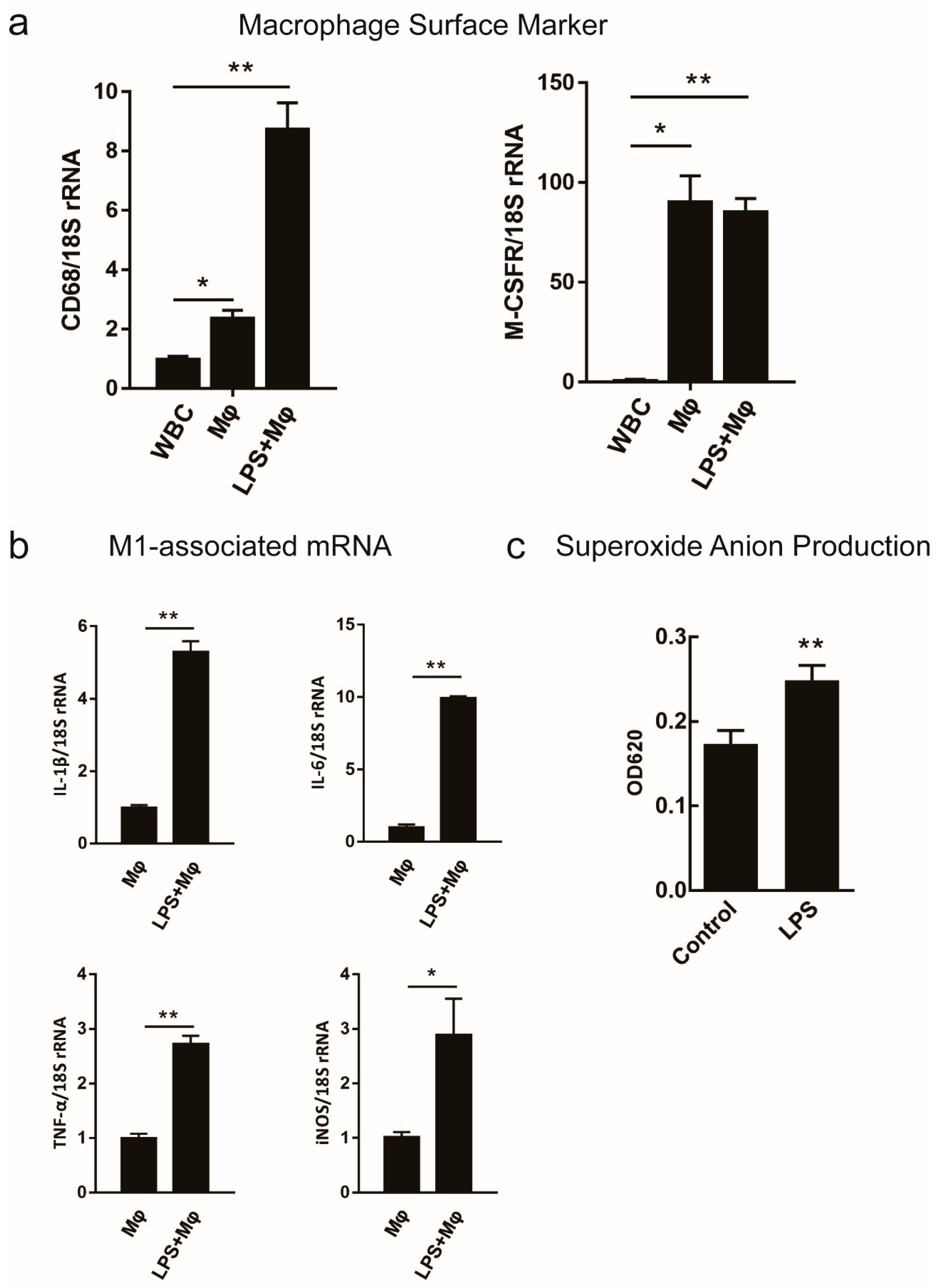
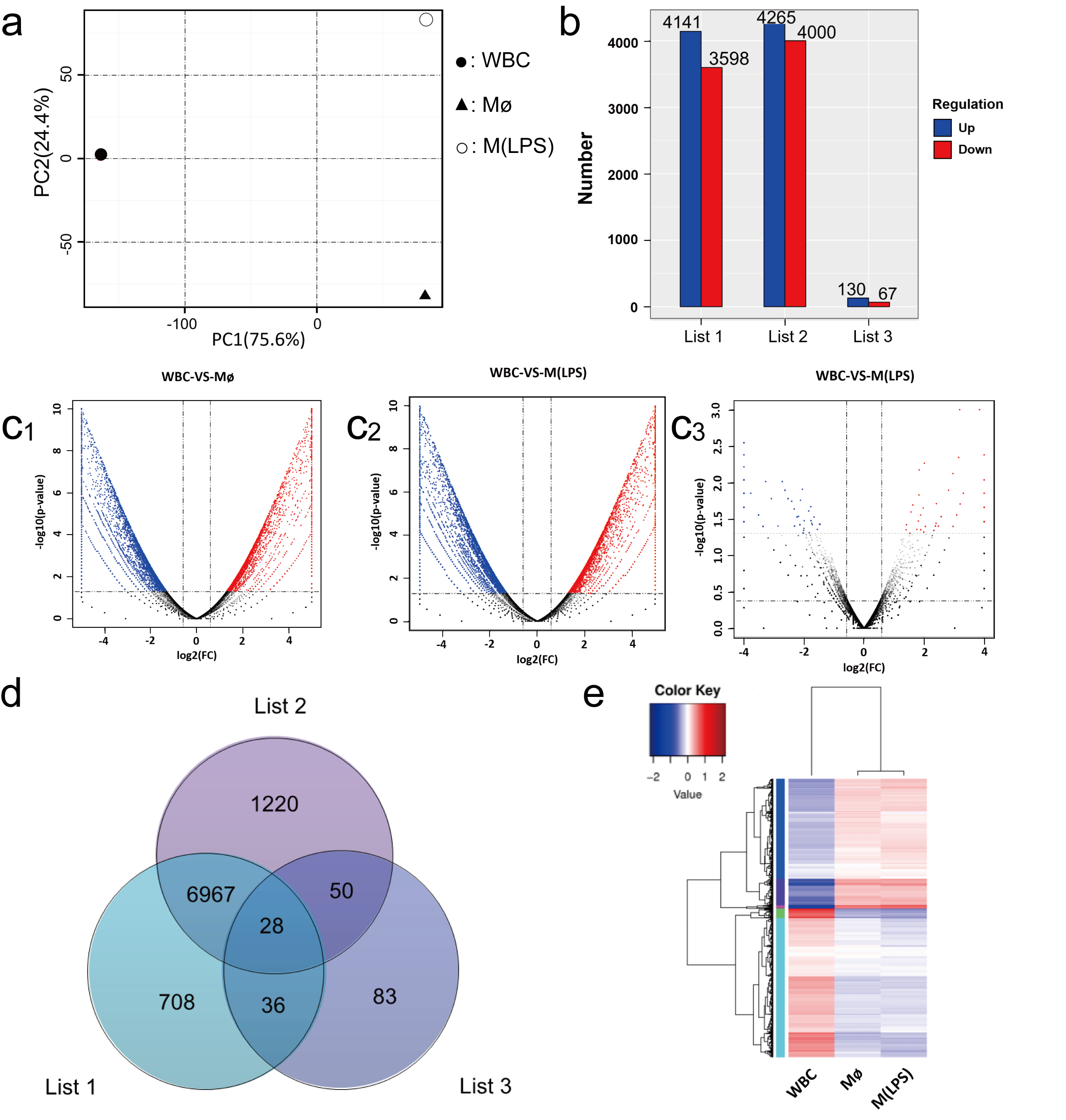
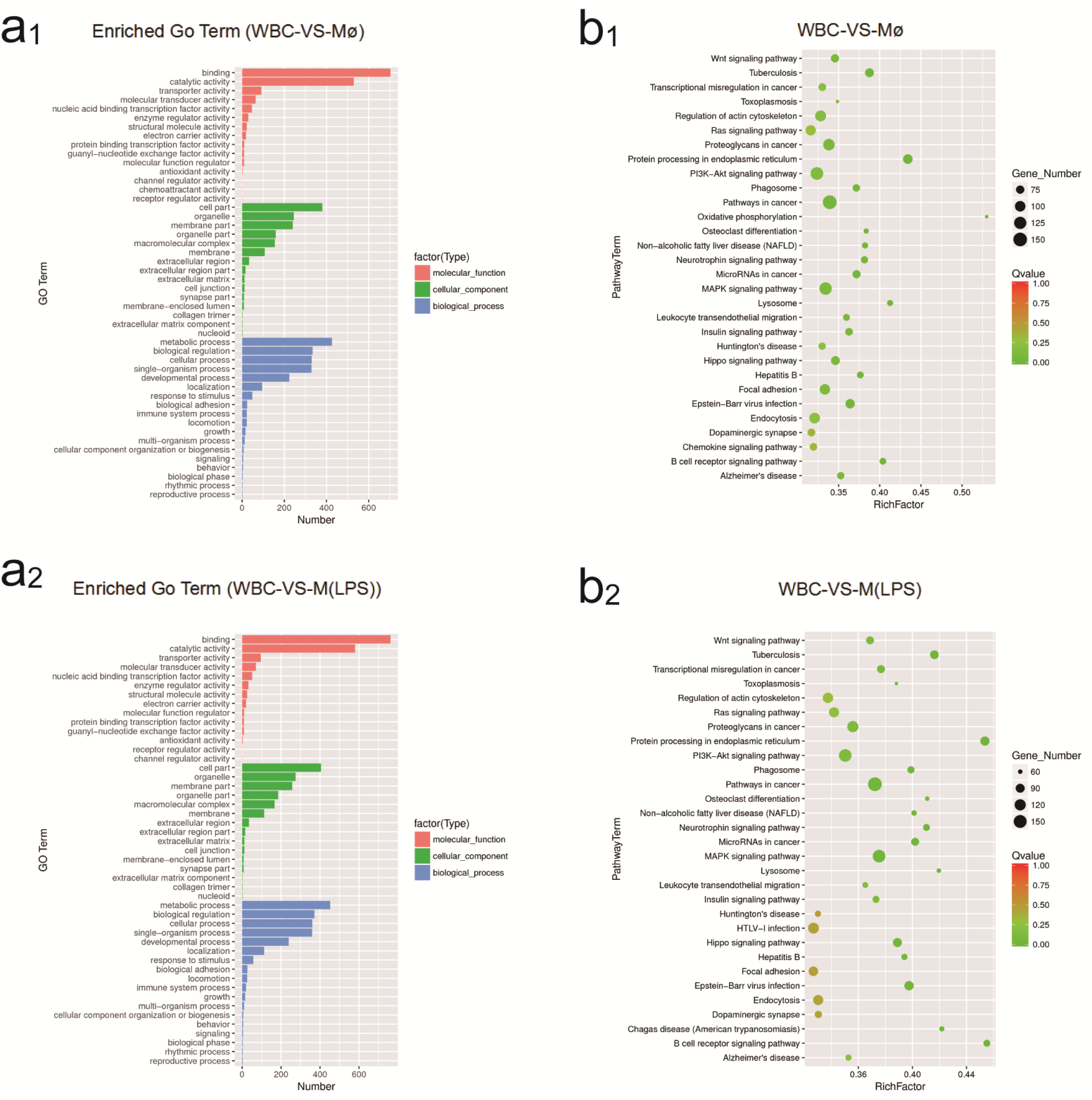
| Transcriptome Analysis Statistics | WBC | Mø | M(LPS) |
|---|---|---|---|
| MiSeq statistics | |||
| Raw reads | 55,358,880 | 42,691,264 | 54,911,804 |
| Average read length (bp) | 150 | 150 | 150 |
| Total base pairs (bp) | 8,303,832,000 | 6,403,689,600 | 8,236,770,600 |
| ≥ Q20 of clean reads (%) | 91.42 | 91.8 | 92.8 |
| Clean reads | 53,792,212 | 41,600,582 | 53,849,176 |
| Average read length (bp) | 146 | 146 | 147 |
| Total base pairs (bp) | 7,871,999,232 | 6,089,281,739 | 7,904,942,691 |
| ≥ Q20 of clean reads (%) | 93.39 | 93.64 | 94.36 |
| Mapping (Analysis of comparison of clean data with reference genomes of C. idella) | |||
| Total mapped (%) | 85.16 | 85.73 | 86.86 |
| Multiple mapped (%) | 4.63 | 4.51 | 4.49 |
| Uniquely mapped (%) | 80.53 | 81.22 | 82.36 |
| Reads map to ‘+’ | 21,615,571 | 16,841,994 | 22,110,713 |
| Reads map to ‘−’ | 21,706,658 | 16,946,649 | 22,243,457 |
| Non_splice reads | 25,894,935 | 18,912,591 | 24,597,501 |
| Splice reads | 17,427,294 | 14,876,052 | 19,756,669 |
| Reads mapped in proper pairs | 38,542,714 | 30,245,418 | 40,189,646 |
| Gene Number | Gene | Primer Name | Forward Primer (5′→3′) | Primer Name | Reverse Primer (5′→3′) |
|---|---|---|---|---|---|
| EU047719 | 18S rRNA | 18F99 | ATTTCCGACAC GGAGAGG | 18R100 | CATGGGTTTAGGATACGCTC |
| KF444352.1 | M-CSFR | M-CSFR-F | CGCTCGACATGGACGACTTA | M-CSFR-R | ACTCGACTGTTGGTGAGCAG |
| JQ040498.1 | TNF-α | TNF-α-F | GCTGCTGTCTGCTTCACGC | TNF-α-R | AGCCTGGTCCTGGTTCACTCT |
| JQ692172.1 | IL-1β | IL-1β-F | TTGGAAATGTGGAGGC ATTCT | IL-1β-R | GATGTTGAGCACCTCTTCTTCA |
| KC535507.1 | IL-6 | IL-6-F | CTCAACCCTGGTCAACGACA | IL-6-R | GCATCCATGCGGATTTGACC |
| HQ589354.1 | iNOS | iNOS-F | TTCACATGGAGCACCCACAA | iNOS-R | TCAGTGCCCATGTACCAACC |
| JN255694 | CXCL8a | CXCL8a-F | TCGTTGGCAGAATGAACTGC | CXCL8a-R | TAATGCAGCGACAGCGTAG |
| MF783120* | CCL39.3 | CCL39.3-F | GAAGCCTGATGTTTCTGCTGG | CCL39.3-R | AGACTTCACCAGTTTCACAGGAA |
| MF783159* | CXCL-like | CXCL-like-F | GTTTGCTGGCTGTCAACCTC | CXCL-like-R | AGCAGTTAGGTCCTTTTGGAGT |
| MF783144* | XCL32a.2 | XCL32a.2-F | TCCTCACCATGGGCTGTTAAT | XCL32a.2-R | TTTGACATTTTCTTAGGAGCCGC |
| MF783098* | CCL20a.3 | CCL20a.3-F | TCGTGATCGTGCTGATGGTT | CCL20a.3-R | GATGTGGCAGTTTCTTGTCATGT |
| CI01000344_01208746_01214936 | CD68 | CD68-F | TATGGGGAACGGTGTGAGTC | CD68-R | CGATAGCGACACGGTAGTCA |
| CI01000001_13329841_13334948 | CD63 | CD63-F | TTGGAGCTTTGGGAGATGACA | CD63-R | TGATCTGCACAAACGCGATG |
| CI01000343_00067132_00076892 | CD48 | CD48-F | CGTGGCCCTTAATTGCACTG | CD48-R | CGAAGAGTCGACAGCATTTCC |
| CI01000219_00233215_00238585 | CD22-like | CD22-like-F | CAGCTCTGACGGAAACCACT | CD22-like-R | CAGCTCTGACGGAAACCACT |
| CI01000017_00722020_00762518 | CD276 | CD276-F | GGCTCTGTTTGGGATGGACA | CD276-R | GGCTCTGCCGATAACTGTGTA |
| CI01000059_00691603_00693842 | CD180 | CD180-F | TGTCCACCTTGTCCCATCTG | CD180-R | CTGAATCCGAGTTGCTTCGC |
| CI01000365_00126423_00134788 | CD11-1 | CD11-1-F | CGATCTCAACGCTCAGAAAGC | CD11-1-R | ACCCAATGATGTAGCGGAGG |
| CI01000365_00275086_00325578 | CD11-2 | CD11-2-F | TCGGGTCATATTTCGGAGCG | CD11-2-R | AGACGTACAGCCTCCCTTCT |
| CI01000027_02147653_02152768 | CD22 | CD22-F | AATATACTGGACCGGGTGGC | CD22-R | TCCGGGGTACAGTTTTCTGTG |
| CI01092720_00000024_00000324 | CD69 | CD69-F | CAACATGAACGACACGAACGA | CD69-R | ACCTGAAGACCACTGCCATTT |
| CI01000057_01361967_01364527 | CD27 | CD27-F | TATTTGTGGGGGCGCTTAGT | CD27-R | TCGGAGCAGTTCTTGGTAACAT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Wei, X.; Liao, Z.; Gao, Y.; Liu, X.; Su, J.; Yuan, G. Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella). Int. J. Mol. Sci. 2018, 19, 3562. https://doi.org/10.3390/ijms19113562
Hu Y, Wei X, Liao Z, Gao Y, Liu X, Su J, Yuan G. Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella). International Journal of Molecular Sciences. 2018; 19(11):3562. https://doi.org/10.3390/ijms19113562
Chicago/Turabian StyleHu, Yazhen, Xiaolei Wei, Zhiwei Liao, Yu Gao, Xiaoling Liu, Jianguo Su, and Gailing Yuan. 2018. "Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella)" International Journal of Molecular Sciences 19, no. 11: 3562. https://doi.org/10.3390/ijms19113562
APA StyleHu, Y., Wei, X., Liao, Z., Gao, Y., Liu, X., Su, J., & Yuan, G. (2018). Transcriptome Analysis Provides Insights into the Markers of Resting and LPS-Activated Macrophages in Grass Carp (Ctenopharyngodon idella). International Journal of Molecular Sciences, 19(11), 3562. https://doi.org/10.3390/ijms19113562






