Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics
Abstract
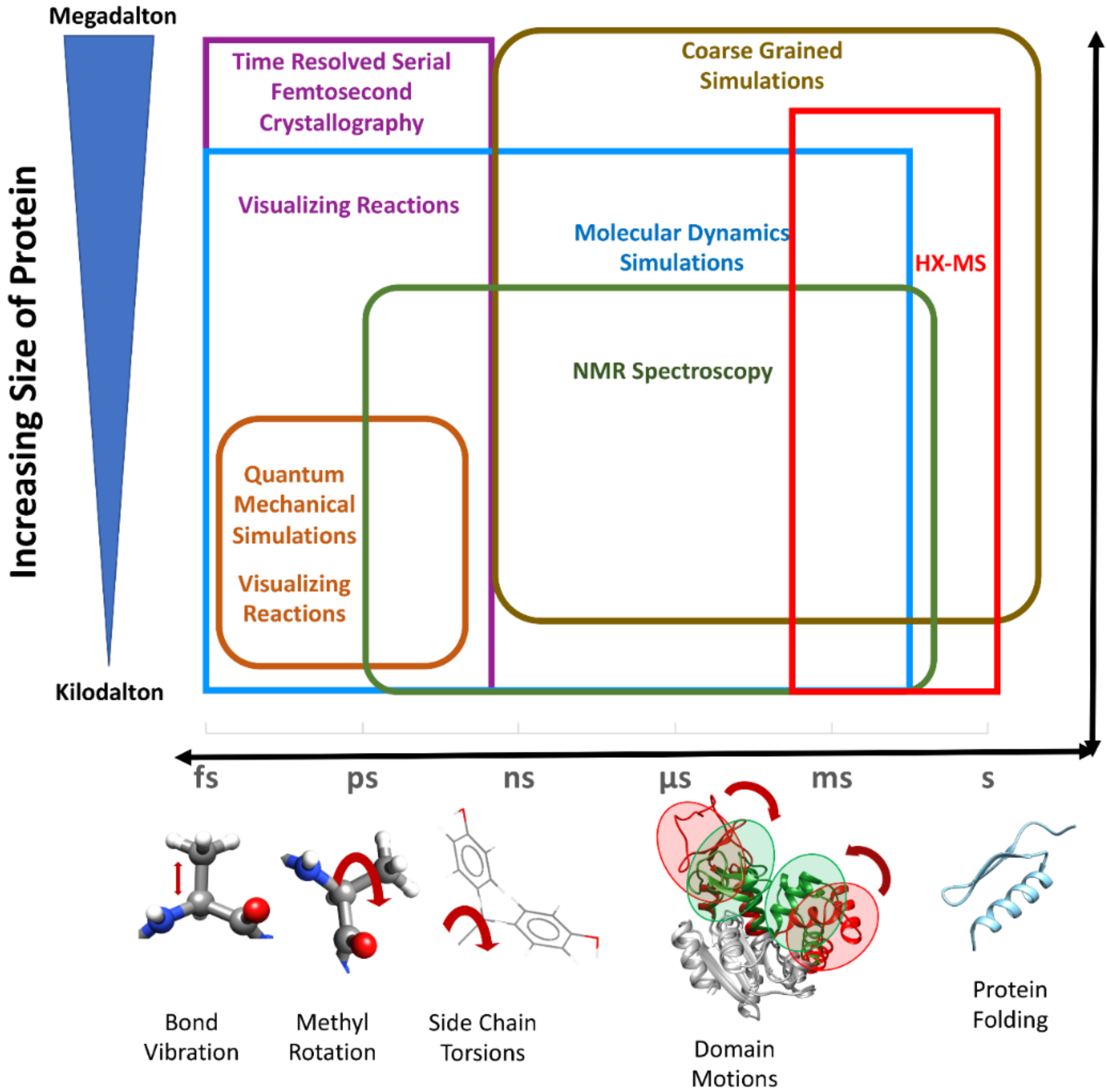
1. Introduction
2. Constraints in Crystallography for Studying Protein Structure and Dynamics
2.1. Challenges in Data Collection and Interpretation of Diffraction Data
2.2. Challenges in Retrieving Biochemical Information from Crystal Structures
2.2.1. Crystal Environment Artefacts
2.2.2. Cryo-Cooling Effects
2.2.3. Missing Residues, High-Flexibility Regions
2.2.4. Missing Water Molecules and Solvent Information
3. Recent Experimental Advances for Enhancing/Complementing Crystallography
3.1. Advancements of Crystallography Methods
3.1.1. Crystallographic Refinement Methods
3.1.2. Serial Femtosecond Crystallography and Single-Particle Experiments with X-ray Free-Electron Laser (XFEL) Sources
3.1.3. Multi-Temperature/Room-Temperature Crystallography
3.1.4. Crystal Contact Free Space
3.2. Other Biophysical Techniques
4. Computational Methods for Complementing/Supplementing Experiments
4.1. Protein Structural Modeling
4.2. Hybrid Methods for Studying Protein Complexes
4.3. Molecular Mechanics Methods
4.3.1. Coarse-Grained Modeling
4.3.2. Enhanced MD Simulations
4.4. Crystal MD Simulations
5. Novel Insights from Computational Methods
5.1. Exploring the Structure and Dynamics of Large Macromolecular Complexes
5.2. Structural and Dynamical Effects of Post-Translational Modifications
5.3. Structure-Based Drug Design
5.4. Understanding the Role of Water in Protein Structure and Function
6. Limitations of Computational Methods
6.1. Force Fields
6.2. Sampling
7. Summary and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Kendrew, J.C.; Bodo, G.; Dintzis, H.M.; Parrish, R.G.; Wyckoff, H.; Phillips, D.C. A three-dimensional model of the Myoglobin molecule obtained by X-ray analysis. Nature 1958, 181, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. A Glimpse of Structural Biology through X-ray Crystallography. Cell 2014, 159, 995–1014. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.; Kaltenbach, M.; Correy, G.J.; Carr, P.D.; Porebski, B.T.; Livingstone, E.K.; Afriat-Jurnou, L.; Buckle, A.M.; Weik, M.; Hollfelder, F.; et al. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 2016, 12, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Sailer, Z.R.; Harms, M.J. Molecular ensembles make evolution unpredictable. Proc. Natl. Acad. Sci. USA 2017, 114, 11938–11943. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rosell, G.; Giorgino, T.; Harvey, M.J.; De Fabritiis, G. Drug Discovery and Molecular Dynamics: Methods, Applications and Perspective beyond the Second Timescale. Curr. Top. Med. Chem. 2017, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chruszcz, M.; Wlodawer, A.; Minor, W. Determination of Protein Structures—A Series of Fortunate Events. Biophys. J. 2008, 95, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R. A historical perspective on protein crystallization from 1840 to the present day. FEBS J. 2013, 280, 6456–6497. [Google Scholar] [CrossRef] [PubMed]
- Taberman, H. Radiation Damage in Macromolecular Crystallography—An Experimentalist’s View. Crystals 2018, 8, 157. [Google Scholar] [CrossRef]
- Yano, J.; Kern, J.; Irrgang, K.-D.; Latimer, M.J.; Bergmann, U.; Glatzel, P.; Pushkar, Y.; Loll, B.; Sauer, K.; Messinger, J.; et al. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. USA 2005, 102, 12047–12052. [Google Scholar] [CrossRef] [PubMed]
- Pozharski, E.; Weichenberger, C.X.; Rupp, B. Techniques, tools and best practices for ligand electron-density analysis and results from their application to deposited crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Weichenberger, C.X.; Pozharski, E.; Rupp, B. Visualizing ligand molecules in twilight electron density. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Depristo, M.A.; De Bakker, P.I.W.; Blundell, T.L. Heterogeneity and Inaccuracy in Protein Structures Solved by X-ray Crystallography. Structure 2004, 12, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E., Jr. Conformational changes: How small is big enough? Nat. Med. 1998, 4, 1112–1114. [Google Scholar] [CrossRef] [PubMed]
- Gutteridge, A.; Thornton, J. Conformational Changes Observed in Enzyme Crystal Structures upon Substrate Binding. J. Mol. Biol. 2005, 346, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.; del Sol, A.; Nussinov, R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol. 2009, 378, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Nakamura, H.; Kinoshita, K. Discrimination between biological interfaces and crystal-packing contacts. Adv. Appl. Bioinform. Chem. 2008, 1, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Janin, J.; Rodier, F. Protein–Protein interaction at Crystal Contacts. Proteins Struct. Funct. Genet. 1995, 23, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Bahadur, R.P.; Chakrabarti, P.; Rodier, F.; Janin, J. A Dissection of Specific and Non-specific Protein-Protein Interfaces. J. Mol. Biol. 2004, 336, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O.; Argos, P. Protein-protein crystal-packing contacts. Protein Sci. 1997, 6, 2261–2263. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, L.S.; Miyashita, O. Molecular Simulation Uncovers the Conformational Space of the λ Cro Dimer in Solution. Biophys. J. 2011, 101, 2516–2524. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, L.S.; Miyashita, O. Packing interface energetics in different crystal forms of the λ Cro dimer. Proteins Struct. Funct. Bioinform. 2014, 82, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Andrec, M.; Snyder, D.A.; Hou, Z.; Young, J.; Montelione, G.T.; Levy, R.M. A large data set comparison of protein structures determined by crystallography and NMR: Statistical test for structural differences and the effect of crystal packing Michael. Proteins Struct. Funct. Bioinform. 2007, 69, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-J.; Wozniak, J.A.; Matthews, B.W. Protein Flexibility and Adaptability Seen in 25 Crystal Forms of T4 Lysozyme. J. Mol. Biol. 1995, 250, 527–552. [Google Scholar] [CrossRef] [PubMed]
- Klopffleisch, K.; Issinger, O.G.; Niefind, K. Low-density crystal packing of human protein kinase CK2 catalytic subunit in complex with resorufin or other ligands: A tool to study the unique hinge-region plasticity of the enzyme without packing bias. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Hirota, T.; Irle, S.; Tama, F. Conformational dynamics of human protein kinase CK2α and its effect on function and inhibition. Proteins Struct. Funct. Bioinform. 2018, 86, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, D62, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanic, A.; Pannu, N.S.; Zagrovic, B. X-ray refinement significantly underestimates the level of microscopic heterogeneity in biomolecular crystals. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Tilton, R.F.; Dewan, J.C.; Petsko, G.A. Effects of Temperature on Protein Structure and Dynamics: X-ray Crystallographic Studies of the Protein Ribonuclease-A at Nine Different Temperatures from 98 to 320. Biochemistry 1992, 31, 2469–2481. [Google Scholar] [CrossRef] [PubMed]
- Halle, B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc. Natl. Acad. Sci. USA 2004, 101, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- Djinovic-Carugo, K.; Carugo, O. Missing strings of residues in protein crystal structures. Intrinsically Disord. Proteins 2015, 3, e1095697. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef] [PubMed]
- Linke, K.; Ho, F.M. Water in Photosystem II: Structural, functional and mechanistic considerations. BBA Bioenerg. 2014, 1837, 14–32. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. Water is an active matrix of life for cell and molecular biology. Proc. Natl. Acad. Sci. USA 2017, 114, 13327–13335. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.; Singh, A.K.; Bhasne, K.; Dogra, P.; Datta, A.; Das, P.; Mukhopadhyay, S. Femtosecond Hydration Map of Intrinsically Disordered α-Synuclein. Biophys. J. 2018, 114, 2540–2551. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-D.; Zhang, H.; Terwilliger, T.C.; Liljas, A.; Xiao, J.; Dong, Y. Protein Crystallography from the Perspective of Technology Developments. Crystallogr. Rev. 2015, 21, 122–153. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Hirata, K.; Hasegawa, K.; Ueno, G.; Yamamoto, M. Synchrotron radiation dose dependence of radiation damage for protein crystals studied at various X-ray energies. J. Synchrotron Radiat. 2007, 14, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Rosenbaum, G.; Dauter, M.; Dauter, Z. Radiation decay of thaumatin crystals at three X-ray energies. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Holton, J.M. A beginner’s guide to radiation damage. J. Synchrotron Radiat. 2009, 16, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.T.; Ng, H.-L.; Fraser, J.S.; Corn, J.E.; Echols, N.; Sales, M.; Holton, J.M.; Alber, T. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 2010, 19, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.J.; Kondrashov, D.A.; Wesenberg, G.E.; Phillips, G.N. Ensemble Refinement of Protein Crystal Structures: Validation and Application. Structure 2007, 15, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Adams, P.D.; Moriarty, N.W.; Zwart, P.; Read, R.J.; Turk, D.; Hung, L.-W. Interpretation of ensembles created by multiple iterative rebuilding of macromolecular models. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bedem, H.; Dhanik, A.; Latombe, J.-C.; Deacon, A.M. Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Tom Burnley, B.; Afonine, P.V.; Adams, P.D.; Gros, P. Modelling dynamics in protein crystal structures by ensemble refinement. Elife 2012, 1, 311. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, F.; Echols, N.; Headd, J.J.; Terwilliger, T.C.; Adams, P.D.; Baker, D. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods 2013, 10, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Pearce, N.M.; Krojer, T.; Bradley, A.R.; Collins, P.; Nowak, R.P.; Talon, R.; Marsden, B.D.; Kelm, S.; Shi, J.; Deane, C.M.; et al. A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.C.; Stauch, B.; Ishchenko, A.; Cherezov, V. A Bright Future for Serial Femtosecond Crystallography with XFELs. Trends Biochem. Sci. 2017, 42, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Martin-Garcia, J.M.; Conrad, C.E.; Coe, J.; Roy-Chowdhury, S.; Fromme, P. Serial femtosecond crystallography: A revolution in structural biology. Arch. Biochem. Biophys. 2016, 602, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Neutze, R.; Wouts, R.; van der Spoel, D.; Weckert, E.; Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wacker, D.; Gati, C.; Han, G.W.; James, D.; Wang, D.; Nelson, G.; Weierstall, U.; Katritch, V.; Barty, A.; et al. Serial Femtosecond Crystallography of G Protein—Coupled Receptors. Science 2013, 342, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- Tosha, T.; Nomura, T.; Nishida, T.; Saeki, N.; Okubayashi, K.; Yamagiwa, R.; Sugahara, M.; Nakane, T.; Yamashita, K.; Hirata, K.; et al. Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate. Nat. Commun. 2017, 8, 1585. [Google Scholar] [CrossRef] [PubMed]
- Shimada, A.; Kubo, M.; Baba, S.; Yamashita, K.; Hirata, K.; Ueno, G.; Nomura, T.; Kimura, T.; Shinzawa-Itoh, K.; Baba, J.; et al. A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome C oxidase. Sci. Adv. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- Nogly, P.; Weinert, T.; James, D.; Carbajo, S.; Ozerov, D.; Furrer, A.; Gashi, D.; Borin, V.; Skopintsev, P.; Jaeger, K.; et al. Retinal isomerization in bacteriorhodopsin captured by a femtosecond X-ray laser. Science 2018, 361, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nango, E.; Royant, A.; Kubo, M.; Nakane, T.; Wickstrand, C.; Kimura, T.; Tanaka, T.; Tono, K.; Song, C.; Tanaka, R.; et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 2016, 354, 1552–1557. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.J.D. Femtosecond Crystallography with Ultrabright Electrons and X-rays: Capturing Chemistry in Action. Science 2014, 343, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, O.; Joti, Y. X-ray free electron laser single-particle analysis for biological systems. Curr. Opin. Struct. Biol. 2017, 43, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Fan, J.; Li, H.; Jiang, H. Current Status of Single Particle Imaging with X-ray Lasers. Appl. Sci. 2018, 8, 132. [Google Scholar] [CrossRef]
- Aquila, A.; Barty, A.; Bostedt, C.; Boutet, S.; Carini, G.; Deponte, D.; Drell, P.; Doniach, S.; Downing, K.H.; Earnest, T.; et al. The linac coherent light source single particle imaging road map. Struct. Dyn. 2015, 2, 41701. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Miyashita, O.; Jonic, S.; Tokuhisa, A.; Tama, F. Single-particle XFEL 3D reconstruction of ribosome-size particles based on Fourier slice matching: Requirements to reach subnanometer resolution. J. Synchrotron Radiat. 2018, 25, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Hantke, M.F.; Ekeberg, T.; Maia, F.R.N.C. Condor: A simulation tool for flash X-ray imaging. J. Appl. Cryst. 2016, 49, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Joti, Y.; Shibuya, A.; Song, C.; Kim, S.; Tono, K.; Yabashi, M.; Tamakoshi, M.; Moriya, T.; Oshima, T.; et al. Imaging live cell in micro-liquid enclosure by X-ray laser diffraction. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]
- Loh, N.-T.D.; Elser, V. Reconstruction algorithm for single-particle diffraction imaging experiments. Phys. Rev. E 2009, 80, 026705. [Google Scholar] [CrossRef] [PubMed]
- Ekeberg, T.; Svenda, M.; Abergel, C.; Maia, F.R.N.C.; Seltzer, V.; Claverie, J.-M.; Hantke, M.; Jönsson, O.; Nettelblad, C.; Van Der Schot, G.; et al. Three-Dimensional Reconstruction of the Giant Mimivirus Particle with an X-ray Free-Electron Laser. Phys. Rev. Lett. 2015, 114, 098102. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Mochizuki, Y.; Joti, Y.; Tama, F.; Miyashita, O. Gaussian mixture model for coarse-grained modeling from XFEL. Opt. Express 2018, 26, 26734. [Google Scholar] [CrossRef]
- Fraser, J.S.; Clarkson, M.W.; Degnan, S.C.; Erion, R.; Kern, D.; Alber, T. Hidden alternative structures of proline isomerase essential for catalysis. Nature 2009, 462, 669–673. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.S.; van den Bedem, H.; Samelson, A.J.; Lang, P.T.; Holton, J.M.; Echols, N.; Alber, T. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. USA 2011, 108, 16247–16252. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, R.B.; van den Bedem, H.; Fraser, J.S.; Wright, P.E. Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR. Proc. Natl. Acad. Sci. USA 2014, 111, E445–E454. [Google Scholar] [CrossRef] [PubMed]
- Sierra, R.G.; Gati, C.; Laksmono, H.; Dao, E.H.; Gul, S.; Fuller, F.; Kern, J.; Chatterjee, R.; Ibrahim, M.; Brewster, A.S.; et al. Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II. Nat. Methods 2015, 13, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, R.; Shimada, A.; Komuro, Y.; Sugita, Y.; Kohda, D. Rational design of crystal contact-free space in protein crystals for analyzing spatial distribution of motions within protein molecules. Protein Sci. 2016, 25, 754–768. [Google Scholar] [CrossRef] [PubMed]
- Becker, W.; Bhattiprolu, K.C.; Gubensäk, N.; Zangger, K. Investigating Protein–Ligand Interactions by Solution Nuclear Magnetic Resonance Spectroscopy. ChemPhysChem 2018, 19, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Teilum, K.; Kunze, M.B.A.; Erlendsson, S.; Kragelund, B.B. (S)Pinning down protein interactions by NMR. Protein Sci. 2017, 26, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Chiliveri, S.C.; Deshmukh, M.V. Structure of RDE-4 dsRBDs and mutational studies provide insights into dsRNA recognition in the Caenorhabditis elegans RNAi pathway. Biochem. J. 2014, 458, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Chaitanya Chiliveri, S.; Deshmukh, M.V. Recent excitements in protein NMR: Large proteins and biologically relevant dynamics. J. Biosci. 2016, 41, 787–803. [Google Scholar] [CrossRef]
- Huang, C.; Kalodimos, C.G. Structures of Large Protein Complexes Determined by Nuclear Magnetic Resonance Spectroscopy. Annu. Rev. Biophys. 2017, 46, 317–336. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E. New Views of Functionally Dynamic Proteins by Solution NMR Spectroscopy. J. Mol. Biol. 2016, 428, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Becker-Baldus, J.; Bamann, C.; Saxena, K.; Gustmann, H.; Brown, L.J.; Brown, R.C.D.; Reiter, C.; Bamberg, E.; Wachtveitl, J.; Schwalbe, H.; et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 9896–9901. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J.; Marassi, F.M. Applications of NMR to membrane proteins. Arch. Biochem. Biophys. 2017, 628, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Jaswal, S.S. Biological insights from hydrogen exchange mass spectrometry. BBA Proteins Proteom. 2013, 1834, 1188–1201. [Google Scholar] [CrossRef] [PubMed]
- Lorenz Eisinger, M.; Dörrbaum, A.R.; Michel, H.; Padan, E.; Langer, J.D.; Silberman, A.; Borchers, C.H.; Kaback, R.; Rand, K.D. Ligand-induced conformational dynamics of the Escherichia coli Na+/H+ antiporter NhaA revealed by hydrogen/deuterium exchange mass spectrometry. Proc. Natl. Acad. Sci. USA 2017, 114, 11691–11696. [Google Scholar] [CrossRef] [PubMed]
- Offenbacher, A.R.; Iavarone, A.T.; Klinman, J.P. Hydrogen-deuterium exchange reveals long-range dynamical allostery in soybean lipoxygenase. J. Biol. Chem. 2018, 293, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Van Erp, P.B.G.; Patterson, A.; Kant, R.; Berry, L.; Golden, S.M.; Forsman, B.L.; Carter, J.; Jackson, R.N.; Bothner, B.; Wiedenheft, B. Conformational Dynamics of DNA Binding and Cas3 Recruitment by the CRISPR RNA-Guided Cascade Complex. ACS Chem. Biol. 2017, 13, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Faini, M.; Stengel, F.; Aebersold, R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem. Sci. 2016, 41. [Google Scholar] [CrossRef] [PubMed]
- Elmlund, D.; Le, S.N.; Elmlund, H. High-resolution cryo-EM: The nuts and bolts. Curr. Opin. Struct. Biol. 2017, 46, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Egelman, E.H. The Current Revolution in Cryo-EM. Biophys. J. 2016, 110, 1008–1012. [Google Scholar] [CrossRef] [PubMed]
- Haselbach, D.; Komarov, I.; Agafonov, D.E.; Kastner, B.; Lü Hrmann, R.; Stark, H.; Hartmuth, K.; Graf, B.; Dybkov, O.; Urlaub, H.; et al. Structure and Conformational Dynamics of the Human Spliceosomal B act Complex. Cell 2018, 172, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Baker, M.L.; Wang, Z.; Baker, M.R.; Sinyagovskiy, P.A.; Chiu, W.; Ludtke, S.J.; Serysheva, I.I. Gating machinery of InsP 3 R channels revealed by electron cryomicroscopy. Nature 2015, 527, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Merk, A.; Bartesaghi, A.; Banerjee, S.; Earl, L.A.; Milne, J.L.S.; Falconieri, V.; Rao, P.; Davis, M.I.; Pragani, R.; Boxer, M.B.; et al. Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery. Cell 2016, 165, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- des Georges, A.; Clarke, O.B.; Zalk, R.; Yuan, Q.; Condon, K.J.; Grassucci, R.A.; Hendrickson, W.A.; Marks, A.R.; Frank, J. Structural Basis for Gating and Activation of RyR1. Cell 2016, 167, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Meisburger, S.P.; Thomas, W.C.; Watkins, M.B.; Ando, N. X-ray Scattering Studies of Protein Structural Dynamics. Chem. Rev. 2017, 117, 7615–7672. [Google Scholar] [CrossRef] [PubMed]
- Tuukkanen, A.T.; Spilotros, A.; Svergun, D.I. Progress in small-angle scattering from biological solutions at high-brilliance synchrotrons. IUCrJ 2017, 4, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Rambo, R.P.; Tainer, J.A. Super-Resolution in Solution X-ray Scattering and Its Applications to Structural Systems Biology. Annu. Rev. Biophys. 2013, 42, 415–441. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Tanner, J.J. Determination of protein oligomeric structure from small-angle X-ray scattering. Protein Sci. 2018, 27, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Boon, P.L.S.; Saw, W.G.; Lim, X.X.; Raghuvamsi, P.V.; Huber, R.G.; Marzinek, J.K.; Holdbrook, D.A.; Anand, G.S.; Grüber, G.; Bond, P.J. Partial Intrinsic Disorder Governs the Dengue Capsid Protein Conformational Ensemble. ACS Chem. Biol. 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, T.N.; Herranz-Trillo, F.; Urbanek, A.; Estaña, A.; Cortés, J.; Sibille, N.; Bernadó, P. Small-angle scattering studies of intrinsically disordered proteins and their complexes. Curr. Opin. Struct. Biol. 2017, 42, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Josts, I.; Nitsche, J.; Maric, S.; Mertens, H.D.; Moulin, M.; Haertlein, M.; Prevost, S.; Svergun, D.I.; Busch, S.; Forsyth, V.T.; et al. Conformational States of ABC Transporter MsbA in a Lipid Environment Investigated by Small-Angle Scattering Using Stealth Carrier Nanodiscs. Structure 2018, 26, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Schwede, T.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—Round XII. Proteins Struct. Funct. Bioinform. 2018, 86, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.D.; Das, S.; Sillitoe, I.; Orengo, C. An overview of comparative modelling and resources dedicated to large-scale modelling of genome sequences. Acta Crystallogr. Sect. D Struct. Biol. 2017, D73, 628–640. [Google Scholar] [CrossRef] [PubMed]
- Schaarschmidt, J.; Monastyrskyy, B.; Kryshtafovych, A.; Bonvin, A.M.J.J. Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age. Proteins Struct. Funct. Bioinform. 2018, 86, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Bausewein, T.; Mills, D.J.; Langer, J.D.; Nitschke, B.; Nussberger, S.; Kühlbrandt, W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell 2017, 170, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, A.; Aufderheide, A.; Rudack, T.; Beck, F.; Pfeifer, G.; Plitzko, J.M.; Sakata, E.; Schulten, K.; Forster, F.; Baumeister, W. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc. Natl. Acad. Sci. USA 2016, 113, 7816–7821. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, X.; Ma, J.; Li, Z.; You, L.; Wang, J.; Wang, M.; Zhang, X.; Wang, Y. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 2017, 170, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Jasnovidova, O.; Klumpler, T.; Kubicek, K.; Kalynych, S.; Plevka, P.; Stefl, R. Structure and dynamics of the RNAPII CTDsome with Rtt103. Proc. Natl. Acad. Sci. USA 2017, 114, 11133–11138. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Kim, S.J.; An, S.; Cho, S.; Leitner, A.; Jung, T.; Aebersold, R.; Hebert, H.; Cho, U.S.; Song, J.J. Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding. J. Mol. Biol. 2018, 430, 822–841. [Google Scholar] [CrossRef] [PubMed]
- Kosinski, J.; Mosalaganti, S.; Von Appen, A.; Teimer, R.; Diguilio, A.L.; Wan, W.; Bui, K.H.; Hagen, W.J.H.; Briggs, J.A.G.; Glavy, J.S.; et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 2016, 352, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Nam Kim, D.; Sanbonmatsu, K.Y. Tools for the cryo-EM gold rush: Going from the cryo-EM map to the atomistic model. Biosci. Rep. 2017, 37, 20170072. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Miyashita, O.; Brooks, C.L., III. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 2004, 147, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Miyashita, O.; Brooks, C.L., III. Flexible Multi-scale Fitting of Atomic Structures into Low-resolution Electron Density Maps with Elastic Network Normal Mode Analysis. J. Mol. Biol. 2004, 337, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Grubisic, I.; Shokhirev, M.N.; Orzechowski, M.; Miyashita, O.; Tama, F. Biased coarse-grained molecular dynamics simulation approach for flexible fitting of X-ray structure into cryo electron microscopy maps. J. Struct. Biol. 2009, 169, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Whitford, P.C.; Noel, J.K.; Gosavi, S.; Schug, A.; Sanbonmatsu, K.Y.; Onuchic, J.N. An all-atom structure-based potential for proteins: Bridging minimal models with all-atom empirical forcefields. Proteins Struct. Funct. Bioinform. 2009, 75, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Trabuco, L.G.; Villa, E.; Mitra, K.; Frank, J.; Schulten, K. Flexible Fitting of Atomic Structures into Electron Microscopy Maps Using Molecular Dynamics. Structure 2008, 16, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Schröder, G.F.; Brunger, A.T.; Levitt, M. Combining Efficient Conformational Sampling with a Deformable Elastic Network Model Facilitates Structure Refinement at Low Resolution. Structure 2007, 15, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, F.; Song, Y.; Li, X.; Brunner, M.J.; Xu, C.; Conticello, V.; Egelman, E.; Marlovits, T.C.; Cheng, Y.; Baker, D. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 2015, 12, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Topf, M.; Lasker, K.; Webb, B.; Wolfson, H.; Chiu, W.; Sali, A. Protein Structure Fitting and Refinement Guided by Cryo-EM Density. Structure 2008, 16, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Hammel, M.; Tainer, J.A.; Sali, A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Hub, J.S. Interpreting solution X-ray scattering data using molecular simulations. Curr. Opin. Struct. Biol. 2018, 49, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Gorba, C.; Miyashita, O.; Tama, F. Normal-Mode Flexible Fitting of High-Resolution Structure of Biological Molecules toward One-Dimensional Low-Resolution Data. Biophys. J. 2008, 94, 1589–1599. [Google Scholar] [CrossRef] [PubMed]
- Tria, G.; Mertens, H.D.T.; Kachala, M.; Svergun, D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2015, 2, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Russel, D.; Lasker, K.; Webb, B.; Velázquez-Muriel, J.; Tjioe, E.; Schneidman-Duhovny, D.; Peterson, B.; Sali, A. Putting the Pieces Together: Integrative Modeling Platform Software for Structure Determination of Macromolecular Assemblies. PLoS Biol. 2012, 10, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, A.; Herzog, F.; Leitner, A.; Rosenberger, G.; Aebersold, R. Cross-Link Guided Molecular Modeling with ROSETTA. PLoS ONE 2013, 8, e73411. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; van Dijk, M.; Bonvin, A.M.J.J. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.M.A.; Sen, N.; Thalassinos, K.; Topf, M. Modeling Protein Complexes Using Restraints from Crosslinking Mass Spectrometry. Structure 2018, 26, 1015–1024.e2. [Google Scholar] [CrossRef] [PubMed]
- Fritz, B.G.; Roberts, S.A.; Ahmed, A.; Breci, L.; Li, W.; Weichsel, A.; Brailey, J.L.; Wysocki, V.H.; Tama, F.; Montfort, W.R. Molecular model of a soluble guanylyl cyclase fragment determined by small-angle X-ray scattering and chemical cross-linking. Biochemistry 2013, 52, 1568–1582. [Google Scholar] [CrossRef] [PubMed]
- Tokuhisa, A.; Jonic, S.; Tama, F.; Miyashita, O. Hybrid approach for structural modeling of biological systems from X-ray free electron laser diffraction patterns. J. Struct. Biol. 2016, 194, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Quesne, M.G.; Borowski, T.; De Visser, S.P. Quantum Mechanics/Molecular Mechanics Modeling of Enzymatic Processes: Caveats and Breakthroughs. Chem. A Eur. J. 2016, 22, 2562–2581. [Google Scholar] [CrossRef] [PubMed]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Alder, B.J.; Wainwright, T.E. Phase Transition for a Hard Sphere System. J. Chem. Phys. 1957, 27, 1208. [Google Scholar] [CrossRef]
- Kamerlin, S.C.L.; Vicatos, S.; Dryga, A.; Warshel, A. Coarse-Grained (Multiscale) Simulations in Studies of Biophysical and Chemical Systems. Annu. Rev. Phys. Chem. 2011, 62, 41–64. [Google Scholar] [CrossRef] [PubMed]
- Go, N. Theoretical studies of protein folding. Ann. Rev. Biophys. Bioeng. 1983, 12, 183–210. [Google Scholar] [CrossRef] [PubMed]
- Tirion, M.M. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys. Rev. Lett. 1996, 77, 1905–1908. [Google Scholar] [CrossRef] [PubMed]
- Fuglebakk, E.; Tiwari, S.P.; Reuter, N. Comparing the intrinsic dynamics of multiple protein structures using elastic network models. BBA Gen. Subj. 2015, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Yang, L.-W.; Eyal, E. Global Dynamics of Proteins: Bridging Between Structure and Function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Sinha, S. Uncoupling of an ammonia channel as a mechanism of allosteric inhibition in anthranilate synthase of Serratia marcescens: Dynamic and graph theoretical analysis. Mol. BioSyst. 2017, 13, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Sanejouand, Y.-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bagler, G.; Sinha, S. Assortative mixing in Protein Contact Networks and protein folding kinetics. Bioinformatics 2007, 23, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Vishveshwara, S. A Study of Communication Pathways in Methionyl- tRNA Synthetase by Molecular Dynamics Simulations and Structure Network Analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 15711–15716. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, L.; De Ruvo, M.; Paci, P.; Santoni, D.; Giuliani, A. Protein Contact Networks: An emerging paradigm in chemistry. Chem. Rev. 2013, 113, 1598–1613. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.C.; Melo, M.C.R.R.; Schulten, K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Mitsutake, A.; Sugita, Y.; Okamoto, Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers 2001, 60, 96–123. [Google Scholar] [CrossRef]
- Valsson, O.; Tiwary, P.; Parrinello, M. Enhancing Important Fluctuations: Rare Events and Metadynamics from a Conceptual Viewpoint. Annu. Rev. Phys. Chem. 2016, 67, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Sugita, Y.; Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Berg, B.; Neuhaus, T. Multicanonical ensemble: A new approach to simulate first-order phase transitions. Phys. Rev. Lett. 1992, 68, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Hansmann, U.H.E.; Okamoto, Y.; Eisenmenger, F. Molecular dynamics, Langevin and hydrid Monte Carlo simulations in a multicanonical ensemble. Chem. Phys. Lett. 1996, 259, 321–330. [Google Scholar] [CrossRef]
- McGee, T.D.; Edwards, J.; Roitberg, A.E. pH-REMD Simulations Indicate That the Catalytic Aspartates of HIV-1 Protease Exist Primarily in a Monoprotonated State. J. Phys. Chem. B 2014, 118, 12577–12585. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.J.; Tirado-Rives, J.; Jorgensen, W.L. Enhanced Monte Carlo Sampling through Replica Exchange with Solute Tempering. J. Chem. Theory Comput. 2014, 10, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Palmer, J.C.; Debenedetti, P.G. Computational investigation of cold denaturation in the Trp-cage miniprotein. Proc. Natl. Acad. Sci. USA 2016, 113, 8991–8996. [Google Scholar] [CrossRef] [PubMed]
- Stelter, D.; Keyes, T. Enhanced Sampling of Phase Transitions in Coarse-Grained Lipid Bilayers. J. Phys. Chem. B 2017, 121, 5770–5780. [Google Scholar] [CrossRef] [PubMed]
- Stelzl, L.S.; Hummer, G. Kinetics from Replica Exchange Molecular Dynamics Simulations. J. Chem. Theory Comput. 2017, 13, 3927–3935. [Google Scholar] [CrossRef] [PubMed]
- Isralewitz, B.; Gao, M.; Schulten, K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001, 11, 224–230. [Google Scholar] [CrossRef]
- Wang, F.; Landau, D.P. Determining the density of states for classical statistical models: A random walk algorithm to produce a flat histogram. Phys. Rev. E 2001, 64, 056101. [Google Scholar] [CrossRef] [PubMed]
- Laio, A.; Parrinello, M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA 2002, 99, 12562–12566. [Google Scholar] [CrossRef] [PubMed]
- Kitson, D.H.; Hagler, A.T. Theoretical Studies of the Structure and Molecular Dynamics of a Peptide Crystal. Biochemistry 1988, 27, 5246–5257. [Google Scholar] [CrossRef] [PubMed]
- Avbelj, F.; Moult, J.; Kitson, D.H.; James, M.N.G.; Hagler, A.T. Molecular dynamics study of the structure and dynamics of a protein molecule in a crystalline ionic environment, Streptomyces griseus protease A. Biochemistry 1990, 29, 8658–8676. [Google Scholar] [CrossRef] [PubMed]
- Janowski, P.A.; Liu, C.; Deckman, J.; Case, D.A. Molecular dynamics simulation of triclinic lysozyme in a crystal lattice. Protein Sci. 2016, 25, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Janowski, P.A.; Cerutti, D.S.; Holton, J.; Case, D.A. Peptide crystal simulations reveal hidden dynamics. J. Am. Chem. Soc. 2013, 135, 7938–7948. [Google Scholar] [CrossRef] [PubMed]
- Ahlstrom, L.S.; Vorontsov, I.I.; Shi, J.; Miyashita, O. Effect of the Crystal Environment on Side-Chain Conformational Dynamics in Cyanovirin-N Investigated through Crystal and Solution Molecular Dynamics Simulations. PLoS ONE 2017, 12, e0170337. [Google Scholar] [CrossRef] [PubMed]
- Vorontsov, I.I.; Miyashita, O. Solution and Crystal Molecular Dynamics Simulation Study of m4-Cyanovirin-N Mutants Complexed with Di-Mannose. Biophys. J. 2009, 97, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Bond, P.J.; Faraldo-Gomez, J.D.; Deol, S.S.; Sansom, M.S.P. Membrane protein dynamics and detergent interactions within a crystal: A simulation study of OmpA. Proc. Natl. Acad. Sci. USA 2006, 103, 9518–9523. [Google Scholar] [CrossRef] [PubMed]
- Malek, K.; Coppens, M.-O. Molecular Simulations of Solute Transport in Xylose Isomerase Crystals. J. Phys. Chem. B 2008, 112, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Jiang, J. Molecular Dynamics Simulations for Water and Ions in Protein Crystals. Langmuir 2008, 24, 4215–4223. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Steitz, T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008, 9, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Whitford, P.C.; Ahmed, A.; Yu, Y.; Hennelly, S.P.; Tama, F.; Spahn, C.M.T.; Onuchic, J.N.; Sanbonmatsu, K.Y. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc. Natl. Acad. Sci. USA 2011, 108, 18943–18948. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.V.; Kolář, M.H.; Grubmüller, H. Molecular simulations of the ribosome and associated translation factors. Curr. Opin. Struct. Biol. 2018, 49, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Palermo, G.; Spinello, A.; Rothlisberger, U.; Magistrato, A. All-atom simulations disentangle the functional dynamics underlying gene maturation in the intron lariat spliceosome. Proc. Natl. Acad. Sci. USA 2018, 115, 6584–6589. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.; Mori, T.; Ando, T.; Harada, R.; Jung, J.; Sugita, Y.; Feig, M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. Elife 2016, 5, e19274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, F.; Liu, J.; Gao, B.; Liu, Y.; Zhai, Y.; Ma, J.; Zhang, K.; Baker, T.S.; Schulten, K.; et al. Atomic Model of Rabbit Hemorrhagic Disease Virus by Cryo-Electron Microscopy and Crystallography. PLoS Pathog. 2013, 9, e1003132. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Tsai, C.-J.; Xin, F.; Radivojac, P. Allosteric post-translational modification codes. Trends Biochem. Sci. 2012, 37, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Craveur, P.; Rebehmed, J.; De Brevern, A.G. PTM-SD: A database of structurally resolved and annotated posttranslational modifications in proteins. Database 2014, 2014, 41. [Google Scholar] [CrossRef] [PubMed]
- Polyansky, A.A.; Zagrovic, B.; Shemyakin, M.M.; Ovchinnikov, Y.A. Protein Electrostatic Properties Predefining the Level of Surface Hydrophobicity Change upon Phosphorylation. J. Phys. Chem. Lett. 2012, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Moffett, A.S.; Bender, K.W.; Huber, S.C.; Shukla, D. Allosteric Control of a Plant Receptor Kinase through S-Glutathionylation. Biophys. J. 2017, 113, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L. The Many Roles of Computation in Drug Discovery. Science 2004, 303, 1813–1818. [Google Scholar] [CrossRef] [PubMed]
- Feixas, F.; Lindert, S.; Sinko, W.; Mccammon, J.A. Exploring the role of receptor flexibility in structure-based drug discovery. Biophys. Chem. 2014, 186, 31–45. [Google Scholar] [CrossRef] [PubMed]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef] [PubMed]
- Antolin, A.A.; Carotti, A.; Nuti, R.; Hakkaya, A.; Camaioni, E.; Mestres, J.; Pellicciari, R.; Macchiarulo, A. Exploring the effect of PARP-1 flexibility in docking studies. J. Mol. Graph. Model. 2013, 45, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Seidel, T.; khan Ibis, G.; Bendix, F.; Wolber, G. Strategies for 3D pharmacophore-based virtual screening. Drug Discov. Today Technol. 2010, 7, e221–e228. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, C.; Priyakumar, U.D.; Sastry, G.N. Dynamics Based Pharmacophore Models for Screening Potential Inhibitors of Mycobacterial Cyclopropane Synthase. J. Chem. Inf. Model. 2015, 55, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Wieder, M.; Garon, A.; Perricone, U.; Boresch, S.; Seidel, T.; Almerico, A.M.; Langer, T. Common Hits Approach: Combining Pharmacophore Modeling and Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Sinko, W.; de Oliveira, C.; Williams, S.; Van Wynsberghe, A.; Durrant, J.D.; Cao, R.; Oldfield, E.; Mccammon, J.A. Applying molecular dynamics simulations to identify rarely sampled ligand-bound conformational states of undecaprenyl pyrophosphate synthase, an antibacterial target. Chem. Biol. Drug Des. 2011, 77, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Perricone, U.; Wieder, M.; Seidel, T.; Langer, T.; Padova, A.; Almerico, A.M.; Tutone, M. A Molecular Dynamics–Shared Pharmacophore Approach to Boost Early-Enrichment Virtual Screening: A Case Study on Peroxisome Proliferator-Activated Receptor α. ChemMedChem 2017, 12, 1399–1407. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bradley, P. Probing the role of interfacial waters in protein-DNA recognition using a hybrid implicit/explicit solvation model. Proteins Struct. Funct. Bioinform. 2013, 81, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Breiten, B.; Lockett, M.R.; Sherman, W.; Fujita, S.; Al-Sayah, M.; Lange, H.; Bowers, C.M.; Heroux, A.; Krilov, G.; Whitesides, G.M. Water Networks Contribute to Enthalpy/Entropy Compensation in Protein−Ligand Binding. J. Am. Chem. Soc. 2013, 135, 15579–15584. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Bondar, A.-N. Mechanism by which water and protein electrostatic interactions control proton transfer at the active site of channelrhodopsin. PLoS ONE 2018, 13, e0201298. [Google Scholar] [CrossRef] [PubMed]
- Conti Nibali, V.; Havenith, M.; Nibali, V.C.; Havenith, M. New insights into the role of water in biological function: Studying solvated biomolecules using terahertz absorption spectroscopy in conjunction with molecular dynamics simulations. J. Am. Chem. Soc. 2014, 136, 12800–12807. [Google Scholar] [CrossRef] [PubMed]
- Schirò, G.; Fichou, Y.; Gallat, F.-X.; Wood, K.; Gabel, F.; Moulin, M.; Härtlein, M.; Heyden, M.; Colletier, J.-P.; Orecchini, A.; et al. Translational diffusion of hydration water correlates with functional motions in folded and intrinsically disordered proteins. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Mackerell, A.D. Empirical Force Fields for Biological Macromolecules: Overview and Issues. J. Comput. Chem. 2004, 25, 1584–1604. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Maragakis, P.; Piana, S.; Eastwood, M.P.; Dror, R.O. Systematic Validation of Protein Force Fields against Experimental Data. PLoS ONE 2012, 7, e32131. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zheng, W.; Mittal, J. Balanced Protein−Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. J. Chem. Theory Comput 2014, 10, 5113–5124. [Google Scholar] [CrossRef] [PubMed]
- Henriques, J.; Cragnell, C.; Skepö, M. Molecular Dynamics Simulations of Intrinsically Disordered Proteins: Force Field Evaluation and Comparison with Experiment. J. Chem. Theory Comput. 2015, 11, 3420–3431. [Google Scholar] [CrossRef] [PubMed]
- Lemkul, J.A.; Huang, J.; Roux, B.; Mackerell, A.D. An Empirical Polarizable Force Field Based on the Classical Drude Oscillator Model: Development History and Recent Applications. Chem. Rev. 2016, 116, 4983–5013. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.E.; Grossman, J.P.; Bank, J.A.; Batson, B.; Butts, J.A.; Chao, J.C.; Deneroff, M.M.; Dror, R.O.; Even, A.; Fenton, C.H.; et al. Anton 2: Raising the Bar for Performance and Programmability in a Special-Purpose Molecular Dynamics Supercomputer. In Proceedings of the SC14: IEEE International Conference for High Performance Computing, Networking, Storage and Analysis, New Orleans, LA, USA, 16–21 November 2014; pp. 41–53. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páall, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Bowman, G.R.; Pande, V.S.; Noé, F. (Eds.) An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2014; Volume 797, ISBN 978-94-007-7605-0. [Google Scholar]
- Sali, A.; Berman, H.M.; Schwede, T.; Trewhella, J.; Kleywegt, G.; Burley, S.K.; Markley, J.; Nakamura, H.; Adams, P.; Bonvin, A.M.J.J.; et al. Meeting Review Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Struct. Des. 2015, 23, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Kurisu, G.; Markley, J.L.; Nakamura, H.; Velankar, S.; Berman, H.M.; Sali, A.; Schwede, T.; Trewhella, J. PDB-Dev: A Prototype System for Depositing Integrative/Hybrid Structural Models. Structure 2017, 25, 1317–1318. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, A.; Nagai, T.; Srivastava, A.; Miyashita, O.; Tama, F. Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics. Int. J. Mol. Sci. 2018, 19, 3401. https://doi.org/10.3390/ijms19113401
Srivastava A, Nagai T, Srivastava A, Miyashita O, Tama F. Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics. International Journal of Molecular Sciences. 2018; 19(11):3401. https://doi.org/10.3390/ijms19113401
Chicago/Turabian StyleSrivastava, Ashutosh, Tetsuro Nagai, Arpita Srivastava, Osamu Miyashita, and Florence Tama. 2018. "Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics" International Journal of Molecular Sciences 19, no. 11: 3401. https://doi.org/10.3390/ijms19113401
APA StyleSrivastava, A., Nagai, T., Srivastava, A., Miyashita, O., & Tama, F. (2018). Role of Computational Methods in Going beyond X-ray Crystallography to Explore Protein Structure and Dynamics. International Journal of Molecular Sciences, 19(11), 3401. https://doi.org/10.3390/ijms19113401




