Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression
Abstract
:1. Introduction
2. Results
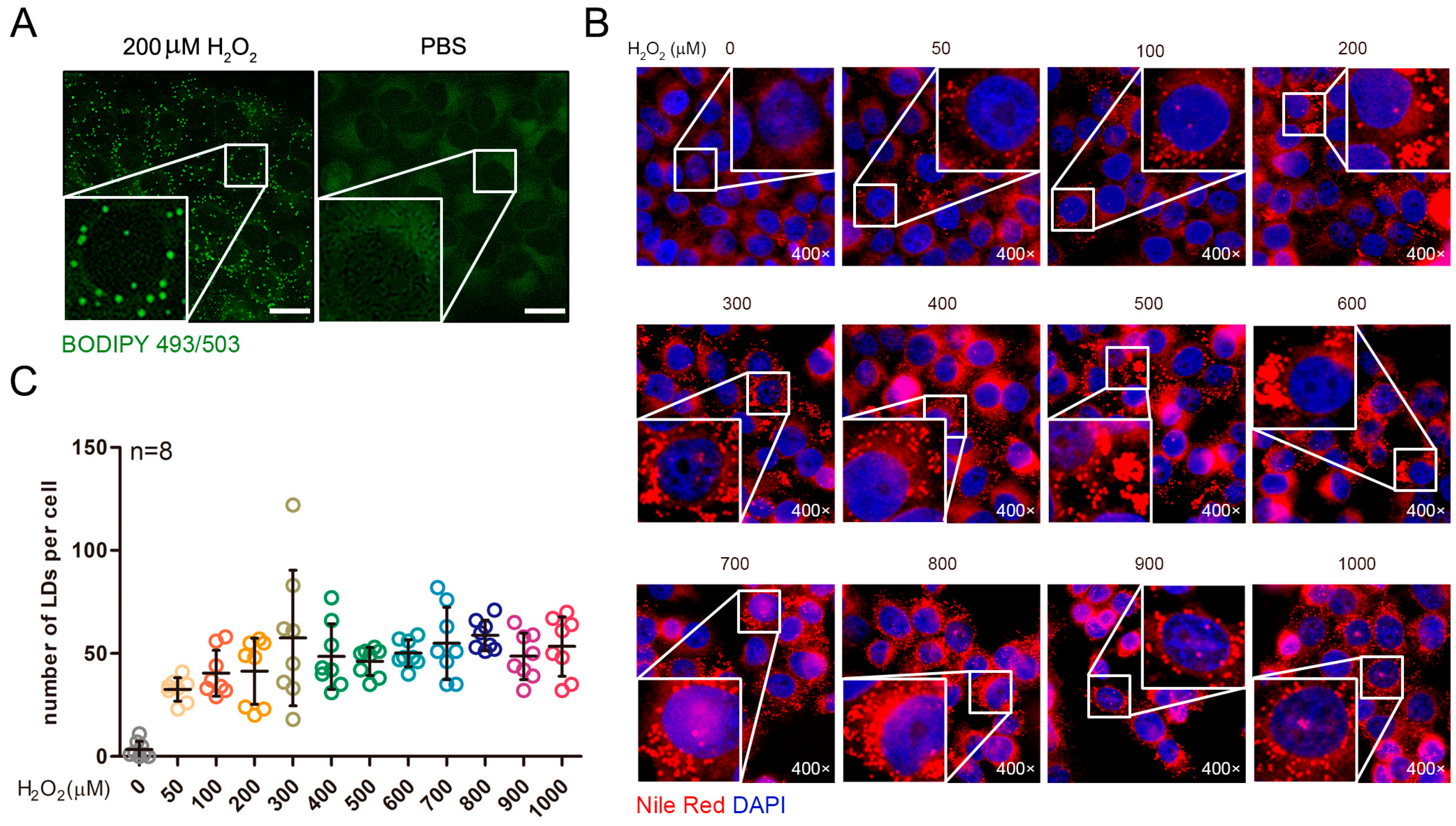
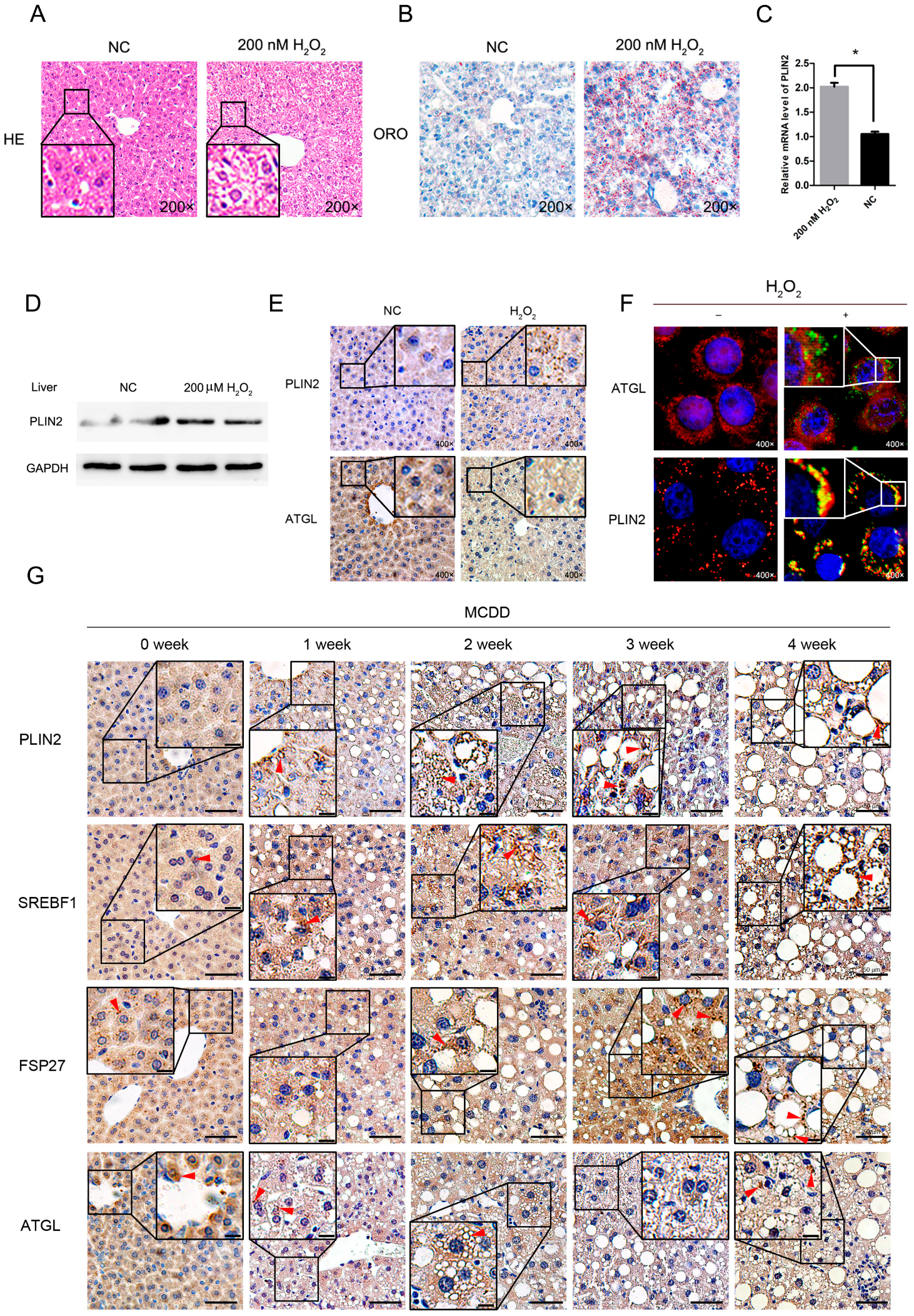
2.1. Exogenous Addition of Hydrogen Peroxide Promotes the Formation of Lipid Droplets in Cells
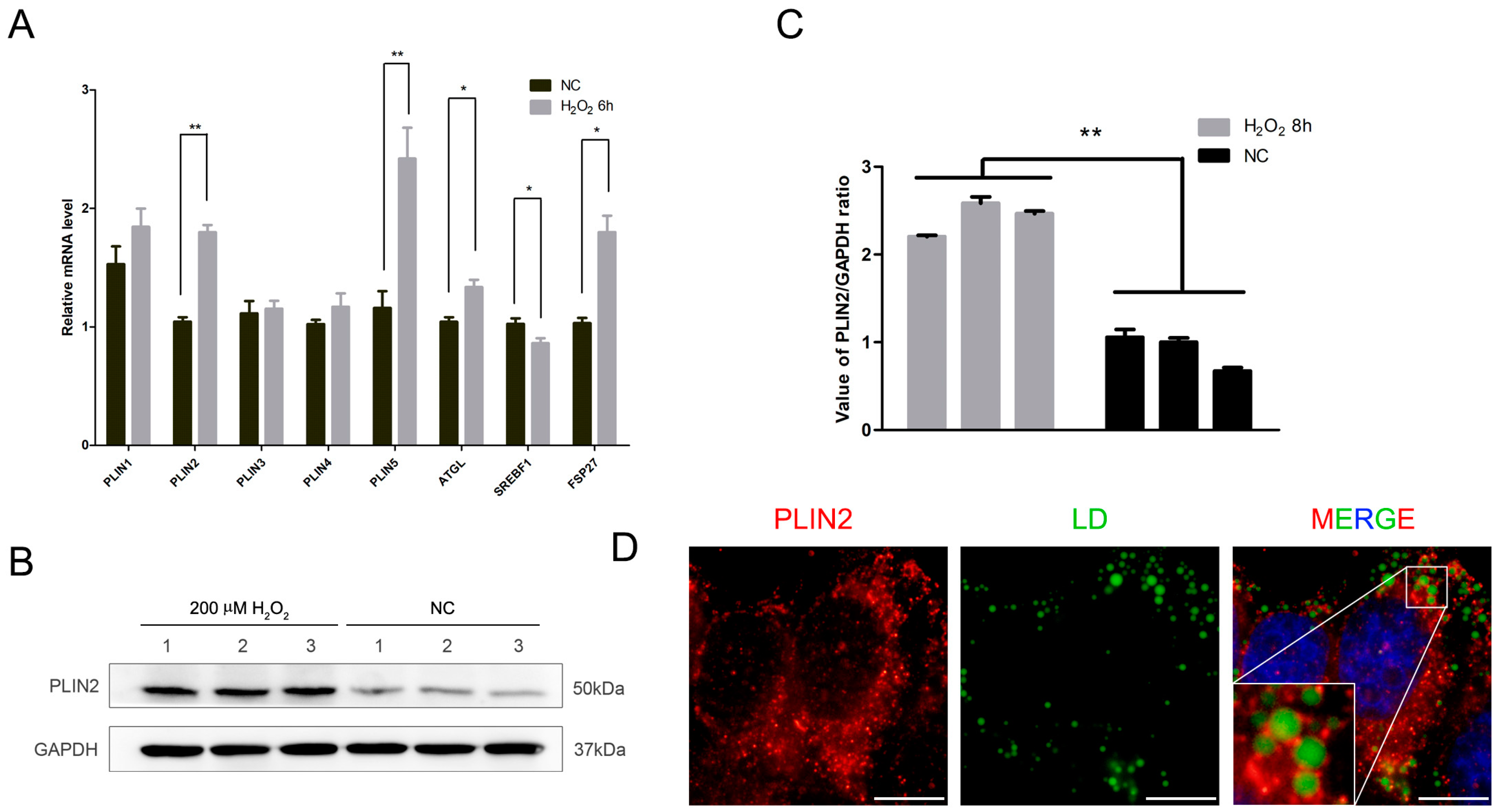
2.2. PLIN2 Expression Level Increased after Treatment by Hydrogen Peroxide
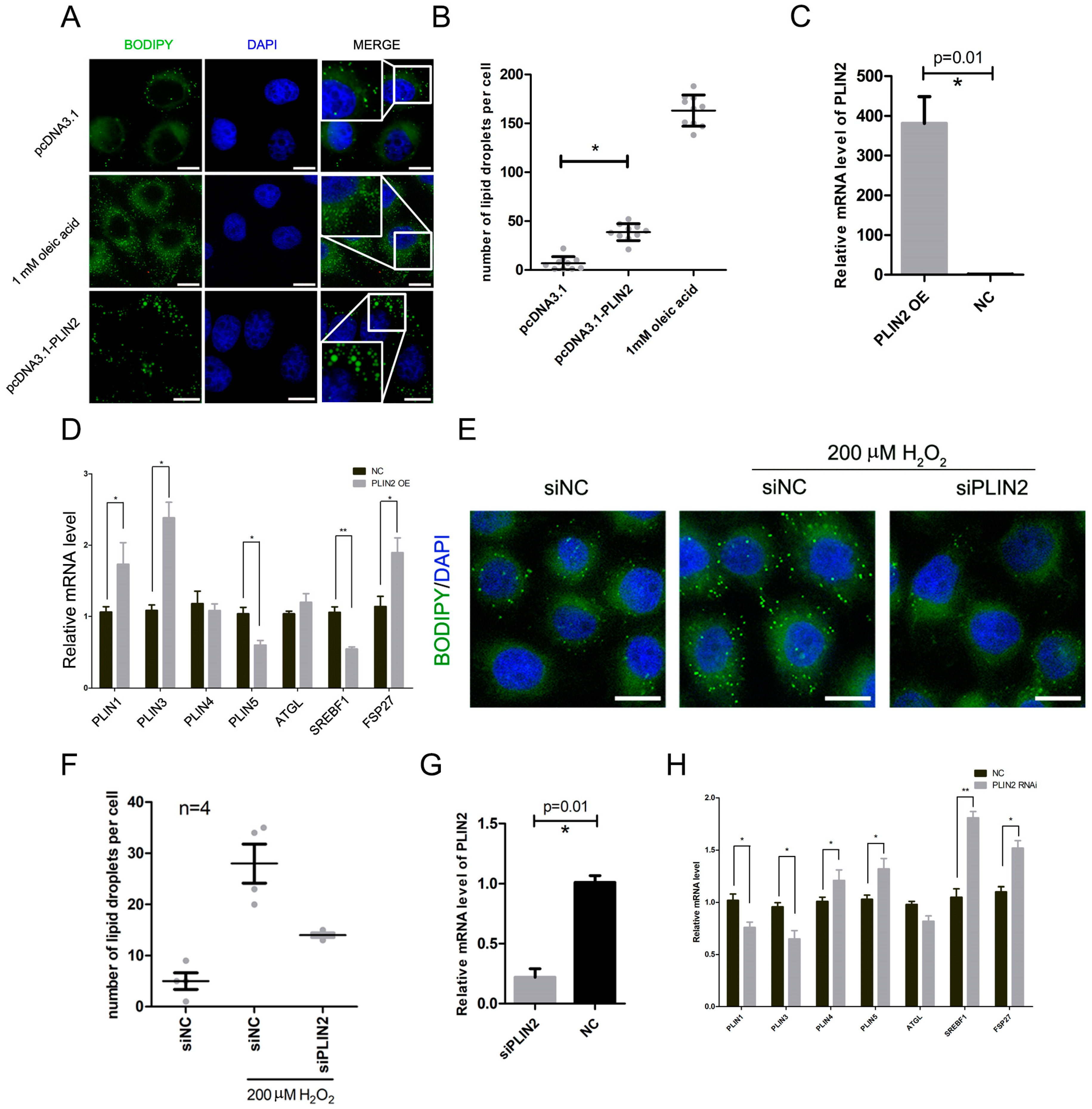
2.3. PLIN2 Overexpression Promotes Lipid Droplets Formation in Cells
2.4. PLIN2 Interference Inhibits Hydrogen Peroxide-Induced Lipid Droplet Accumulation
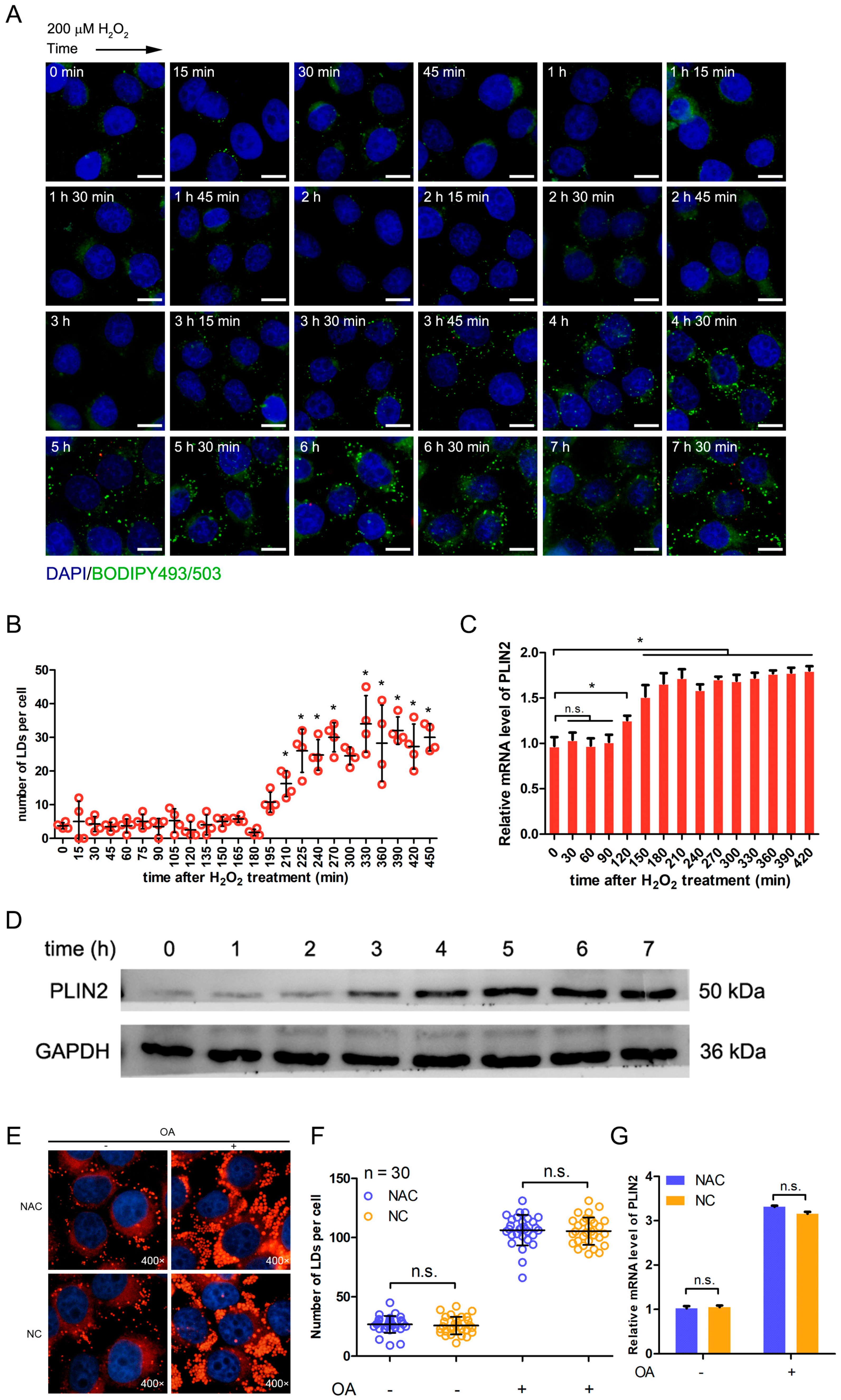
2.5. Hydrogen Peroxide Treatment Promotes the Formation of Lipid Droplets through Up-Regulating PLIN2
2.6. Exogenous Addition of Hydrogen Peroxide Promotes the Formation of Lipid Droplets in Murine Hepatocytes
2.7. Lipid Droplet-Related Proteins Enhanced Expression in Steatotic Liver Tissue
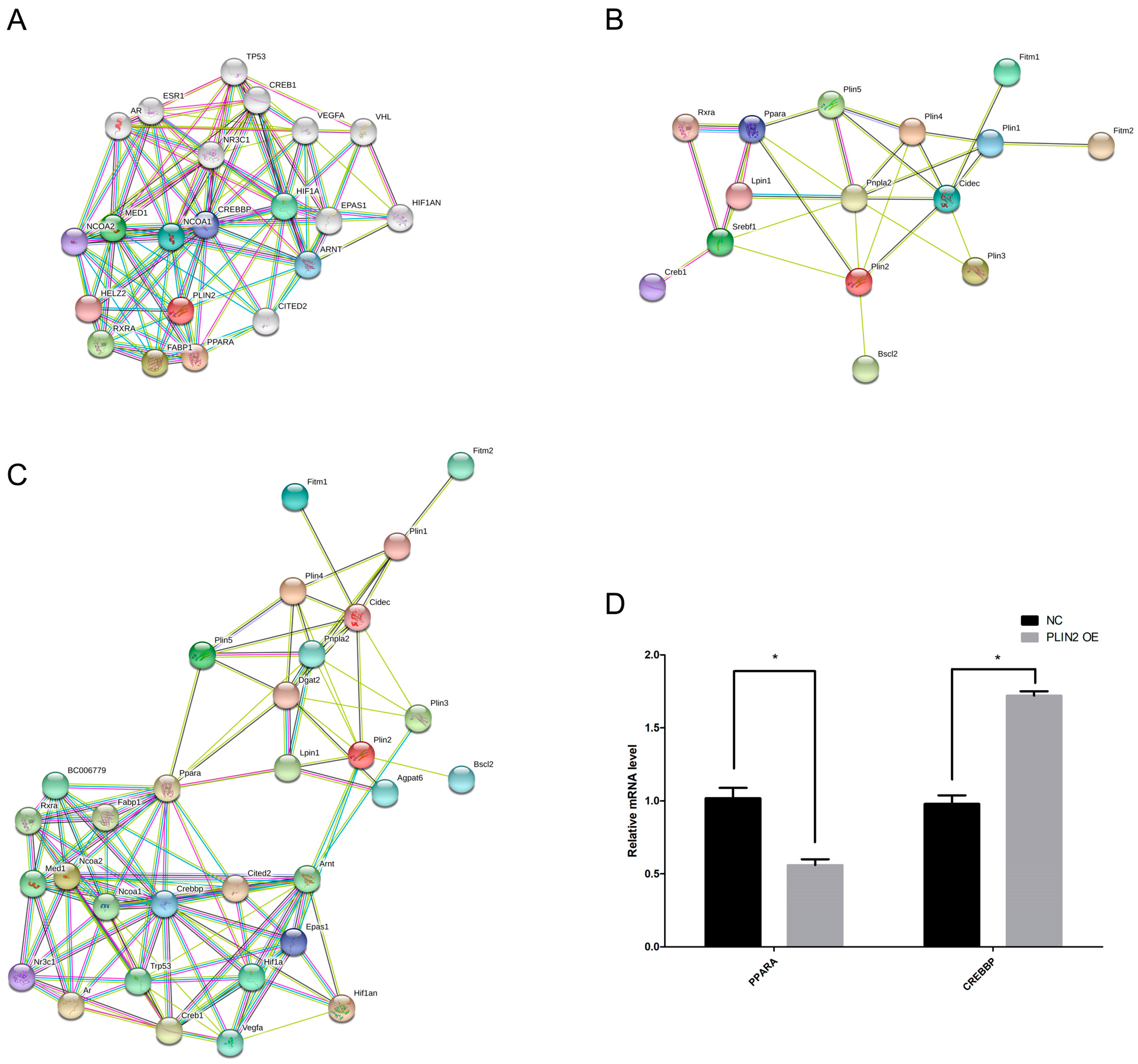
2.8. PLIN2 Modulates Lipid Droplet Formation in Cells via PPAR (Peroxisome Proliferator-Activated Receptor) and CREBBP (CREB Binding Protein) Signaling Pathways
3. Discussion and Conclusions
4. Methods and Materials
4.1. Animals and Cell Lines
4.2. Antibodies
4.3. Plasmid DNA Construction
4.4. Cell Culture and Transfection
4.5. Hydrogen Peroxide Treatment
4.6. Hematoxylin-Eosin Staining and Oil Red O Staining of Histological Sections
4.7. Lipid Droplets Marking and Observation
4.8. Western Blot and Real-Time PCR
4.9. Immunofluorescence Assay
4.10. Intraperitoneal Injection of Hydrogen Peroxide in Mice
4.11. Immunohistochemistry Assay
4.12. Bioinformatics and Data Analysis
4.13. Statistical Analyses
Authors Contribution
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | reactive oxygen species |
| NAFLD | non-alcoholic fatty liver disease |
| NASH | non-alcoholic steatohepatitis |
| PLIN1 | perilipin1 |
| PLIN2 | perilipin2 |
| PLIN3 | perilipin3 |
| PLIN4 | perilipin4 |
| PLIN5 | perilipin5 |
| FSP27/CIDEC | cell death-inducing DFFA-like effector c |
| SREBF1 | sterol regulatory element binding transcription factor 1 |
| SREBP1c | sterol regulatory element binding protein 1c |
| PPAR | peroxisome proliferator activated receptor alpha |
| RXRA | retinoid X receptor alpha |
| CREB | cAMP responsive element binding protein |
| CREBBP | CREB binding protein |
| SOD1 | superoxide dismutase 1 |
| JNK/MAPK8 | mitogen-activated protein kinase 8 |
| AMPK | adenosine 5‘-monophosphate (AMP)-activated protein kinase |
| OA | oleic acid |
| CITED2 | Cbp/P300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 2 |
| NCOA1 | nuclear receptor coactivator 1 |
| NCOA2 | nuclear receptor coactivator 2 |
| HIF1 | hypoxia inducible factor 1 |
| HIF1AN | hypoxia inducible factor 1 alpha subunit inhibitor |
| TP53 | tumor protein p53 |
| ARNT | aryl hydrocarbon receptor nuclear translocator |
| VEGFA | vascular endothelial growth factor A |
| EPAS1 | endothelial PAS domain protein 1 |
| AR | androgen receptor |
| MED1 | mediator complex subunit 1 |
| FABP1 | fatty acid binding protein 1 |
| HELZ2 | helicase with zinc finger 2 |
| NR3C | nuclear receptor subfamily 3 group C member |
| SEIPIN/BSCL2 | seipin lipid droplet biogenesis associated |
| FITM1 | fat storage inducing transmembrane protein 1 |
| FITM2 | fat storage inducing transmembrane protein 2 |
| ATGL/PNPLA2 | patatin like phospholipase domain containing 2 |
References
- Anderson, N.; Borlak, J. Molecular Mechanisms and Therapeutic Targets in Steatosis and Steatohepatitis. Pharmacol. Rev. 2008, 60, 311–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malaguarnera, M.; Di, R.M.; Nicoletti, F.; Malaguarnera, L. Molecular mechanisms involved in NAFLD progression. J. Mol. Med. 2009, 87, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, A.S.; Oben, J. Nonalcoholic fatty liver disease and lipids. Curr. Opin. Lipidol. 2012, 23, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Videla, L.A.; Rodrigo, R.; Araya, J.; Poniachik, J. Insulin resistance and oxidative stress interdependency in non-alcoholic fatty liver disease. Trends Mol. Med. 2006, 12, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Gawrieh, S.; Opara, E.C.; Koch, T.R. Oxidative stress in nonalcoholic fatty liver disease: Pathogenesis and antioxidant therapies. J. Investig. Med. Offic. Publ. Am. Fed. Clin. Res. 2004, 52, 506. [Google Scholar]
- Videla, L.A.; Rodrigo, R.; Orellana, M.; Fernandez, V.; Tapia, G.; Quiñones, L.; Varela, N.; Contreras, J.; Lazarte, R.; Csendes, A. Oxidative stress-related parameters in the liver of non-alcoholic fatty liver disease patients. Clin. Sci. 2004, 106, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, K.; Sandoval, H.; Yamamoto, S.; Jaiswal, M.; Sanz, E.; Li, Z.; Hui, J.; Graham, B.H.; Quintana, A. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell 2015, 160, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Müller, G.; Wied, S.; Jung, C.; Over, S. Hydrogen peroxide-induced translocation of glycolipid-anchored (c)AMP-hydrolases to lipid droplets mediates inhibition of lipolysis in rat adipocytes. Br. J. Pharmacol. 2010, 154, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Blasgarcía, A.; Apostolova, N.; Ballesteros, D.; Monleón, D.; Morales, J.M.; Rocha, M.; Victor, V.M.; Esplugues, J.V. Inhibition of mitochondrial function by efavirenz increases lipid content in hepatic cells. Hepatology 2010, 52, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiya, M.; Hiraishi, A.; Touyama, M.; Sakamoto, K. Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochem. Biophys. Res. Commun. 2008, 375, 602–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhao, S.; Yao, Z.; Wang, L.; Shao, J.; Chen, A.; Zhang, F.; Zheng, S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox Biol. 2017, 11, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, S.A.; Haller, J.F.; Ferrante, T.; Zoeller, R.A.; Corkey, B.E. Reactive Oxygen Species Facilitate Translocation of Hormone Sensitive Lipase to the Lipid Droplet during Lipolysis in Human Differentiated Adipocytes. PLoS ONE 2012, 7, e34904. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.U.; Sheikh, T.A. Endoplasmic reticulum stress and Oxidative stress in the pathogenesis of Non-alcoholic fatty liver disease. Free Radic Res. 2015, 49, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Ceni, E.; Mello, T.; Galli, A. Pathogenesis of alcoholic liver disease: Role of oxidative metabolism. World J. Gastroenterol. 2014, 20, 17756–17772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigouroux, C.; Caron-Debarle, M.; Le Dour, C.; Magre, J.; Capeau, J. Molecular mechanisms of human lipodystrophies: From adipocyte lipid droplet to oxidative stress and lipotoxicity. Int. J. Biochem. Cell Biol. 2011, 43, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Copic, A.; Antoinebally, S.; Gimenezandres, M.; Garay, C.L.T.; Antonny, B.; Manni, M.M.; Pagnotta, S.; Guihot, J.; Jackson, C.L. A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat. Commun. 2018, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, R.E.K.; Ramos, S.V.; Vandenboom, R.; Roy, B.D.; Peters, S.J. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am. J. Physiol. Regulat. Integr. Comp. Physiol. 2013, 304, R644–R650. [Google Scholar] [CrossRef] [PubMed]
- Boutant, M.; Kulkarni, S.S.; Joffraud, M.; Ratajczak, J.; Valera-Alberni, M.; Combe, R.; Zorzano, A.; Cantã³, C. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 2017, 36, 1543–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covington, J.D.; Galgani, J.E.; Moro, C.; Lagrange, J.M.; Zhang, Z.; Rustan, A.C.; Ravussin, E.; Bajpeyi, S. Skeletal Muscle Perilipin 3 and Coatomer Proteins Are Increased Following Exercise and Are Associated with Fat Oxidation. PLoS ONE 2014, 9, e91675. [Google Scholar] [CrossRef] [PubMed]
- Covington, J.D.; Noland, R.C.; Hebert, R.C.; Masinter, B.S.; Smith, S.R.; Rustan, A.C.; Ravussin, E.; Bajpeyi, S. Perilipin 3 Differentially Regulates Skeletal Muscle Lipid Oxidation in Active, Sedentary, and Type 2 Diabetic Males. J. Clin. Endocrinol. Metabol. 2015, 100, 3683. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Minnaard, R.; Sparks, L.M.; Schaart, G.; Losen, M.; Baets, M.H.D.; Duimel, H.; Kersten, S.; Bickel, P.E.; Schrauwen, P. The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 2012, 137, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Sparks, L.M.; Hooiveld, G.J.; Jorgensen, J.A.; Houten, S.M.; Schrauwen, P.; Kersten, S.; Hesselink, M.K.C. Overexpression of PLIN5 in skeletal muscle promotes oxidative gene expression and intramyocellular lipid content without compromising insulin sensitivity. BBA-Mol. Cell Biol. Lipids 2013, 1831, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.-A.L.S.; Skinner, J.R.; Shew, T.M.; Pietka, T.A.; Abumrad, N.A.; Wolins, N.E. Perilipin 5–Driven Lipid Droplet Accumulation in Skeletal Muscle Stimulates the Expression of Fibroblast Growth Factor 21. Diabetes 2015, 64, 2757–2768. [Google Scholar] [CrossRef] [PubMed]
- Rambold, A.S.; Cohen, S.; Lippincottschwartz, J. Fatty Acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Develop. Cell 2015, 32, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.L.; Yang, Z.H.; Wang, X.C.; Liu, Y.; Yang, Y.H.; Li, L.X.; Liang, W.C.; Zhou, W.B.; Hu, R.M. Adipophilin affects the expression of TNF-alpha, MCP-1, and IL-6 in THP-1 macrophages. Mol. Cell. Biochem. 2010, 337, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Listenberger, L.L.; Ostermeyerfay, A.G.; Goldberg, E.B.; Brown, W.J.; Brown, D.A. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 2007, 48, 2751–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, B.; Wang, H.; Chen, H.; Mclenithan, J.C.; Gong, D.W.; Yang, R.Z.; Yu, D.; Fried, S.K.; Quon, M.J.; Londos, C. Consequences of Lipid Droplet Coat Protein Downregulation in Liver Cells Abnormal Lipid Droplet Metabolism and Induction of Insulin Resistance. Diabetes 2008, 57, 2037–2045. [Google Scholar]
- Chang, B.H.J.; Li, L.; Saha, P.; Chan, L. Absence of adipose differentiation related protein upregulates hepatic VLDL secretion, relieves hepatosteatosis, and improves whole body insulin resistance in leptin-deficient mice. J. Lipid Res. 2010, 51, 2132–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, R.M.; Peralta, G.; Yin, X.; Ahima, R.S. Absence of Perilipin 2 Prevents Hepatic Steatosis, Glucose Intolerance and Ceramide Accumulation in Alcohol-Fed Mice. PLoS ONE 2014, 9, e97118. [Google Scholar] [CrossRef] [PubMed]
- Mcmanaman, J.L.; Bales, E.S.; Orlicky, D.J.; Jackman, M.; Maclean, P.S.; Cain, S.; Crunk, A.E.; Mansur, A.; Graham, C.E.; Bowman, T.A. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J. Lipid Res. 2013, 54, 1346. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.H.; Li, L.; Paul, A.; Taniguchi, S.; Nannegari, V.; Heird, W.C.; Chan, L. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 2006, 26, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Ikura, Y.; Caldwell, S.H. Lipid droplet-associated proteins in alcoholic liver disease: A potential linkage with hepatocellular damage. Int. J. Clin. Exp. Pathol. 2015, 8, 8699–8708. [Google Scholar] [PubMed]
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 2000, 279, L1005. [Google Scholar] [CrossRef] [PubMed]
- Herms, A.; Bosch, M.; Ariotti, N.; Reddy, B.J.N.; Fajardo, A.; Fernándezvidal, A.; Alvarezguaita, A.; Fernándezrojo, M.A.; Rentero, C.; Tebar, F. Cell-to-Cell Heterogeneity in Lipid Droplets Suggests a Mechanism to Reduce Lipotoxicity. Curr. Biol. 2013, 23, 1489–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnova, E.; Goldberg, E.B.; Makarova, K.S.; Lin, L.; Brown, W.J.; Jackson, C.L. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006, 7, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.W.; Preuss, C.; Shu, P.W.; Hao, Y.; Bo, J.; Carter, G.W.; Gladdy, R.; Andelfinger, G.; Mitchell, G.A. Epistatic interaction between the lipase-encoding genes Pnpla2 and Lipe causes liposarcoma in mice. PLoS Genet. 2017, 13, e1006716. [Google Scholar]
- Bosma, M.; Hesselink, M.K.C.; Sparks, L.M.; Timmers, S.; Ferraz, M.J.; Mattijssen, F.; Beurden, D.V.; Schaart, G.; Baets, M.H.D.; Verheyen, F.K. Perilipin 2 Improves Insulin Sensitivity in Skeletal Muscle Despite Elevated Intramuscular Lipid Levels. Diabetes 2012, 61, 2679. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Cook, W.S.; Qi, C.; Yeldandi, A.V.; Reddy, J.K.; Rao, M.S. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 2000, 275, 28918–28928. [Google Scholar] [CrossRef] [PubMed]
- Kliewer, S.A.; Forman, B.M.; Blumberg, B.; Ong, E.S.; Borgmeyer, U.; Mangelsdorf, D.J.; Umesono, K.; Evans, R.M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 7355–7359. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Jia, Y.; Yang, G.; Zhang, X.; Boddu, P.C.; Petersen, B.; Narsingam, S.; Zhu, Y.J.; Thimmapaya, B.; Kanwar, Y.S. PPARα-Deficient ob/ob Obese Mice Become More Obese and Manifest Severe Hepatic Steatosis Due to Decreased Fatty Acid Oxidation. Am. J. Pathol. 2015, 185, 1396–1408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.W.; Klemm, D.J.; Vinson, C.; Lane, M.D. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J. Biol. Chem. 2004, 279, 4471–4478. [Google Scholar] [CrossRef] [PubMed]
- Reusch, J.E.B.; Colton, L.A.; Klemm, D.J. CREB Activation Induces Adipogenesis in 3T3-L1 Cells. Mol. Cell. Biol. 2000, 20, 1008–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Deb, I.; Chakraborty, J.; Mukhopadhyay, S.; Das, S. A polymorphism of the CREB binding protein (CREBBP) gene is a risk factor for addiction. Brain Res. 2011, 1406, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.K.; Olivieri, C.; Walker, C.; Manu, V.S.; Gao, J.; Bernlohr, D.A.; Tonelli, M.; Markley, J.L.; Veglia, G. Probing Protein-Protein Interactions Using Asymmetric Labeling and Carbonyl-Carbon Selective Heteronuclear NMR Spectroscopy. Molecules 2018, 23, 1937. [Google Scholar] [CrossRef] [PubMed]
| Gene Symbol | Primer Sequence 5′-3′ | Product Size (bp) | Accession |
|---|---|---|---|
| PLIN1 (perilipin1) | Forward: TGGGTGGTGTGGCACATAC | 143 | XM_005254934.4 |
| Reverse: CCTCCCCTTGGTTGAGGAGA | |||
| PLIN2 (perilipin2) | Forward: TTGCAGTTGCCAATACCTATGC | 148 | XM_017014259.2 |
| Reverse: CCAGTCACAGTAGTCGTCACA | |||
| PLIN3 (perilipin3) | Forward: GCCCAAGAGATGGTGTCTAGC | 119 | NM_005817.4 |
| Reverse: CCGGTCACTACGGACTTTGT | |||
| PLIN4 (perilipin4) | Forward: GGAGCTGCAACCTTCGGAAA | 131 | NM_001080400.1 |
| Reverse: GGACCACTCCCTTAGCCAC | |||
| PLIN5 (perilipin5) | Forward: AAGGCCCTGAAGTGGGTTC | 192 | NM_001013706.2 |
| Reverse: GCATGTGGTCTATCAGCTCCA | |||
| SREBP1 (sterol regulatory element binding protein 1c) | Forward: ACAGTGACTTCCCTGGCCTAT | 222 | XM_024450893.1 |
| Reverse: CATGGACGGGTACATCTTCAA | |||
| FSP27 | Forward: ATTGATGTGGCCCGTGTAACG | 122 | NM_001199623.1 |
| Reverse: CAGCAGTGCAGATCATAGGAAA | |||
| ATGL | Forward: ATGGTGGCATTTCAGACAACC | 89 | NM_020376.3 |
| Reverse: CGGACAGATGTCACTCTCGC | |||
| CREBBP | Forward: CGGCTCTAGTATCAACCCAGG | 237 | NM_004380.2 |
| Reverse: TTTTGTGCTTGCGGATTCAGT | |||
| PPARα | Forward: TTCGCAATCCATCGGCGAG | 146 | NM_005036.5 |
| Reverse: CCACAGGATAAGTCACCGAGG | |||
| GAPDH | Forward: CTGGGCTACACTGAGCACC | 101 | NM_002046.6 |
| Reverse: AAGTGGTCGTTGAGGGCAATG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Tan, Y.; Chen, L.; Liu, Y.; Ren, Z. Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression. Int. J. Mol. Sci. 2018, 19, 3445. https://doi.org/10.3390/ijms19113445
Jin Y, Tan Y, Chen L, Liu Y, Ren Z. Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression. International Journal of Molecular Sciences. 2018; 19(11):3445. https://doi.org/10.3390/ijms19113445
Chicago/Turabian StyleJin, Yi, Yanjie Tan, Lupeng Chen, Yan Liu, and Zhuqing Ren. 2018. "Reactive Oxygen Species Induces Lipid Droplet Accumulation in HepG2 Cells by Increasing Perilipin 2 Expression" International Journal of Molecular Sciences 19, no. 11: 3445. https://doi.org/10.3390/ijms19113445





