Abstract
CircRNAs have particular biological structure and have proven to play important roles in diseases. It is time-consuming and costly to identify circRNA-disease associations by biological experiments. Therefore, it is appealing to develop computational methods for predicting circRNA-disease associations. In this study, we propose a new computational path weighted method for predicting circRNA-disease associations. Firstly, we calculate the functional similarity scores of diseases based on disease-related gene annotations and the semantic similarity scores of circRNAs based on circRNA-related gene ontology, respectively. To address missing similarity scores of diseases and circRNAs, we calculate the Gaussian Interaction Profile (GIP) kernel similarity scores for diseases and circRNAs, respectively, based on the circRNA-disease associations downloaded from circR2Disease database (http://bioinfo.snnu.edu.cn/CircR2Disease/). Then, we integrate disease functional similarity scores and circRNA semantic similarity scores with their related GIP kernel similarity scores to construct a heterogeneous network made up of three sub-networks: disease similarity network, circRNA similarity network and circRNA-disease association network. Finally, we compute an association score for each circRNA-disease pair based on paths connecting them in the heterogeneous network to determine whether this circRNA-disease pair is associated. We adopt leave one out cross validation (LOOCV) and five-fold cross validations to evaluate the performance of our proposed method. In addition, three common diseases, Breast Cancer, Gastric Cancer and Colorectal Cancer, are used for case studies. Experimental results illustrate the reliability and usefulness of our computational method in terms of different validation measures, which indicates PWCDA can effectively predict potential circRNA-disease associations.
1. Introduction
In recent years, an increasing number of circRNAs [1] have been uncovered and have drawn more attention than before. CircRNA is a newly discovered category of non-coding RNAs. Non-coding RNAs also include a large number of different RNAs, such as miRNAs, lncRNAs, piRNAs [2]. The first discovery of circular RNA was in the Tetrahymena cell [3]. There is an obvious difference between circular RNAs and common linear RNAs. That is, circRNA has a circular closed loop RNA structure, yet have no free 5’ and 3’ compared with linear RNAs [4]. In addition, circRNAs can also be classified into 4 categories as follows: Exonic circRNAs, intronic circRNAs, exonintron circRNAs and intergenic circRNAs [4,5]. Because of such a closed loop structures, they are usually stable, abundant, conserved, and tissue-specifically expressed [5].
With the progress of high throughput sequencing technology [6], more and more circRNAs have been confirmed to play significant roles in different biological processes [7]. According to many experiments, a large amount of circRNAs functions have been found to work as a scaffold in the assembly of protein complexes [8], and local subcellular positions [9], and so on. They also regulate the expression of their ancestor genes [10] and acts as a microRNA (miRNA) sponge [11,12]. Especially, many studies have proved that circRNA can be biomarkers of tumors [13,14,15].
Recently, a sharply increasing number of circRNAs have been discovered and there are also some circRNA-disease databases being developed, such as circR2Disease [16], Circ2Traits [17] and Circ2Disease [18]. Simultaneously, circRNAs-related diseases also have been verified by classic biological experiments. However, they are both time-consuming and expensive. Therefore, it is appealing to develop computational methods that can produce reliable prediction results and reduce both time and cost. Although, some computational methods have been proposed for predicting miRNA-disease associations [19,20,21], lncRNA-disease associations [22,23] and drug-target associations [18,24,25], there is no computational method for predicting circRNA-disease associations yet.
In this study, we propose the first computational method, Path Weighed method for predicting CircRNA-Disease Associations (PWCDA). After building a heterogeneous network consisting of three sub-networks, the disease similarity network, the circRNA similarity network and circRNA-disease association network, we calculate an association score for each circRNA-disease pair based on the paths connecting them in the heterogeneous network to determine whether a circRNA-disease pair is associated. Our method is evaluated with leave one out cross validation (LOOCV) and five-fold cross validation. The average AUC (Area Under roc Curve) of LOOCV is 0.900, while the AUC value of five-fold cross validation is 0.890. For further investigating the performance of our proposed model, we conduct several case studies of some common cancers. What’s more, we compare our method with some other computational prediction methods. The results show that our method outperforms other methods, which indicates that our proposed model has the better capability to predict potential circRNA-disease associations.
2. Results and Discussion
2.1. Effect of Parameter
Based on the previous study [26], we fix the maximum path length as 3. If the maximum path length is more than 3, not only do the running time of the method increases, but our method also takes some noisy information. In this study, we give a comprehensive analysis for the parameter α in our decaying function. After we calculate scores for each disease-circRNA pair, we can obtain a disease-circRNA association score matrix. Based on the scores matrix, we calculate the AUC. The results are represented in Table 1. It’s obvious that the effect of different values of α on the final AUC value is quite small and it can take value from 1 to 3. Therefore, we adopt the best result setting the value of α as 1. In order to reduce the running time, we don’t use any cross validation in this experiment. Furthermore, we also carry out an experiment to analyze another parameter, the threshold γ, which is represented in Table 2. For the sake of reducing the running time, any cross validation is not adopted. The result shows that the parameter γ might have tiny effect on the final AUC value. Thus, we set the γ value as 0.5, which gets the greatest AUC value.

Table 1.
The Area Under roc Curve (AUC) value based on changing α and fixed pathway maximum length.

Table 2.
The AUC value based on changing γ and fixed pathway maximum length.
2.2. LOOCV
For a given particular disease i, there are some associations between disease i and a number of circRNAs. In LOOCV, during each computational iteration, we leave one association out as a test data and use the remaining associations as a training dataset. If there is just one association between disease i and circRNAs in our dataset, we do not adopt LOOCV for this kind of disease. In LOOCV, we obtain an association score for each circRNA-disease pair and then rank all the prediction association scores. If a score value is greater than the pre-set threshold, we determine that the corresponding disease-circRNA is associated. With the change of the threshold, we can get a variety of true positive rates (TPRs) and false positive rates (FPRs), which can be used to draw the Receiver Operating Characteristic Curve (ROC) curve. In the end, we have compared our prediction method with other computational prediction methods [27,28]. The results can be found in Figure 1 and show that our proposed method outperforms the existing prediction methods.
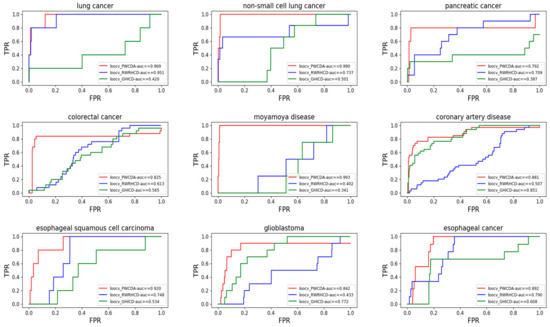
Figure 1.
Comparison of Path Weighed method for predicting CircRNA-Disease Associations (PWCDA) with other models by leave one out cross validation (LOOCV). FPR, false positive rate.
2.3. Five-Fold Cross Validation
In order to further illustrate the performance of our proposed method, we have adopted five-fold cross validation verification method as well for investigating the prediction performance. In our study, we divide all disease-circRNA associations into 5 parts. Each time we pick up one part as the test dataset and the remaining four parts consist of the training set. Then we can obtain the scores of all circRNA-disease associations. Similarly, we follow the same procedure as LOOCV to draw the AUC curve based on five-fold cross validation. What’s more, we have compared our proposed computational method with other prediction methods [27,28]. Our method gets more outstanding result than other methods, which is shown in the Figure 2.
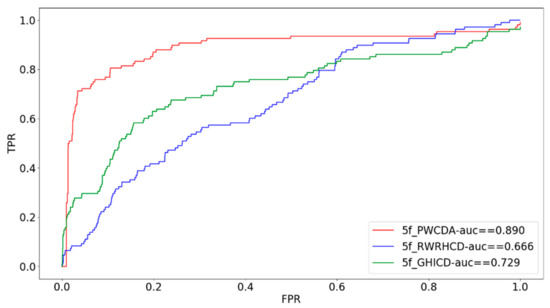
Figure 2.
Comparison of PWCDA with other computational methods via five-fold cross validation.
2.4. Case Studies
Here, we also have conducted some case studies, which can help us further understand the associations between circRNAs and diseases. In this study, we choose three common diseases as prediction targets of our case studies, which are Breast Cancer [29], Gastric Cancer [30] and Colorectal Cancer [31]. In order to prove the prediction accuracy of our proposed method, we have used circRNA-disease database, and associations between circRNAs and diseases—which have been experimentally verified in the published articles [32].
Breast cancer is one the common cancers all over the world now [33], and breast cancer causes thousands of deaths every year. With the development of deep sequencing technology, circRNAs are confirmed to be biomarkers for diagnosing breast cancer. Based on our computational method, we have succeeded in predicting 29 of top 30 candidate circRNAs. For example, circpvt1 (top1) can be worked as miRNA spouse to regulate miRNA by moderating let-7 activity selected [30], and circRNA hsa_circ_104689 wasn’t predicted by our method and the predicting result have been presented in Table 3.

Table 3.
The top 30 breast cancer related candidates circRNAs.
Gastric cancer [34] causes a high mortality rate in human. It can be produced in any tissue of the human stomach. These tumors in the stomach are usually malignant tumors, and they can also destroy the surrounding nervous tissue. With our computational method, there are 25 of top 30 candidate circRNAs that have been confirmed by another database, circRNA disease. For example, hsa_circ_0076304 (top1) and hsa_circ_0076305 (top2) are identified to downregulate in a group of gastric cancer [35]. circpvt1 (top3) can be regarded as the sponge of the miR-125 family [13], which can upregulate in the gastric cells. The more details of results are shown in Table 4.

Table 4.
The top 30 gastric cancer related candidates circRNAs.
Colorectal cancer [36] is one of the three most frequent cancers for women. Even though the incidence of colorectal cancer has been declined for a long time, a large proportion of patients die each year from colorectal cancer. In this study, we have succeeded in predicting 24 of top 30 candidate circRNAs. For example, hsa_circ_0001649 (top1) [31] has been identified to downregulate in colorectal cancer tissue. hsa_circ_0007534 (top2) [37] can upregulate in the different colorectal cancer cells. The more details of results are presented in Table 5.

Table 5.
The top 30 colorectal cancer related candidates circRNAs.
3. Materials and Methods
3.1. Human circRNA-Disease Associations Network
All the circRNA-disease associations are downloaded from the website of circR2Disease database [16] (http://bioinfo.snnu.edu.cn/CircR2Disease/). This initial dataset contains 739 associations between 661 circRNA entities and 100 disease entities that are found based on three main species—human, mouse and rat. In this study, we select 541 circRNA entities and 83 human disease entities from our initial dataset, which includes Gastric cancer, Breast cancer, Colorectal cancer, etc. Finally, we obtain 592 circRNA-disease associations, which have experimentally been verified. These make up our circRNA-disease association network with adjacency matrix M. If there is a verified association between disease i and circRNA j, the entry M(i, j) is equal to 1, otherwise it is equal to 0.
3.2. CircRNA Semantic Similarity
For calculating circRNA semantic similarity, we download circRNA and its related gene targets dataset from circR2Disease. To measure circRNA semantic similarities, we also need to obtain gene related annotation terms that can be downloaded from Human Protein Reference Database (HPRD) database [38] (http://www.hprd.org/). Reviewing previous literature [39,40,41], there are some methods that can be referred to calculate the circRNA-related gene GO terms semantic similarities, including path-length-based methods, information-content-based methods, common-term-based methods and hybrid methods. In this study, we utilize a common-term-based method to measure circRNA similarity scores based on JACCARD index. In the previous studies [21,42], genes have been widely adopted to infer RNA similarity. Thus, the more gene related terms were shared by two circRNA Ci and Cj, the higher the similarity score they get. Denote CS as the circRNA semantic similarity matrix, and its entry CS(i, j) can be calculated by the following formula:
where Gi/Gj denotes the GO terms that circRNA Ci/Cj target genes related.
3.3. Disease Functional Similarity
We adopt disease related gene annotations to measure disease functional similarities. These gene annotations are being extracted from two online databases. The first one is DisGeNET [43] (http://www.disgenet.org/web/DisGeNET/menu), which collects 381,056 gene-disease associations (GDAs) between 16,666 genes and 13,172 diseases. In addition, we also download disease phenotype data from OMIM [44]—Online Mendelian Inheritance in Man. OMIM is a biological database that is updated daily. We use the OMIM_2018_04_24 version. Then we integrate multiple annotation resources of diseases related genes, which help us get a more reliable performance.
There are also some methods for calculating disease similarities from previous studies[45]. The common methods include annotation-based measurements, function-based measurements and topology-based measurements [46,47,48,49]. We have adopted annotation-based methods to obtain disease similarities. We apply the JACCARD index, which is a standard method for computing similarities based on two collections of finite numbers of elements so as to estimate the similarity scores between diseases. Let gdi be a collection of annotations of a gene associated with disease di. We calculate the functional similarity score of two diseases di and dj based on the JACCARD similarity coefficient score of gdi and gdi. Denote DS as the disease functional similarity matrix, then its entry DS(i, j) can be calculated by the following formula:
We have constructed circRNA semantic similarity matrix based on their related GO terms and disease functional similarity based on its related annotating genes. However, one essential weakness that cannot be ignored is that the aforementioned similarity matrices are sparse, which indicates similarity of many pairs of diseases (or circRNAs) are unable to be calculated in their functional (or semantic) similarity matrices. To alleviate this weakness, the Gaussian interaction profile (GIP) kernel similarity [50,51] is adopted in this study to get additional information about the similarity of diseases and circRNAs.
3.4. CircRNA GIP Kernel Similarity
There is an assumption that the more similar the circRNA is, the more likely similar patterns of association and non-association with diseases. The GIP kernel similarity is adopted to calculate similarity based on the topological features of the known associations network widely, such miRNA-disease associations network [52], lncRNA-disease associations networks [53] and drug-target association network [54]. Accordingly, GIP kernel similarity is also used in this study to calculate the similarity of circRNA and disease. According to previous literature [54], we use a binary vector C(i) to indicate whether circRNA i is associated with diseases. The GIP kernel similarity between circRNA C(i) and C(j) can be computed by the following formula:
To overcome the shortcomings that the disease functional similarity matrix and circRNA semantic matrix are sparse matrices, the parameter is to adjust the kernel bandwidth, which can be calculated by the following formula:
where nc is the number of circRNAs in our finial dataset. The parameter γ’c is set as 1 based on the previous study [54], which has obtained a better performance.
3.5. Disease GIP Kernel Similarity
We also calculate the GIP kernel similarity score between disease i and j as follows:
where d(i) and d(j) are the association profiles of diseases i and j, respectively, nd is the number of diseases in our finial dataset, γ’d is also set to 1 based on previous studies.
3.6. Combine Multiple Similarity (circRNA and Disease)
We integrate the GIP kernel similarity for circRNAs with the semantic similarity of circRNAs to construct the circRNA similarity network. Specifically, the elements of the adjacency matrix of this network is calculated as follows:
We also integrate the GIP kernel similarity for diseases with the functional similarity diseases to construct the diseases similarity network. Specifically, the elements of the adjacency matrix of this network is calculated as follows:
3.7. Constructing Heterogeneous Network
After we obtain the final disease similarity scores and circRNA similarity scores. We can construct an initial heterogeneous network, which is composed of disease similarity network, circRNA network and disease-circRNA associations network.
In this initial heterogeneous network, there are some small weighted edges, which may represent noises. Therefore, to weaken the effect of those unimportant or noisy edges, we set a threshold γ (γ is equal to 0.5 based on previous studies [26] and our experiment) to remove them. Specifically, let Pfinal and Pinitial be the adjacency matrices of the final and heterogeneous network, respectively, then we have:
3.8. Perfomance Metrics
In this study, we adopt the AUC value to measure the prediction results. The AUC is the area under the ROC curve, which depicts the true positive rate (TPR) verse the false positive rate (FPR). The following equations are adopted to calculate the TPR and FPR:
where TP are positive samples (known associations), which are identified correctly, and TN are negative samples (unknown associations), which are identified correctly. FP are positive samples which are identified incorrectly while FN are negative samples, which are identified incorrectly.
3.9. PWCDA
In this study, we proposed a novel computational model called PWCDA (a Path-Weighted CircRNA-Disease Associations method) to predict potential associations between circRNAs and diseases. The framework of our method is depicted in Figure 3. The computational method PWCDA traverses each node in each pathway without repeating based on heterogeneous network. To avoid traversing the same node repeatedly, we adopt the depth-first search (DFS) algorithm and mark the traversed nodes during each turn. Depth first search is implemented as a recursive function traversing the graph moving along the edge. We modify it to mark nodes, because they are accessed in recursion, and then delete tags before returning from recursive calls. In this study, we set the maximum searching length η as 3 steps according to previous studies [26], i.e., for circRNA i and disease j, there are several pathways, such as circRNA i connecting disease j directly, circRNA i’s neighbor circRNA connecting with disease j or circRNA i connecting with disease j’s neighbor diseases, circRNA i’s neighbor circRNAs connecting with disease j’s neighbor diseases directly. The choice of these paths is based on a hypothesis that the larger similarity score is between two circRNAs, the higher probability that they have the same associations is. Thus, after the weight of each circRNA-disease pair within all three paths are summed up. We can obtain the final scores between each circRNA-disease pair.
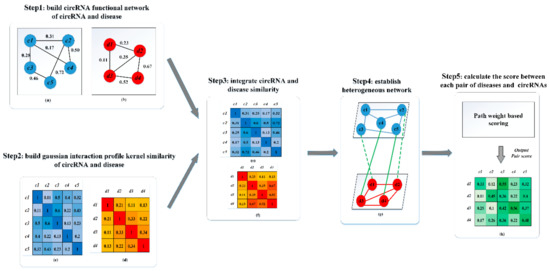
Figure 3.
The flowchart of PWCDA is illustrated by five main steps. Step 1: Calculate circRNA semantic similarity and disease similarity scores, respectively. Step 2: Calculate GIP Kernel similarity scores for circRNAs and diseases. Step 3: Integrate circRNA (disease) semantic (functional) similarity with circRNA/disease GIP Kernel similarity, respectively. Step 4: Construct the heterogeneous network. Step 5: Calculate an association score for each circRNA-disease pair.
The more the number of paths between circRNA j and disease i exists, the greater the predictive score they obtain. Accordingly, the path set that connects circRNA Cj to disease di can be represented as {p1, p2, …, pm}, where m is the number of the paths that connect disease di and circRNA Cj with the length less than η. The final predictive scores of Cj and di can be calculated as follows:
where Spath(Pk) is the score of the path pk = {e1, e2, …, en} [42] can be calculated as follows:
The longer the path is, the smaller the contribution it is made, which means that the longer path would have less effect on predicting potential circRNA-disease associations than the shorter one. Therefore, the decaying function is an exponential function to reduce the influence of long path on final prediction scores, which can be represented as Equation (14):
where α is a constraint factor and len(pk) is the length of path pk.
An example for calculating the score between circRNA c1 and disease d2 is shown in Figure 4. In the Figure 4, three paths {c1-c4-d2}, {c1-c3-d1-d2} and {c1-c5-d3-d2}, which are marked as red, are used to calculate the score between c1 and d2. Therefore, the score of c1 and d2 can be calculated as follows: Score (c1, d2) = {c1-c4-d2} (w2 × w5)3*exp(2) + {c1-c3-d1-d2} (w1 × w4 × w7)3*exp(3) + {c1-c5-d3-d2} (w3 × w6 × w8)3*exp(3). There are also some other paths that can connect c1 with d2. Because the length of those paths, such as {c1-c2-c5-d3-d2}, are more than 3, we don’t consider this path.
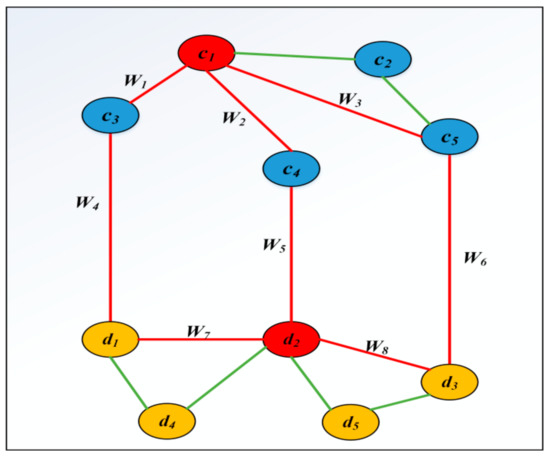
Figure 4.
The path between c1 and d2 is within the maximum path length.
4. Conclusions
With the increasing number of diseases related to circRNAs being discovered, more and more researchers have been paying attention to investigate diseases-related circRNAs. Although, experimental methods can find potential circRNA-disease associations with a high precision, the process is not only time-consuming, but also expensive. Here, we have proposed an effective computational method called PWCDA, which can predict potential circRNA-disease associations. Firstly, we calculate disease/circRNA similarities by combining their functional/semantic similarity and GIP kernel similarity. Secondly, we build a heterogeneous network, including the circRNA-disease association sub-network, the disease similarity sub-network and the circRNA similarity sub-network. PWCDA searches all the paths within three steps to compute an association score for each circRNA-disease pair to determine if a circRNA-disease pair is associated.
To thoroughly investigate the performance of our proposed method, we adopt LOOCV and five-fold cross validation. Furthermore, we have also compared our method with two state-of-the-art prediction methods. The comparison results illustrate that our methods work much better than other methods. The AUC value of five-fold cross validation is 0.884. Moreover, we apply our method to three diseases: Breast Cancer, Gastric Cancer, Colorectal Cancer for case studies.
There are several significant factors, which may explain why our proposed method can get a better performance than other computational models. Firstly, we have taken into account the sparsity of disease/circRNA similarity sub-networks. Thus, we have integrated disease functional similarity scores and circRNA semantic similarity scores with their corresponding GIP kernel similarity scores. Secondly, according to previous studies, we just use the paths within three steps, which can reduce the noisy information. Although we have combined different similarity scores, there is still some information unavailable. Therefore, we set a threshold to remove those edges whose weights are less than the predefined threshold.
Although we get a much better performance than other computational models, we can’t ignore the limitation. The prediction of associations between circRNAs and diseases is a relatively new research field, and the amount of data that we can use is limited. The ratio of positive samples to negative samples of circRNA-disease association is seriously unbalanced. To solve this problem, we may have two main solutions. One is that we can update the circRNA-disease database to obtain new data. The other is that we can extract the same number of positive samples as that of negative samples. Furthermore, our computational method tends to predict those circRNA-disease associations that are covered in the known associations’ dataset, and it just predicts fewer novel circRNA-disease associations. Thus, we will adopt more biological data to overcome this weakness. As a future topic, we can apply this work to the disease diagnosis based on network biomarkers [55,56,57] and disease prediction based on dynamic network biomarkers [58,59,60] in an accurate and reliable manner.
Author Contributions
X.L. conceptualized the draft and proposed the methodology; Z.F. designed the software; L.C. and F.W. gave the validation and formal analysis; Z.F. performed the investigation and analyzed the resources and data curation; X.L. and Z.F. drafted the manuscript; L.C. and F.W. reviewed and edited the manuscript; F.W. polished the English expression; Z.F. visualized and X.L. supervised; X.L. is the project administrator and has acquired the funding. All the authors read and approved the manuscript.
Acknowledgments
This work was supported by the funding from National Natural Science Foundation of China (61672334, 61502290, 61401263, 31771476, 91529303), and the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB13040700).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| PWCDA | Path Weighed method, for predicting CircRNA-Disease Associations |
| LOOCV | leave one out cross validation |
| GIP | Gaussian Interaction Profile |
| GO | Gene Ontology |
| AUC | Area Under roc Curve |
| ROC | Receiver Operating Characteristic Curve |
| TPR | True Positive Rate |
| FPR | False Positive Rate |
References
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, P.J.; Zaug, A.J.; Cech, T.R. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell 1981, 23, 467–476. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176. [Google Scholar] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 426. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Bao, Y.; Yee, M.-C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Armakola, M.; Higgins, M.J.; Figley, M.D.; Barmada, S.J.; Scarborough, E.A.; Diaz, Z.; Fang, X.; Shorter, J.; Krogan, N.J.; Finkbeiner, S.; et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat. Genet. 2012, 44, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, A. Circular RNAs as miRNA sponges. Nat. Rev. Mol. Cell Boil. 2013, 14, 195. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Sève, P.; Reiman, T.; Dumontet, C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer 2010, 67, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Xu, X.; Ouyang, Y.; Wang, Y.; Yang, J.; Yin, L.; Ge, J.; Wang, H. Microarray expression profile analysis of circular RNAs in pancreatic cancer. Mol. Med. Rep. 2018, 17, 7661–7671. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Lei, X.; Fang, Z.; Jiang, Q.; Wu, F.-X. CircR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Das, S.; Sen, R.; Basak, P.; Chakrabarti, J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013, 4, 283. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Zhang, L.; Zheng, M.; Sun, X.; Lu, Y.; Liu, P. Circ2Disease: A manually curated database of experimentally validated circRNAs in human disease. Sci. Rep. 2018, 8, 11018. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Liu, B.; Yan, C. SACMDA: MiRNA-Disease Association Prediction with Short Acyclic Connections in Heterogeneous Graph. Neuroinformatics 2018, 16, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, L.-Y.; Huang, L. NDAMDA: Network distance analysis for MiRNA-disease association prediction. J. Cell. Mol. Med. 2018, 22, 2884–2895. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, X.; He, Z.; Zou, Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Z.; Chen, Z.; Deng, L. Integrating Multiple Heterogeneous Networks for Novel LncRNA-disease Association Inference. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Wang, J.; Domeniconi, C.; Yu, G. Matrix factorization-based data fusion for the prediction of lncRNA-disease associations. Bioinformatics 2018, 34, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, N.; Chen, P.; Zhang, J.; Wang, B. DrugECs: An Ensemble System with Feature Subspaces for Accurate Drug-Target Interaction Prediction. Biomed. Res. Int. 2017, 2017, 6340316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, Y.; Li, D. Drug-Target Interaction Prediction through Label Propagation with Linear Neighborhood Information. Molecules 2017, 22, 2056. [Google Scholar] [CrossRef] [PubMed]
- Ba-Alawi, W.; Soufan, O.; Essack, M.; Kalnis, P.; Bajic, V.B. DASPfind: New efficient method to predict drug-target interactions. J. Cheminform. 2016, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Patra, J.C. Genome-wide inferring gene-phenotype relationship by walking on the heterogeneous network. Bioinformatics (Oxf. Engl.) 2010, 26, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.-H.; Huang, Y.-A.; Yan, G.-Y. HGIMDA: Heterogeneous graph inference for miRNA-disease association prediction. Oncotarget 2016, 7, 65257–65269. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yang, Y.; Xu, J.; Bai, W.; Ren, X.; Wu, H. CircRNAs as biomarkers of cancer: A meta-analysis. BMC Cancer 2018, 18, 303. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C.; Grammatikakis, I.; Kim, K.M.; De, S.; Martindale, J.L.; Munk, R.; Yang, X.; Abdelmohsen, K.; Gorospe, M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017, 45, 4021–4035. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, L.; Su, Y.; Zhang, X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol. Rep. 2018, 39, 2499–2512. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, K.; Wu, F.; Wang, W.; Zhang, K.; Hu, H.; Liu, Y.; Jiang, T. circRNA disease: A manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Reis-Filho, J.S.; Baehner, F.; Dabbs, D.J.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010, 12, 207. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Lan, F.; Ouyang, X.; Wang, K.; Lin, Y.; Yu, Y.; Wang, L.; Wang, Y.; Huang, Q. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J. Surg. Oncol. 2015, 13, 302. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Ouyang, X.; Zhang, F.; Wang, K.; Lin, Y.; Sun, B.; Wang, Y.; Wang, L.; Huang, Q. Circular RNAs expression profiles in human gastric cancer. Sci. Rep. 2017, 7, 9060. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, J.; Zhao, J.; Wang, X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 118–126. [Google Scholar] [PubMed]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Price, T.; Peña, F.I.; Cho, Y.-R. Survey: Enhancing protein complex prediction in PPI networks with GO similarity weighting. Interdiscip. Sci. 2013, 5, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, T.; Pakhomov, S.V.S.; Patwardhan, S.; Chute, C.G. Measures of semantic similarity and relatedness in the biomedical domain. J. Biomed. Inf. 2007, 40, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, P.H.; Mina, M.; Guerra, C.; Cannataro, M. Semantic similarity analysis of protein data: Assessment with biological features and issues. Brief. Bioinform. 2012, 13, 569–585. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-A.; Chan, K.C.C.; You, Z.-H. Constructing prediction models from expression profiles for large scale lncRNA-miRNA interaction profiling. Bioinformatics 2018, 34, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Pinero, J.; Queralt-Rosinach, N.; Bravo, A.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Oyston, J. Online Mendelian Inheritance in Man. Anesthesiology 1998, 89, 811–812. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Yang, M.; Luo, F.; Wu, F.-X.; Li, M.; Pan, Y.; Li, Y.; Wang, J. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics 2018, 34, 3357–3364. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Gonçalves, J.P.; Larminie, C.; Przulj, N. Predicting disease associations via biological network analysis. BMC Bioinform. 2014, 15, 304. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, M.; Shi, H.; Ju, H.; Jiang, Q.; Cheng, L. Measuring disease similarity and predicting disease-related ncRNAs by a novel method. BMC Med. Genom. 2017, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Jiang, Y.; Wang, Z.; Shi, H.; Sun, J.; Yang, H.; Zhang, S.; Hu, Y.; Zhou, M. DisSim: An online system for exploring significant similar diseases and exhibiting potential therapeutic drugs. Sci. Rep. 2016, 6, 30024. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhao, L.; Liu, Z.; Ju, H.; Shi, H.; Xu, P.; Wang, Y.; Cheng, L. DisSetSim: An online system for calculating similarity between disease sets. J. Biomed. Semant. 2017, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, M.-X.; Yan, G.-Y. RWRMDA: Predicting novel human microRNA-disease associations. Mol. Biosyst. 2012, 8, 2792–2798. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, G.-Y. Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 2013, 29, 2617–2624. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, A.; Feng, H.; Wang, M. NTSMDA: Prediction of miRNA-disease associations by integrating network topological similarity. Mol. Biosyst. 2016, 12, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. KATZLDA: KATZ measure for the lncRNA-disease association prediction. Sci. Rep. 2015, 5, 16840. [Google Scholar] [CrossRef] [PubMed]
- van Laarhoven, T.; Nabuurs, S.B.; Marchiori, E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics 2011, 27, 3036–3043. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zeng, T.; Liu, X.; Chen, L. Diagnosing phenotypes of single-sample individuals by edge biomarkers. J. Mol. Cell Biol. 2015, 7, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, J.; Sun, S.; Zhou, X.; Zeng, T.; Chen, L. Individual-specific edge-network analysis for disease prediction. Nucleic Acids Res. 2017, 45, e170. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, Y.; Zhang, X.J.; Chen, L. Part mutual information for quantifying direct associations in networks. Proc. Natl. Acad. Sci. USA 2016, 113, 5130–5135. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, R.; Liu, Z.P.; Li, M.; Aihara, K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2012, 2, 342. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, M.; Tang, W.; Liu, W.; Zhang, S.; Chen, L.; Xia, J. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 2018, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, C.; Liu, W.; Liu, C.; Cui, J.; Li, Q.; Ni, H.; Yang, Y.; Wu, C.; Chen, C.; et al. Dysfunction of PLA2G6 and CYP2C44 associated network signals imminent carcinogenesis from chronic inflammation to hepatocellular carcinoma. J. Mol. Cell Biol. 2018, 9, 489–503. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).