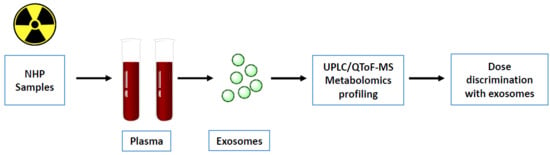
Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates
Abstract
1. Introduction
2. Results
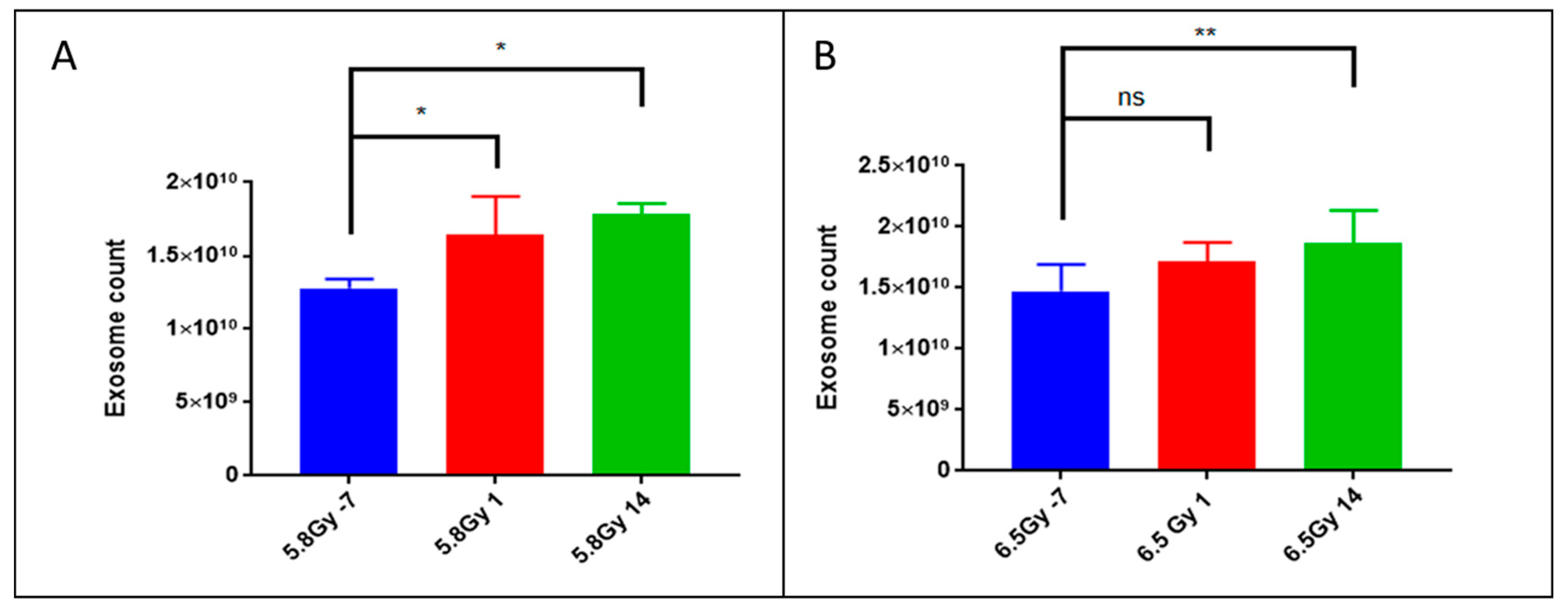
2.1. Exposure to Ionizing Radiation Enhances Exosomal Shedding in Circulation
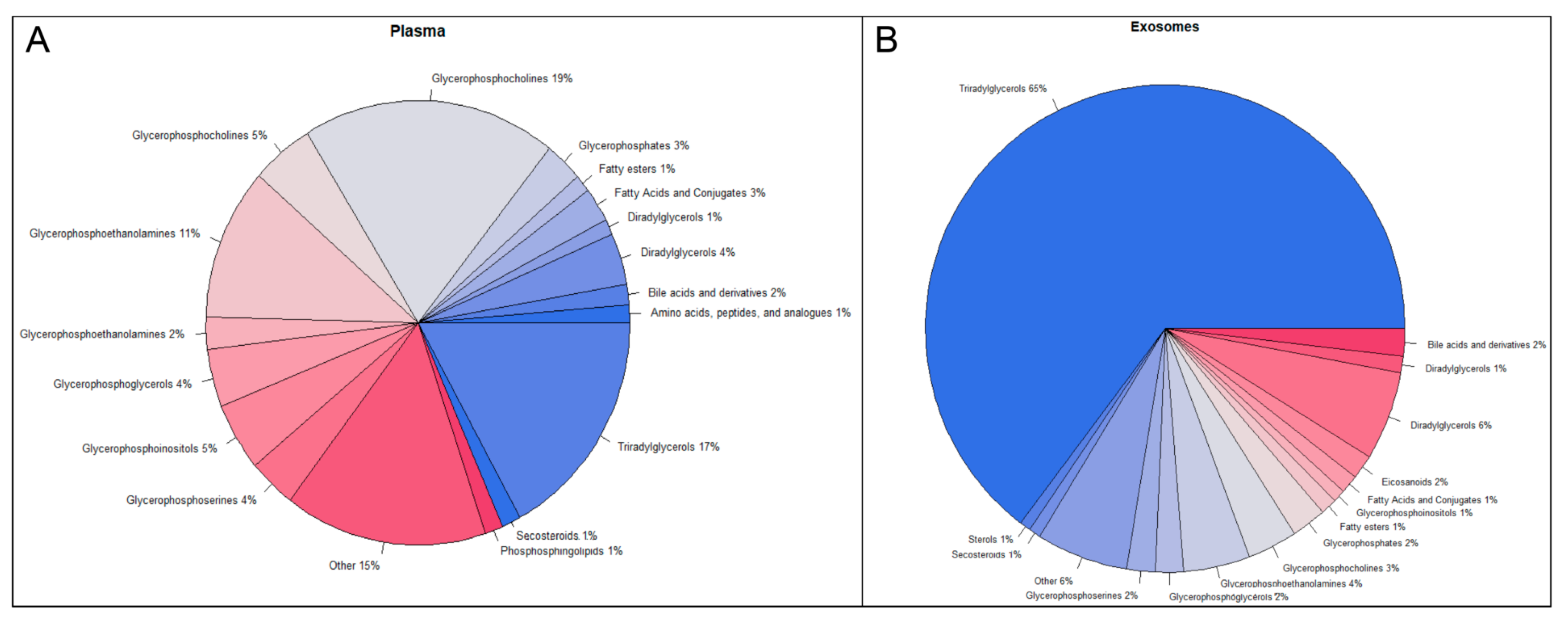
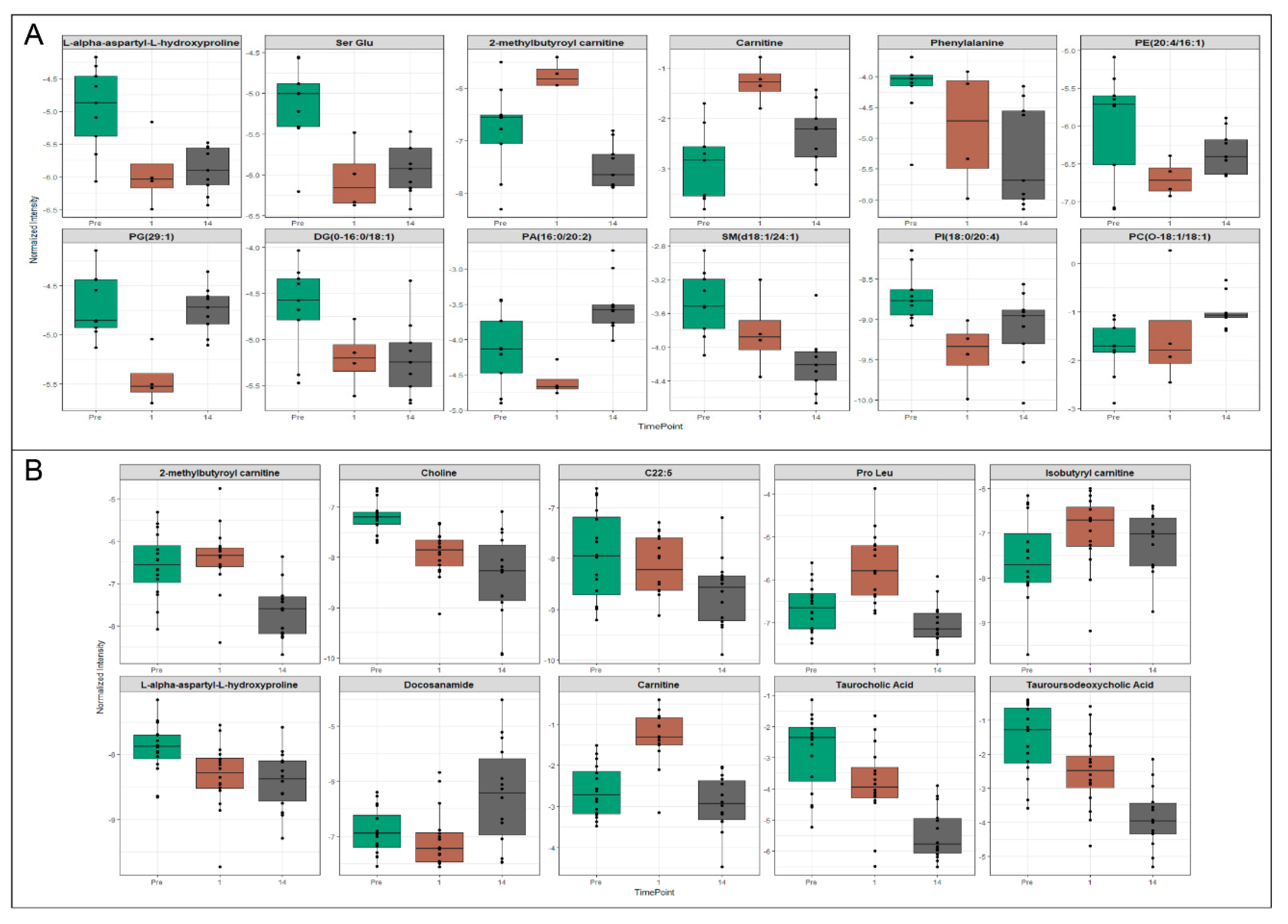
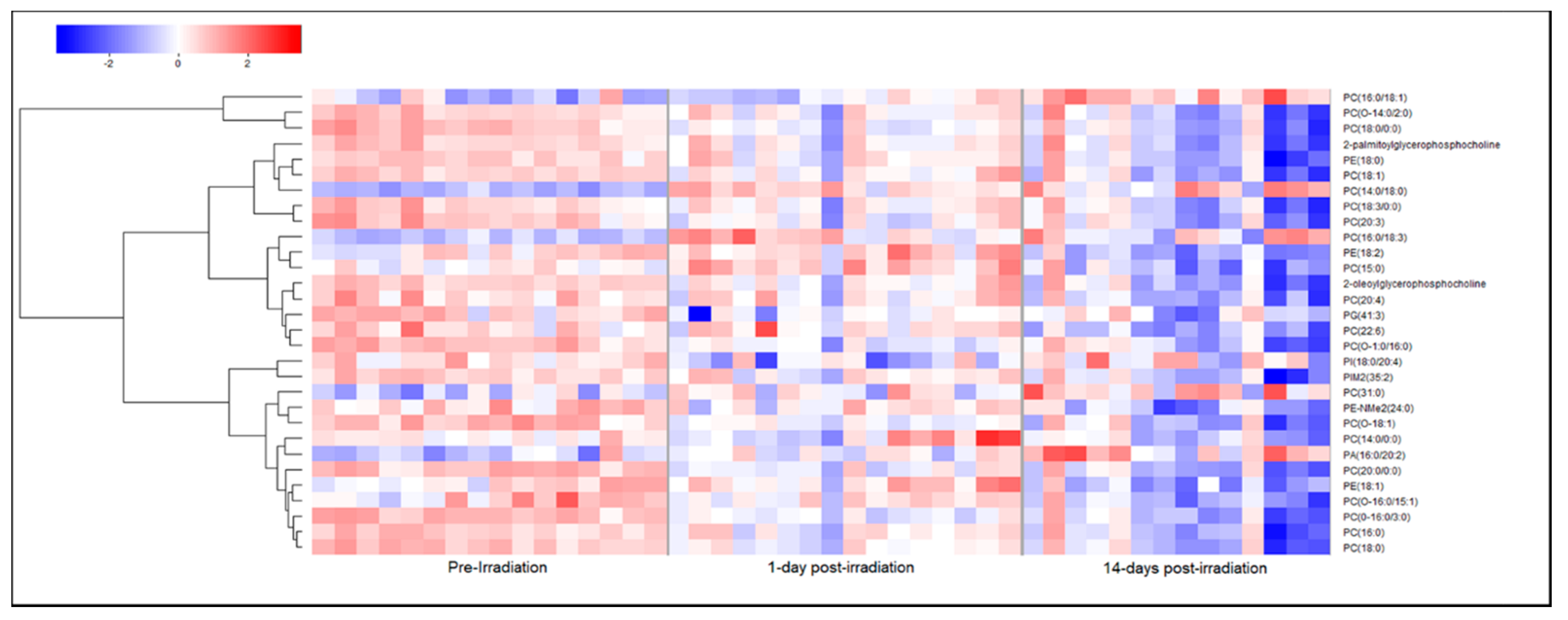
2.2. Metabolomic and Lipidomic Profiles of Plasma and Plasma-Derived Exosomes
2.3. Exosomes Yield Metabolic Information that May Be Missed from Plasma Profiling
3. Discussion
4. Materials and Methods
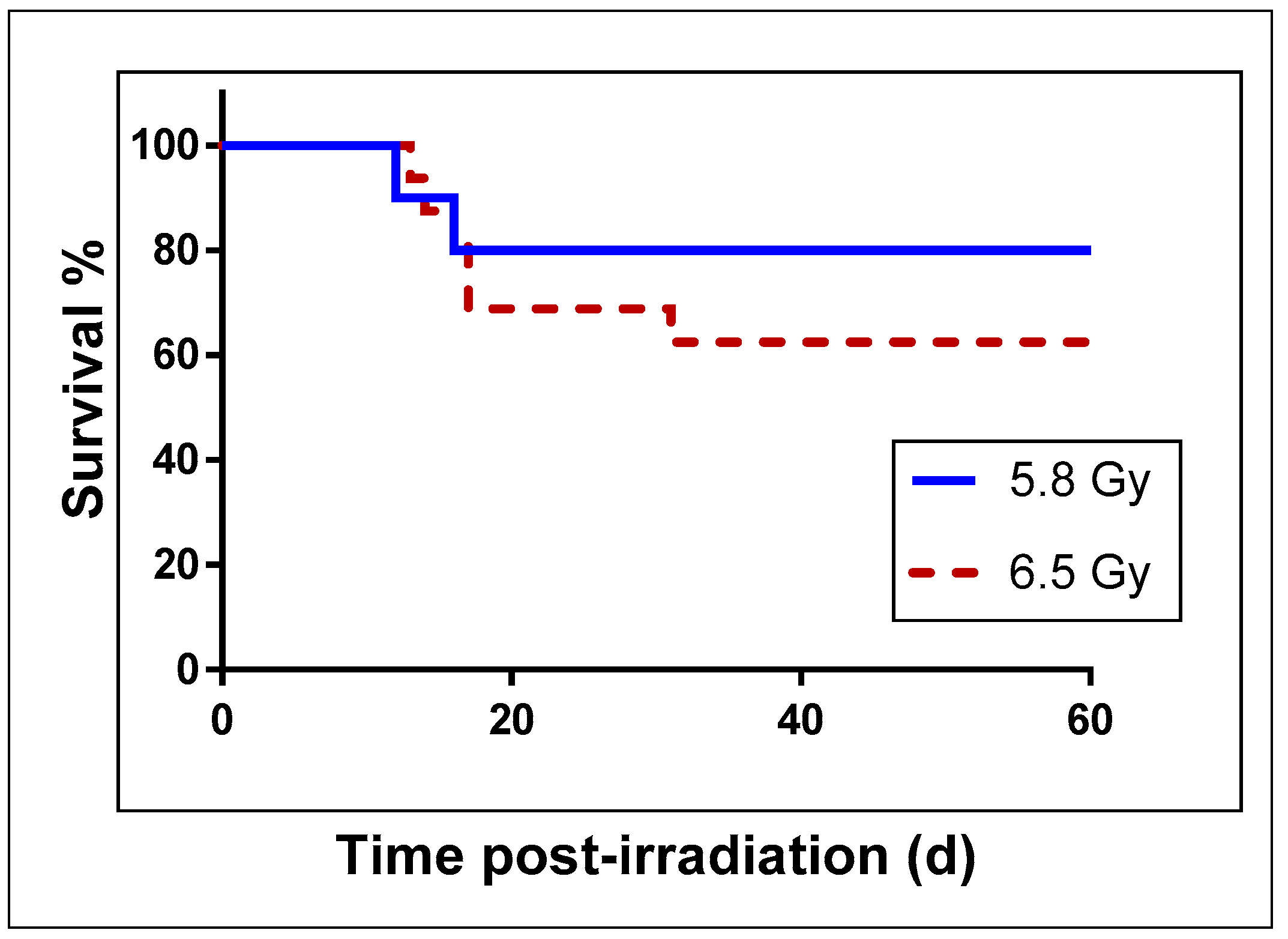
4.1. Animals and Animal Care
4.2. Radiation Exposure
4.3. Plasma Sample Collection
4.4. Exosome Isolation and Characterization
4.5. Sample Preparation for Mass Spectrometry (MS)
4.6. UPLC/Q-TOF-MS Data Acquisition and Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coleman, C.N.; Hrdina, C.; Bader, J.L.; Norwood, A.; Hayhurst, R.; Forsha, J.; Yeskey, K.; Knebel, A. Medical response to a radiologic/nuclear event: Integrated plan from the Office of the Assistant Secretary for Preparedness and Response, Department of Health and Human Services. Ann. Emerg. Med. 2009, 53, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Newman, V.L.; Romaine, P.L.; Hauer-Jensen, M.; Pollard, H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn. 2016, 16, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Fornace, A.J., Jr.; Laiakis, E.C. Metabolomic applications in radiation biodosimetry: Exploring radiation effects through small molecules. Int. J. Radiat. Biol. 2017, 93, 1151–1176. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. Guidance for Industry and FDA Staff: Qualification Process for Drug Development Tools. Available online: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm230597.pdf (accessed on 10 July 2018).
- U.S. Food and Drug Administration. Guidance on Pharmacogenomic Data Submissions Examples of Voluntary Submissions or Submissions Required under 21 CFR 312, 314, or 601. Available online: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm079851.pdf (accessed on 23 July 2018).
- Singh, V.K.; Newman, V.L.; Berg, A.N.; MacVittie, T.J. Animal models for acute radiation syndrome drug discovery. Expert Opin. Drug Discov. 2015, 10, 497–517. [Google Scholar] [CrossRef] [PubMed]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Pocsfalvi, G.; Stanly, C.; Vilasi, A.; Fiume, I.; Capasso, G.; Turiak, L.; Buzas, E.I.; Vekey, K. Mass spectrometry of extracellular vesicles. Mass Spectrom. Rev. 2016, 35, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Nazimek, K.; Bryniarski, K.; Santocki, M.; Ptak, W. Exosomes as mediators of intercellular communication: Clinical implications. Pol. Arch. Med. Wewn. 2015, 125, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Altadill, T.; Campoy, I.; Lanau, L.; Gill, K.; Rigau, M.; Gil-Moreno, A.; Reventos, J.; Byers, S.; Colas, E.; Cheema, A.K. Enabling metabolomics based biomarker discovery studies using molecular phenotyping of exosome-like vesicles. PLoS ONE 2016, 11, e0151339. [Google Scholar] [CrossRef] [PubMed]
- Varghese, R.S.; Cheema, A.; Cheema, P.; Bourbeau, M.; Tuli, L.; Zhou, B.; Jung, M.; Dritschilo, A.; Ressom, H.W. Analysis of LC-MS data for characterizing the metabolic changes in response to radiation. J. Proteome Res. 2010, 9, 2786–2793. [Google Scholar] [CrossRef] [PubMed]
- de la Torre Gomez, C.; Goreham, R.V.; Bech Serra, J.J.; Nann, T.; Kussmann, M. “Exosomics”—A review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front. Genet. 2018, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Mollaei, H.; Safaralizadeh, R.; Pouladi, N. A brief review of exosomes and their roles in cancer. Meta Gene 2017, 11, 70–74. [Google Scholar] [CrossRef]
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New molecular targets of diseases. Acta Pharmacol. Sin. 2018, 39, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome mediated communication within the tumor microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Giricz, Z.; Varga, Z.V.; Baranyai, T.; Sipos, P.; Paloczi, K.; Kittel, A.; Buzas, E.I.; Ferdinandy, P. Cardioprotection by remote ischemic preconditioning of the rat heart is mediated by extracellular vesicles. J. Mol. Cell. Cardiol. 2014, 68, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Miousse, I.R.; Kutanzi, K.R.; Koturbash, I. Effects of ionizing radiation on DNA methylation: From experimental biology to clinical applications. Int. J. Radiat. Biol. 2017, 93, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lau, K.K.; Zhou, X.; Wu, J.; Yang, J.; Wang, C. Urinary metabolic signatures and early triage of acute radiation exposure in rat model. Mol. Biosyst. 2017, 13, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.S.; Uppal, M.; Randhawa, S.; Cheema, M.S.; Aghdam, N.; Usala, R.L.; Ghosh, S.P.; Cheema, A.K.; Dritschilo, A. Radiation metabolomics: Current status and future directions. Front. Oncol. 2016, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Coy, S.L.; Cheema, A.K.; Tyburski, J.B.; Laiakis, E.C.; Collins, S.P.; Fornace, A., Jr. Radiation metabolomics and its potential in biodosimetry. Int. J. Radiat. Biol. 2011, 87, 802–823. [Google Scholar] [CrossRef] [PubMed]
- Boerma, M.; Sridharan, V.; Mao, X.W.; Nelson, G.A.; Cheema, A.K.; Koturbash, I.; Singh, S.P.; Tackett, A.J.; Hauer-Jensen, M. Effects of ionizing radiation on the heart. Mutat. Res. 2016, 770, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Suman, S.; Kaur, P.; Singh, R.; Fornace, A.J., Jr.; Datta, K. Long-term differential changes in mouse intestinal metabolomics after gamma and heavy ion radiation exposure. PLoS ONE 2014, 9, e87079. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Mak, T.D.; Astarita, G.; Authier, S.; Wong, K.; Fornace, A.J., Jr. A lipidomic and metabolomic serum signature from nonhuman primates exposed to ionizing radiation. Metabolomics 2016, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Kulkarni, S.; Fatanmi, O.O.; Wise, S.Y.; Newman, V.L.; Romaine, P.L.; Hendrickson, H.; Gulani, J.; Ghosh, S.P.; Kumar, K.S.; et al. Radioprotective efficacy of gamma-tocotrienol in nonhuman primates. Radiat. Res. 2016, 185, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Mutschelknaus, L.; Azimzadeh, O.; Heider, T.; Winkler, K.; Vetter, M.; Kell, R.; Tapio, S.; Merl-Pham, J.; Huber, S.M.; Edalat, L.; et al. Radiation alters the cargo of exosomes released from squamous head and neck cancer cells to promote migration of recipient cells. Sci. Rep. 2017, 7, 12423. [Google Scholar] [CrossRef] [PubMed]
- Jelonek, K.; Widlak, P.; Pietrowska, M. The influence of ionizing radiation on exosome composition, secretion and intercellular communication. Protein Pept. Lett. 2016, 23, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Koller, A.; Mani, K.M.; Wen, R.; Alfieri, A.; Saha, S.; Wang, J.; Patel, P.; Bandeira, N.; Guha, C.; et al. Identifying Urinary and Serum Exosome Biomarkers for Radiation Exposure Using a Data Dependent Acquisition and SWATH-MS Combined Workflow. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Garcia, M.; Fornace, A.J., Jr.; Singh, V.K. Nonhuman primates with acute radiation syndrome: Results from a global serum metabolomics study after 7.2 Gy total-body irradiation. Radiat. Res. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Singh, V.K.; Fornace, A.J. Lipidomic signatures of nonhuman primates with radiation-induced hematopoietic syndrome. Sci. Rep. 2017, 7, 9777. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, M.; Weber, W.M.; Chung, J.; Doyle-Eisele, M.; Melo, D.R.; Mak, T.D.; Strawn, S.J.; Brenner, D.J.; Guilmette, R.; Fornace, A.J., Jr. Serum dyslipidemia is induced by internal exposure to strontium-90 in mice, lipidomic profiling using a data-independent liquid chromatography-mass spectrometry approach. J. Proteome Res. 2015, 14, 4039–4049. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Fornace, A.J., Jr.; Fatanmi, O.O.; Singh, V.K. A metabolomic serum signature from nonhuman primates treated with a radiation countermeasure, gamma-tocotrienol, and exposed to ionizing radiation. Health Phys. 2018, 115, 3–11. [Google Scholar] [CrossRef] [PubMed]
- National Research Council of the National Academy of Sciences. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Cheema, A.K.; Mehta, K.Y.; Fatanmi, O.O.; Wise, S.Y.; Hinzman, C.P.; Wolff, J.; Singh, V.K. A Metabolomic and lipidomic serum signature from nonhuman primates administered with a promising radiation countermeasure, gamma-tocotrienol. Int. J. Mol. Sci. 2017, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Gil de la Fuente, A.; Godzien, J.; Fernandez Lopez, M.; Ruperez, F.J.; Barbas, C.; Otero, A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheema, A.K.; Hinzman, C.P.; Mehta, K.Y.; Hanlon, B.K.; Garcia, M.; Fatanmi, O.O.; Singh, V.K. Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates. Int. J. Mol. Sci. 2018, 19, 3427. https://doi.org/10.3390/ijms19113427
Cheema AK, Hinzman CP, Mehta KY, Hanlon BK, Garcia M, Fatanmi OO, Singh VK. Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates. International Journal of Molecular Sciences. 2018; 19(11):3427. https://doi.org/10.3390/ijms19113427
Chicago/Turabian StyleCheema, Amrita K., Charles P. Hinzman, Khyati Y. Mehta, Briana K. Hanlon, Melissa Garcia, Oluseyi O. Fatanmi, and Vijay K. Singh. 2018. "Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates" International Journal of Molecular Sciences 19, no. 11: 3427. https://doi.org/10.3390/ijms19113427
APA StyleCheema, A. K., Hinzman, C. P., Mehta, K. Y., Hanlon, B. K., Garcia, M., Fatanmi, O. O., & Singh, V. K. (2018). Plasma Derived Exosomal Biomarkers of Exposure to Ionizing Radiation in Nonhuman Primates. International Journal of Molecular Sciences, 19(11), 3427. https://doi.org/10.3390/ijms19113427