Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review
Abstract
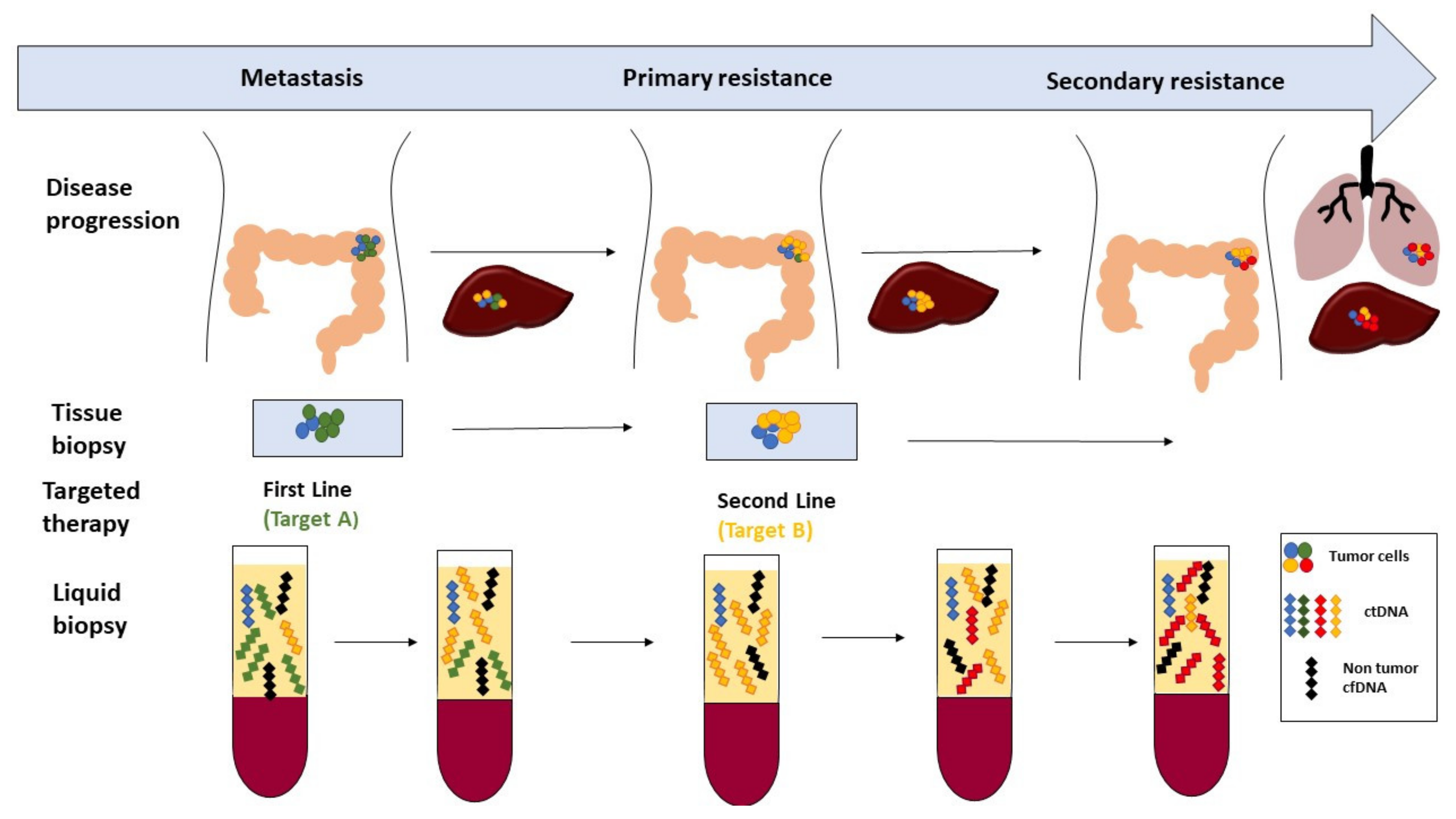
:1. Introduction
1.1. Liquid Biopsy
1.2. Cell-Free DNA
1.3. Preanalytical Considerations
- -
- Plasma is a better source of cfDNA than serum since it avoids blood cell genomic DNA contamination
- -
- EDTA or cell-free DNATM collection tubes prevent blood cell lysis by keeping tubes at 4 °C
- -
- Blood must be processed within a maximum of 4 h following blood drawing to preserve cfDNA concentration and fragmentation
- -
- To ensure any absence of cells in plasma, first, centrifugation is recommended at 1200–1600 g for 10 min and, second, microcentrifugation at 16,000 g for 10 min, (the second step can be indifferently realized before or even after storage of plasma samples)
- -
- Plasma samples must be stored at −80 °C for up to nine months (samples are sensitive to freeze-thaw cycles)
- -
- cfDNA extracts may tolerate a maximum of three freeze-thaw cycles and storage at −20 °C for up to three months
2. Results
2.1. Cell-Free DNA as a Physiological Mobile Genetic Element
2.2. Cell-Free DNA and Colorectal Cancer
2.2.1. Cell-Free DNA as a Diagnostic Biomarker in Colorectal Cancer
2.2.2. Cell-Free DNA as a Predictive Biomarker in Colorectal Cancer
2.2.3. Cell-Free DNA as a Prognostic Biomarker in Colorectal Cancer
2.3. Cell-Free DNA in Other Body Fluids
3. Conclusions
4. Methods
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ARMs | amplification refractory mutations system |
| BEAMing | bead emulsion amplification and magnetics |
| CAPP-Seq | cancer personalized profiling by deep sequencing |
| cSMART | circulating single molecule amplification and re-sequencing technology |
| cfDNA | circulating cell-free DNA |
| COLD-PCR | Co-amplification at lower denaturation temperature |
| CNV | copy number variations |
| CRC | colorectal cancer |
| CRT | chemoradiotherapy |
| CTC | circulating tumor cells |
| ctDNA | circulating tumor DNA |
| ddPCR | droplet digital PCR |
| DDR | DNA-damage response |
| dPCR | digital PCR |
| FISH | fluorescent in situ hybridization |
| MSI | microsatellite instability |
| MS-PCR | methylation specific PCR |
| NGS | next-generation sequencing |
| PNAs-LNA | peptide nucleic acid-locked nucleic acid |
| qPCR | quantitative PCR |
| RC | rectal cancer |
| RT-PCR | real-time PCR |
| Safe-Seq | safe sequencing system |
| SERS | surface-enhanced Raman spectroscopy |
| SNV | single nucleotide variation |
| SOLID | Sequencing by Oligonucleotide Ligation and Detection |
| SSCP | single strand conformation PCR |
| TAM-Seq | tagged-amplicon deep sequencing |
| UltraSEEK | high-throughput multiplex ultrasensitive mutation detection |
| WGS | whole genome sequencing |
References
- Salvi, S.; Gurioli, G.; De Giorgi, U.; Conteduca, V.; Tedaldi, G.; Calistri, D.; Casadio, V. Cell-free DNA as a diagnostic marker for cancer: Current insights. Oncotargets Ther. 2016, 9, 6549–6559. [Google Scholar] [CrossRef] [PubMed]
- Colussi, D.; Brandi, G.; Bazzoli, F.; Ricciardiello, L. Molecular pathways involved in colorectal cancer: Implications for disease behavior and prevention. Int. J. Mol. Sci. 2013, 14, 16365–16385. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T. Colorectal carcinogenesis: Review of human and experimental animal studies. J. Carcinog. 2009, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Frattini, M.; Balestra, D.; Suardi, S.; Oggionni, M.; Alberici, P.; Radice, P.; Costa, A.; Daidone, M.G.; Leo, E.; Pilotti, S.; et al. Different genetic features associated with colon and rectal carcinogenesis. Clin. Cancer Res. 2004, 10, 4015–4021. [Google Scholar] [CrossRef] [PubMed]
- Pramateftakis, M.G.; Kanellos, D.; Tekkis, P.P.; Touroutoglou, N.; Kanellos, I. Rectal cancer: Multimodal treatment approach. Int. J. Surg. Oncol. 2012, 2012, 279341. [Google Scholar] [CrossRef] [PubMed]
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Brandi, G.; De Lorenzo, S.; Nannini, M.; Curti, S.; Ottone, M.; Dall’Olio, F.G.; Barbera, M.A.; Pantaleo, M.A.; Biasco, G. Adjuvant chemotherapy for resected colorectal cancer metastases: Literature review and meta-analysis. World J. Gastroenterol. 2016, 22, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Naccarati, A.; Rosa, F.; Vymetalkova, V.; Barone, E.; Jiraskova, K.; Di Gaetano, C.; Novotny, J.; Levy, M.; Vodickova, L.; Gemignani, F.; et al. Double-strand break repair and colorectal cancer: Gene variants within 3′ utrs and micrornas binding as modulators of cancer risk and clinical outcome. Oncotarget 2016, 7, 23156–23169. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; DeSantis, C.; Virgo, K.; Stein, K.; Mariotto, A.; Smith, T.; Cooper, D.; Gansler, T.; Lerro, C.; Fedewa, S.; et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 2012, 62, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Mroziewicz, M.; Tyndale, R.F. Pharmacogenetics: A tool for identifying genetic factors in drug dependence and response to treatment. Addict. Sci. Clin. Pract. 2010, 5, 17–29. [Google Scholar] [PubMed]
- Balmativola, D.; Marchio, C.; Maule, M.; Chiusa, L.; Annaratone, L.; Maletta, F.; Montemurro, F.; Kulka, J.; Figueiredo, P.; Varga, Z.; et al. Pathological non-response to chemotherapy in a neoadjuvant setting of breast cancer: An inter-institutional study. Breast Cancer Res. Treat. 2014, 148, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Vanderlaan, P.A.; Yamaguchi, N.; Folch, E.; Boucher, D.H.; Kent, M.S.; Gangadharan, S.P.; Majid, A.; Goldstein, M.A.; Huberman, M.S.; Kocher, O.N.; et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014, 84, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybinski, B.; Yun, K. Addressing intra-tumoral heterogeneity and therapy resistance. Oncotarget 2016, 7, 72322–72342. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.H.; Chow, C.; To, K.F. Latest development of liquid biopsy. J. Thorac. Dis. 2018, 10, S1645–S1651. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.B.; Chen, J.S.; Fan, C.W.; Li, Y.S.; Chan, E.C. Comparison of kras mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clin. Chim. Acta 2014, 433, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Burch, J.A.; Soares-Weiser, K.; St John, D.J.; Duffy, S.; Smith, S.; Kleijnen, J.; Westwood, M. Diagnostic accuracy of faecal occult blood tests used in screening for colorectal cancer: A systematic review. J. Med. Screen 2007, 14, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.; Llagostera, M.; Esteva, M.; Cabeza, E.; Cantero, X.; Segarra, M.; Martin-Rabadan, M.; Artigues, G.; Torrent, M.; Taltavull, J.M.; et al. Knowledge and attitudes of primary healthcare patients regarding population-based screening for colorectal cancer. BMC Cancer 2011, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E.; Ahmed, N.C.; Vos, P.W.; Bonnerup, C.; Atkins, J.N.; Casey, M.; Nuovo, G.J.; Naziri, W.; Wiley, J.E.; Mota, H.; et al. Diagnostic microrna markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genom. Proteom. 2013, 10, 93–113. [Google Scholar]
- Ulz, P.; Heitzer, E.; Geigl, J.B.; Speicher, M.R. Patient monitoring through liquid biopsies using circulating tumor DNA. Int. J. Cancer 2017, 141, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Auer, M.; Hoffmann, E.M.; Pichler, M.; Gasch, C.; Ulz, P.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Mohan, S.; et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int. J. Cancer 2013, 133, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of kras and braf mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Les acides nucleiques du plasma sanguin chez 1 homme. C R Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Hsieh, J.S.; Chang, M.Y.; Huang, T.J.; Chen, F.M.; Cheng, T.L.; Alexandersen, K.; Huang, Y.S.; Tzou, W.S.; Lin, S.R. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J. Surg. 2004, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.A.; Smith, B.M.; Walsh, T.; Johnson, S.; Primrose, L.; Slade, M.J.; Walker, R.A.; Coombes, R.C. Microsatellite alterations plasma DNA of primary breast cancer patients. Clin. Cancer Res. 2000, 6, 1119–1124. [Google Scholar] [PubMed]
- Fujiwara, K.; Fujimoto, N.; Tabata, M.; Nishii, K.; Matsuo, K.; Hotta, K.; Kozuki, T.; Aoe, M.; Kiura, K.; Ueoka, H.; et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin. Cancer Res. 2005, 11, 1219–1225. [Google Scholar] [PubMed]
- Brychta, N.; Krahn, T.; von Ahsen, O. Detection of KRAS mutations in circulating tumor DNA by digital PCR in early stages of pancreatic cancer. Clin. Chem. 2016, 62, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, H.; Zang, W.; Li, B.; Rao, G.; Li, L.; Yu, Y.; Li, Z.; Dong, B.; Lu, Z.; et al. Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci. 2017, 108, 1881–1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.C. Scanning for cancer genomic changes in plasma: Toward an era of personalized blood-based tumor markers. Clin. Chem. 2013, 59, 1553–1555. [Google Scholar] [CrossRef] [PubMed]
- Atamaniuk, J.; Kopecky, C.; Skoupy, S.; Saemann, M.D.; Weichhart, T. Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol. Dial. Transpl. 2012, 27, 902–905. [Google Scholar] [CrossRef] [PubMed]
- Tug, S.; Helmig, S.; Menke, J.; Zahn, D.; Kubiak, T.; Schwarting, A.; Simon, P. Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell. Immunol. 2014, 292, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Li, L.H.; Hua, D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett. 2006, 243, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Shin, D.G.; Park, M.K.; Baik, S.H.; Kim, T.H.; Kim, S.; Lee, S. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: Diagnostic validity and significant reduction of cfDNA after surgical resection. Ann. Surg. Treat. Res. 2014, 86, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Caldas, C. Nucleosome mapping in plasma DNA predicts cancer gene expression. Nat. Genet. 2016, 48, 1105–1106. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Chakrabarty, M.; Cohn, E.M.; Leon, S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983, 51, 2116–2120. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.H.; Wang, M.; Brenner, D.E.; Norton, P.A.; Block, T.M. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann. N. Y. Acad. Sci. 2008, 1137, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 2010, 56, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Chan, K.C.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Mouliere, F.; Gongora, C.; Ollier, J.; Robert, B.; Ychou, M.; Del Rio, M.; Molina, F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010, 38, 6159–6175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouliere, F.; Rosenfeld, N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc. Natl. Acad. Sci. USA 2015, 112, 3178–3179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umetani, N.; Kim, J.; Hiramatsu, S.; Reber, H.A.; Hines, O.J.; Bilchik, A.J.; Hoon, D.S. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: Direct quantitative PCR for ALU repeats. Clin. Chem. 2006, 52, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Hwu, H.R.; Roberts, J.W.; Davidson, E.H.; Britten, R.J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc. Natl. Acad. Sci. USA 1986, 83, 3875–3879. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Schiller, J.; Schmitz, A.; Schuetzmann, D.; Schatz, C.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Cancer therapy monitoring in xenografts by quantitative analysis of circulating tumor DNA. Biomarkers 2012, 17, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; El Messaoudi, S.; Gongora, C.; Guedj, A.S.; Robert, B.; Del Rio, M.; Molina, F.; Lamy, P.J.; Lopez-Crapez, E.; Mathonnet, M.; et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl. Oncol. 2013, 6, 319–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, K.; Jiang, P.; Chan, K.C.; Wong, J.; Cheng, Y.K.; Liang, R.H.; Chan, W.K.; Ma, E.S.; Chan, S.L.; Cheng, S.H.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef] [PubMed]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment length of circulating tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.H.; Jiang, P.; Sun, K.; Cheng, Y.K.; Chan, K.C.; Leung, T.Y.; Chiu, R.W.; Lo, Y.M. Noninvasive prenatal testing by nanopore sequencing of maternal plasma DNA: Feasibility assessment. Clin. Chem. 2015, 61, 1305–1306. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; El Messaoudi, S.; Pang, D.; Dritschilo, A.; Thierry, A.R. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol. Oncol. 2014, 8, 927–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef] [PubMed]
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards standardisation of cell-free DNA measurement in plasma: Controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Montalvo, L.; Chrebtow, V.; Busch, M.P. Quantitation of genomic DNA in plasma and serum samples: Higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001, 41, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.; Tein, M.S.; Lau, T.K.; Haines, C.J.; Leung, T.N.; Poon, P.M.; Wainscoat, J.S.; Johnson, P.J.; Chang, A.M.; Hjelm, N.M. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998, 62, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Klotzek, S.; Lewandowski, M.; Fleischhacker, M.; Jung, K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin. Chem. 2003, 49, 1028–1029. [Google Scholar] [CrossRef] [PubMed]
- Board, R.E.; Williams, V.S.; Knight, L.; Shaw, J.; Greystoke, A.; Ranson, M.; Dive, C.; Blackhall, F.H.; Hughes, A. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann. N. Y. Acad. Sci. 2008, 1137, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Nikolaev, S.; Lemmens, L.; Koessler, T.; Blouin, J.L.; Nouspikel, T. Circulating tumoral DNA: Preanalytical validation and quality control in a diagnostic laboratory. Anal. Biochem. 2018, 542, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, N.; Sakaida, I.; Moribe, T.; Fujita, N.; Miura, T.; Stark, M.; Tamatsukuri, S.; Ishitsuka, H.; Uchida, K.; Terai, S.; et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis c virus-associated hepatocellular carcinoma. Anticancer Res. 2006, 26, 4713–4719. [Google Scholar] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra168. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Jiang, P.; Chan, C.W.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.; Ng, S.S.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012, 4, 162ra154. [Google Scholar] [CrossRef] [PubMed]
- Khakoo, S.; Georgiou, A.; Gerlinger, M.; Cunningham, D.; Starling, N. Circulating tumour DNA, a promising biomarker for the management of colorectal cancer. Crit. Rev. Oncol. Hematol. 2018, 122, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Han, W.Y.; Feng, X.; She, Q.X. Reverse gyrase functions in genome integrity maintenance by protecting DNA breaks in vivo. Int. J. Mol. Sci. 2017, 18, 1340. [Google Scholar] [CrossRef] [PubMed]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Mittra, I. Circulating nucleic acids: A new class of physiological mobile genetic elements. F1000Research 2015, 4, 924. [Google Scholar] [CrossRef] [PubMed]
- Mittra, I.; Khare, N.K.; Raghuram, G.V.; Chaubal, R.; Khambatti, F.; Gupta, D.; Gaikwad, A.; Prasannan, P.; Singh, A.; Iyer, A.; et al. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J. Biosci. 2015, 40, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Nevers, P.; Saedler, H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nature 1977, 268, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Bahar, R.; Hartmann, C.H.; Rodriguez, K.A.; Denny, A.D.; Busuttil, R.A.; Dolle, M.E.; Calder, R.B.; Chisholm, G.B.; Pollock, B.H.; Klein, C.A.; et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 2006, 441, 1011–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Olmo, D.C.; Dominguez, C.; Garcia-Arranz, M.; Anker, P.; Stroun, M.; Garcia-Verdugo, J.M.; Garcia-Olmo, D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Dvorakova, M.; Karafiat, V.; Pajer, P.; Kluzakova, E.; Jarkovska, K.; Pekova, S.; Krutilkova, L.; Dvorak, M. DNA released by leukemic cells contributes to the disruption of the bone marrow microenvironment. Oncogene 2013, 32, 5201–5209. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Lefort, F.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Chen, X.Q.; Stroun, M.; Mulcahy, H.E.; Farthing, M.J. K-ras mutations are found in DNA extracted from the plasma of patients with colorectal cancer. Gastroenterology 1997, 112, 1114–1120. [Google Scholar] [CrossRef]
- De Kok, J.B.; van Solinge, W.W.; Ruers, T.J.; Roelofs, R.W.; van Muijen, G.N.; Willems, J.L.; Swinkels, D.W. Detection of tumour DNA in serum of colorectal cancer patients. Scand. J. Clin. Lab. Investig. 1997, 57, 601–604. [Google Scholar] [CrossRef]
- Kopreski, M.S.; Benko, F.A.; Kwee, C.; Leitzel, K.E.; Eskander, E.; Lipton, A.; Gocke, C.D. Detection of mutant K-ras DNA in plasma or serum of patients with colorectal cancer. Br. J. Cancer 1997, 76, 1293–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hibi, K.; Robinson, C.R.; Booker, S.; Wu, L.; Hamilton, S.R.; Sidransky, D.; Jen, J. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Res. 1998, 58, 1405–1407. [Google Scholar] [PubMed]
- Lecomte, T.; Berger, A.; Zinzindohoue, F.; Micard, S.; Landi, B.; Blons, H.; Beaune, P.; Cugnenc, P.H.; Laurent-Puig, P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int. J. Cancer 2002, 100, 542–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, B.M.; Lefort, F.; McManus, R.; Daly, J.; Keeling, P.W.; Weir, D.G.; Kelleher, D. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: Strong prognostic indicator in postoperative follow up. Gut 2003, 52, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.K.; To, K.F.; Man, E.P.; Chan, M.W.; Bai, A.H.; Hui, A.J.; Chan, F.K.; Sung, J.J. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am. J. Gastroenterol. 2005, 100, 2274–2279. [Google Scholar] [CrossRef] [PubMed]
- Lindforss, U.; Zetterquist, H.; Papadogiannakis, N.; Olivecrona, H. Persistence of K-ras mutations in plasma after colorectal tumor resection. Anticancer Res. 2005, 25, 657–661. [Google Scholar] [PubMed]
- Bazan, V.; Bruno, L.; Augello, C.; Agnese, V.; Calo, V.; Corsale, S.; Gargano, G.; Terrasi, M.; Schiro, V.; Di Fede, G.; et al. Molecular detection of TP53, Ki-Ras and p16INK4A promoter methylation in plasma of patients with colorectal cancer and its association with prognosis. Results of a 3-year goim (gruppo oncologico dell’italia meridionale) prospective study. Ann. Oncol. 2006, 17 (Suppl. 7), vii84–vii90. [Google Scholar] [CrossRef] [PubMed]
- Flamini, E.; Mercatali, L.; Nanni, O.; Calistri, D.; Nunziatini, R.; Zoli, W.; Rosetti, P.; Gardini, N.; Lattuneddu, A.; Verdecchia, G.M.; et al. Free DNA and carcinoembryonic antigen serum levels: An important combination for diagnosis of colorectal cancer. Clin. Cancer Res. 2006, 12, 6985–6988. [Google Scholar] [CrossRef] [PubMed]
- Frattini, M.; Gallino, G.; Signoroni, S.; Balestra, D.; Battaglia, L.; Sozzi, G.; Leo, E.; Pilotti, S.; Pierotti, M.A. Quantitative analysis of plasma DNA in colorectal cancer patients: A novel prognostic tool. Ann. N. Y. Acad. Sci. 2006, 1075, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Trevisiol, C.; Di Fabio, F.; Nascimbeni, R.; Peloso, L.; Salbe, C.; Ferruzzi, E.; Salerni, B.; Gion, M. Prognostic value of circulating KRAS2 gene mutations in colorectal cancer with distant metastases. Int. J. Biol. Markers 2006, 21, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Wallner, M.; Herbst, A.; Behrens, A.; Crispin, A.; Stieber, P.; Goke, B.; Lamerz, R.; Kolligs, F.T. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin. Cancer Res. 2006, 12, 7347–7352. [Google Scholar] [CrossRef] [PubMed]
- Boni, L.; Cassinotti, E.; Canziani, M.; Dionigi, G.; Rovera, F.; Dionigi, R. Free circulating DNA as possible tumour marker in colorectal cancer. Surg. Oncol. 2007, 16 (Suppl. 1), S29–S31. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, G.; Hibi, K.; Kodera, Y.; Koike, M.; Fujiwara, M.; Nakao, A. P16 methylation in serum as a potential marker for the malignancy of colorectal carcinoma. Anticancer Res. 2007, 27, 3367–3370. [Google Scholar] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frattini, M.; Gallino, G.; Signoroni, S.; Balestra, D.; Lusa, L.; Battaglia, L.; Sozzi, G.; Bertario, L.; Leo, E.; Pilotti, S.; et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008, 263, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Lofton-Day, C.; Model, F.; Devos, T.; Tetzner, R.; Distler, J.; Schuster, M.; Song, X.; Lesche, R.; Liebenberg, V.; Ebert, M.; et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 2008, 54, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Stoehlmacher, J.; Pantel, K.; Goekkurt, E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann. N. Y. Acad. Sci. 2008, 1137, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Zitt, M.; Muller, H.M.; Rochel, M.; Schwendinger, V.; Zitt, M.; Goebel, G.; Devries, A.; Margreiter, R.; Oberwalder, M.; Zeillinger, R.; et al. Circulating cell-free DNA in plasma of locally advanced rectal cancer patients undergoing preoperative chemoradiation: A potential diagnostic tool for therapy monitoring. Dis. Markers 2008, 25, 159–165. [Google Scholar] [CrossRef] [PubMed]
- De Vos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grutzmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Herbst, A.; Rahmig, K.; Stieber, P.; Philipp, A.; Jung, A.; Ofner, A.; Crispin, A.; Neumann, J.; Lamerz, R.; Kolligs, F.T. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am. J. Gastroenterol. 2011, 106, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating cell-free DNA: A promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted egfr blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.R.; Whiteley, J.; Donald, E.; Smith, J.; Eisenberg, M.T.; Kallam, E.; Kam-Morgan, L. Comparison of kras mutation assessment in tumor DNA and circulating free DNA in plasma and serum samples. Clin. Med. Insights Pathol. 2012, 5, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Philipp, A.B.; Stieber, P.; Nagel, D.; Neumann, J.; Spelsberg, F.; Jung, A.; Lamerz, R.; Herbst, A.; Kolligs, F.T. Prognostic role of methylated free circulating DNA in colorectal cancer. Int. J. Cancer 2012, 131, 2308–2319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spindler, K.L.; Pallisgaard, N.; Vogelius, I.; Jakobsen, A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin. Cancer Res. 2012, 18, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.Q.; Liu, X.J.; Wang, Y.; Ge, F.J.; Zhao, C.H.; Fu, Y.L.; Lin, L.; Xu, J.M. Correlation analysis between abundance of k-ras mutation in plasma free DNA and its correlation with clinical outcome and prognosis in patients with metastatic colorectal cancer. Zhonghua Zhong Liu Za Zhi 2013, 35, 666–671. [Google Scholar] [PubMed]
- Cassinotti, E.; Boni, L.; Segato, S.; Rausei, S.; Marzorati, A.; Rovera, F.; Dionigi, G.; David, G.; Mangano, A.; Sambucci, D.; et al. Free circulating DNA as a biomarker of colorectal cancer. Int. J. Surg. 2013, 11 (Suppl. 1), S54–S57. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Hwang, S.M.; Kim, T.S.; Kim, D.W.; Park, D.J.; Kang, S.B.; Kim, H.H.; Park, K.U. Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl. Oncol. 2013, 6, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.G.; Appelt, A.L.; Pallisgaard, N.; Andersen, R.F.; Jakobsen, A. Kras-mutated plasma DNA as predictor of outcome from irinotecan monotherapy in metastatic colorectal cancer. Br. J. Cancer 2013, 109, 3067–3072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Sun, Y.; Zhu, M.; Wang, Z.; Zhang, H.; Xin, Y.; Jiang, G.; Guo, X.; Zhang, Z.; Liu, Y. The role of plasma cell-free DNA detection in predicting preoperative chemoradiotherapy response in rectal cancer patients. Oncol. Rep. 2014, 31, 1466–1472. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.K.; Lin, P.C.; Lin, C.H.; Jiang, J.K.; Yang, S.H.; Liang, W.Y.; Chen, W.S.; Chang, S.C. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: Based on the mutation spectra detected in primary tumors. Ann. Surg. Oncol. 2014, 21 (Suppl. 4), S680–S686. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Heitzer, E.; Ulz, P.; Lafer, I.; Lax, S.; Auer, M.; Pichler, M.; Gerger, A.; Eisner, F.; Hoefler, G.; et al. Changes in colorectal carcinoma genomes under anti-egfr therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014, 10, e1004271. [Google Scholar] [CrossRef] [PubMed]
- Perrone, F.; Lampis, A.; Bertan, C.; Verderio, P.; Ciniselli, C.M.; Pizzamiglio, S.; Frattini, M.; Nucifora, M.; Molinari, F.; Gallino, G.; et al. Circulating free DNA in a screening program for early colorectal cancer detection. Tumori 2014, 100, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.L.; Pallisgaard, N.; Andersen, R.F.; Jakobsen, A. Changes in mutational status during third-line treatment for metastatic colorectal cancer—Results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int. J. Cancer 2014, 135, 2215–2222. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.L.; Appelt, A.L.; Pallisgaard, N.; Andersen, R.F.; Brandslund, I.; Jakobsen, A. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int. J. Cancer 2014, 135, 2984–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tham, C.; Chew, M.; Soong, R.; Lim, J.; Ang, M.; Tang, C.; Zhao, Y.; Ong, S.Y.; Liu, Y. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 2014, 120, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.M.; Liu, X.J.; Ge, F.J.; Lin, L.; Wang, Y.; Sharma, M.R.; Liu, Z.Y.; Tommasi, S.; Paradiso, A. KRAS mutations in tumor tissue and plasma by different assays predict survival of patients with metastatic colorectal cancer. J. Exp. Clin. Cancer Res. 2014, 33, 104. [Google Scholar] [CrossRef] [PubMed]
- Carpinetti, P.; Donnard, E.; Bettoni, F.; Asprino, P.; Koyama, F.; Rozanski, A.; Sabbaga, J.; Habr-Gama, A.; Parmigiani, R.B.; Galante, P.A.; et al. The use of personalized biomarkers and liquid biopsies to monitor treatment response and disease recurrence in locally advanced rectal cancer after neoadjuvant chemoradiation. Oncotarget 2015, 6, 38360–38371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.C.; Lin, J.K.; Lin, C.H.; Lin, H.H.; Yang, S.H.; Jiang, J.K.; Chen, W.S.; Chou, C.C.; Tsai, S.F.; Chang, S.C. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann. Surg. Oncol. 2015, 22 (Suppl. 3), S1419–S1427. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Scholer, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Sefrioui, D.; Sarafan-Vasseur, N.; Beaussire, L.; Baretti, M.; Gangloff, A.; Blanchard, F.; Clatot, F.; Sabourin, J.C.; Sesboue, R.; Frebourg, T.; et al. Clinical value of chip-based digital-PCR platform for the detection of circulating DNA in metastatic colorectal cancer. Dig. Liver Dis. 2015, 47, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to egfr blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.L.; Pallisgaard, N.; Andersen, R.F.; Brandslund, I.; Jakobsen, A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS ONE 2015, 10, e0108247. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chew, M.H.; Tham, C.K.; Tang, C.L.; Ong, S.Y.; Zhao, Y. Methylation of serum SST gene is an independent prognostic marker in colorectal cancer. Am. J. Cancer Res. 2016, 6, 2098–2108. [Google Scholar] [PubMed]
- Matthaios, D.; Balgkouranidou, I.; Karayiannakis, A.; Bolanaki, H.; Xenidis, N.; Amarantidis, K.; Chelis, L.; Romanidis, K.; Chatzaki, A.; Lianidou, E.; et al. Methylation status of the APC and RASSF1A promoter in cell-free circulating DNA and its prognostic role in patients with colorectal cancer. Oncol. Lett. 2016, 12, 748–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Messaoudi, S.; Mouliere, F.; Du Manoir, S.; Bascoul-Mollevi, C.; Gillet, B.; Nouaille, M.; Fiess, C.; Crapez, E.; Bibeau, F.; Theillet, C.; et al. Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin. Cancer Res. 2016, 22, 3067–3077. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef] [PubMed]
- Agah, S.; Akbari, A.; Talebi, A.; Masoudi, M.; Sarveazad, A.; Mirzaei, A.; Nazmi, F. Quantification of plasma cell-free circulating DNA at different stages of colorectal cancer. Cancer Investig. 2017, 35, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Bhangu, J.S.; Taghizadeh, H.; Braunschmid, T.; Bachleitner-Hofmann, T.; Mannhalter, C. Circulating cell-free DNA in plasma of colorectal cancer patients—A potential biomarker for tumor burden. Surg. Oncol. 2017, 26, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Herbst, A.; Vdovin, N.; Gacesa, S.; Ofner, A.; Philipp, A.; Nagel, D.; Holdt, L.M.; Op den Winkel, M.; Heinemann, V.; Stieber, P.; et al. Methylated free-circulating HPP1 DNA is an early response marker in patients with metastatic colorectal cancer. Int. J. Cancer 2017, 140, 2134–2144. [Google Scholar] [CrossRef] [PubMed]
- Kloten, V.; Ruchel, N.; Bruchle, N.O.; Gasthaus, J.; Freudenmacher, N.; Steib, F.; Mijnes, J.; Eschenbruch, J.; Binnebosel, M.; Knuchel, R.; et al. Liquid biopsy in colon cancer: Comparison of different circulating DNA extraction systems following absolute quantification of KRAS mutations using intplex allele-specific PCR. Oncotarget 2017, 8, 86253–86263. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dittmar, R.L.; Xia, S.; Zhang, H.; Du, M.; Huang, C.C.; Druliner, B.R.; Boardman, L.; Wang, L. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol. Oncol. 2017, 11, 1099–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, A.A.L.; Morelli, M.P.; Overman, M.; Kee, B.; Fogelman, D.; Vilar, E.; Shureiqi, I.; Raghav, K.; Eng, C.; Manuel, S.; et al. Clinical utility of circulating cell-free DNA in advanced colorectal cancer. PLoS ONE 2017, 12, e0183949. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.; Urabe, Y.; Ono, A.; Miki, D.; Ochi, H.; Chayama, K. Serial profiling of circulating tumor DNA for optimization of anti-VEGF chemotherapy in metastatic colorectal cancer patients. Int. J. Cancer 2018, 142, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Toung, J.M.; Jassowicz, A.F.; Vijayaraghavan, R.; Kang, H.; Zhang, R.; Kruglyak, K.M.; Huang, H.J.; Hinoue, T.; Shen, H.; et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018, 29, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Suzuki, K.; Muto, Y.; Ichida, K.; Fukui, T.; Kakizawa, N.; Ishikawa, H.; Watanabe, F.; Hasegawa, F.; Saito, M.; et al. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. Oncotarget 2018, 9, 24398–24413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledo, R.A.; Garralda, E.; Mitsi, M.; Pons, T.; Monsech, J.; Vega, E.; Otero, A.; Albarran, M.I.; Banos, N.; Duran, Y.; et al. Exome sequencing of plasma DNA portrays the mutation landscape of colorectal cancer and discovers mutated VEGFR2 receptors as modulators of antiangiogenic therapies. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Schou, J.V.; Larsen, F.O.; Sorensen, B.S.; Abrantes, R.; Boysen, A.K.; Johansen, J.S.; Jensen, B.V.; Nielsen, D.L.; Spindler, K.L. Circulating cell-free DNA as predictor of treatment failure after neoadjuvant chemo-radiotherapy before surgery in patients with locally advanced rectal cancer. Ann. Oncol. 2018, 29, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Sclafani, F.; Chau, I.; Cunningham, D.; Hahne, J.C.; Vlachogiannis, G.; Eltahir, Z.; Lampis, A.; Braconi, C.; Kalaitzaki, E.; De Castro, D.G.; et al. Kras and braf mutations in circulating tumour DNA from locally advanced rectal cancer. Sci. Rep. 2018, 8, 1445. [Google Scholar] [CrossRef] [PubMed]
- Boysen, A.K.; Sorensen, B.S.; Lefevre, A.C.; Abrantes, R.; Johansen, J.S.; Jensen, B.V.; Schou, J.V.; Larsen, F.O.; Nielsen, D.; Taflin, H.; et al. Methodological development and biological observations of cell free DNA with a simple direct fluorescent assay in colorectal cancer. Clin. Chim. Acta 2018, 487, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Myint, N.N.M.; Verma, A.M.; Fernandez-Garcia, D.; Sarmah, P.; Tarpey, P.S.; Al-Aqbi, S.S.; Cai, H.; Trigg, R.; West, K.; Howells, L.M.; et al. Circulating tumor DNA in patients with colorectal adenomas: Assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Demuth, C.; Spindler, K.G.; Johansen, J.S.; Pallisgaard, N.; Nielsen, D.; Hogdall, E.; Vittrup, B.; Sorensen, B.S. Measuring KRAS mutations in circulating tumor DNA by droplet digital PCR and next-generation sequencing. Transl. Oncol. 2018, 11, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Rokni, P.; Shariatpanahi, A.M.; Sakhinia, E.; Kerachian, M.A. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes Genom. 2018, 40, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Yan, P.; Zhang, S.; Lu, Y.; Pan, L.; Tang, W.; Chen, S.; Chen, S.; Zhang, A.; Liu, W. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis. Markers 2018, 2018, 6437104. [Google Scholar] [CrossRef] [PubMed]
- Molparia, B.; Oliveira, G.; Wagner, J.L.; Spencer, E.G.; Torkamani, A. A feasibility study of colorectal cancer diagnosis via circulating tumor DNA derived CNV detection. PLoS ONE 2018, 13, e0196826. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Gomez, M.; Moran, S.; de la Cadena, M.P.; Martinez-Zorzano, V.S.; Rodriguez-Berrocal, F.J.; Rodriguez-Girondo, M.; Esteller, M.; Cubiella, J.; Bujanda, L.; Castells, A.; et al. A new approach to epigenome-wide discovery of non-invasive methylation biomarkers for colorectal cancer screening in circulating cell-free DNA using pooled samples o. Clin. Epigenet. 2018, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.P.; Moreira-Barbosa, C.; Salta, S.; Palma de Sousa, S.; Pousa, I.; Oliveira, J.; Soares, M.; Rego, L.; Dias, T.; Rodrigues, J.; et al. Cell-free DNA methylation of selected genes allows for early detection of the major cancers in women. Cancers 2018, 10, 357. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Mao, F.; Shi, L.; Xu, X.; Wu, Z.; Zhou, J.; Xiao, M. Urinary measurement of circulating tumor DNA for treatment monitoring and prognosis of metastatic colorectal cancer patients. Clin. Chem. Lab. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Wang, D.; Jin, L.; Yao, H.W.; Zhang, J.H.; Wang, J.; Zhao, X.M.; Shen, C.Y.; Chen, W.; Wang, X.L.; et al. Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, Y.; Hashimoto, S.; Higaki, S.; Fujii, I.; Suzuki, C.; Hoshida, T.; Matsumoto, T.; Yamaoka, Y.; Takami, T.; Sakaida, I.; et al. Blood free-circulating DNA testing by highly sensitive methylation assay to diagnose colorectal neoplasias. Oncotarget 2018, 9, 16974–16987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Huang, T.X.; Cheng, F.S.; Huang, K.B.; Liu, M.; He, W.; Li, M.W.; Zhang, X.N.; Xu, M.Y.; Chen, S.F.; et al. Monitoring colorectal cancer following surgery using plasma circulating tumor DNA. Oncol. Lett. 2018, 15, 4365–4375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomsen, C.B.; Hansen, T.F.; Andersen, R.F.; Lindebjerg, J.; Jensen, L.H.; Jakobsen, A. Monitoring the effect of first line treatment in RAS/RAF mutated metastatic colorectal cancer by serial analysis of tumor specific DNA in plasma. J. Exp. Clin. Cancer Res. 2018, 37, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuki, H.; Yamada, T.; Takahashi, G.; Iwai, T.; Koizumi, M.; Shinji, S.; Yokoyama, Y.; Takeda, K.; Taniai, N.; Uchida, E. Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur. J. Surg. Oncol. 2018, 44, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Klein-Scory, S.; Maslova, M.; Pohl, M.; Eilert-Micus, C.; Schroers, R.; Schmiegel, W.; Baraniskin, A. Significance of liquid biopsy for monitoring and therapy decision of colorectal cancer. Transl. Oncol. 2018, 11, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Scholer, L.V.; Reinert, T.; Orntoft, M.W.; Kassentoft, C.G.; Arnadottir, S.S.; Vang, S.; Nordentoft, I.; Knudsen, M.; Lamy, P.; Andreasen, D.; et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, C.; Kehagias, P.; El Housni, H.; Ameye, L.; Laes, J.F.; Desmedt, C.; Sotiriou, C.; Deleporte, A.; Puleo, F.; Geboes, K.; et al. Circulating tumor DNA in early response assessment and monitoring of advanced colorectal cancer treated with a multi-kinase inhibitor. Oncotarget 2018, 9, 17756–17769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allegra, C.J.; Jessup, J.M.; Somerfield, M.R.; Hamilton, S.R.; Hammond, E.H.; Hayes, D.F.; McAllister, P.K.; Morton, R.F.; Schilsky, R.L. American society of clinical oncology provisional clinical opinion: Testing for kras gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol. 2009, 27, 2091–2096. [Google Scholar] [CrossRef] [PubMed]
- Lievre, A.; Bachet, J.B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.F.; Cote, J.F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef] [PubMed]
- Sidransky, D.; Tokino, T.; Hamilton, S.R.; Kinzler, K.W.; Levin, B.; Frost, P.; Vogelstein, B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 1992, 256, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgard, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Conte, D.; Leon, M.; Ciricione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003, 21, 3902–3908. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, G.; Conte, D.; Mariani, L.; Lo Vullo, S.; Roz, L.; Lombardo, C.; Pierotti, M.A.; Tavecchio, L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001, 61, 4675–4678. [Google Scholar] [PubMed]
- Valtorta, E.; Misale, S.; Sartore-Bianchi, A.; Nagtegaal, I.D.; Paraf, F.; Lauricella, C.; Dimartino, V.; Hobor, S.; Jacobs, B.; Ercolani, C.; et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int. J. Cancer 2013, 133, 1259–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardelli, A.; Corso, S.; Bertotti, A.; Hobor, S.; Valtorta, E.; Siravegna, G.; Sartore-Bianchi, A.; Scala, E.; Cassingena, A.; Zecchin, D.; et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013, 3, 658–673. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Ma, F.; Li, C.; Chen, R.; Yuan, L.; Sun, X.; Guan, X.; Li, L.; Liu, B.; Guan, Y.; et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci. Rep. 2017, 7, 5995. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Zhang, K.; Yang, X.; Ding, J.; Wang, Z.; Li, J. Prognostic value of circulating tumor DNA in patients with colon cancer: Systematic review. PLoS ONE 2017, 12, e0171991. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Barzi, A.; Sartore-Bianchi, A.; Cassingena, A.; Siravegna, G.; Karp, D.D.; Piha-Paul, S.A.; Subbiah, V.; Tsimberidou, A.M.; Huang, H.J.; et al. Mutation-enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin. Cancer Res. 2017, 23, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, D.A.; Skoletsky, J.E.; Boynton, K.A.; Harrington, J.J.; Mahoney, D.W.; Pierceall, W.E.; Thibodeau, S.N.; Shuber, A.P. Colorectal cancer screening by detection of altered human DNA in stool: Feasibility of a multitarget assay panel. Gastroenterology 2000, 119, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, D.A.; Zou, H.; Domanico, M.; Mahoney, D.W.; Yab, T.C.; Taylor, W.R.; Butz, M.L.; Thibodeau, S.N.; Rabeneck, L.; Paszat, L.F.; et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012, 142, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ClinicalTrials.gov. U.S. National Library of Medicine. 2015. Available online: https://clinicaltrials.Gov/ct2./show/nct02284633 (accessed on 6 November 2014).
- ClinicalTrials.gov. U.S. National Library of Medicine. 2016. Available online: https://clinicaltrials.Gov/ct2./show/nct02743910 (accessed on 19 April 2016).

| Advantages | Disadvantages |
|---|---|
| Cost-effective | Lack of standard operating protocol |
| Non-invasive | Released by both healthy and tumor cells |
| Rapid | Requirement of sensitive and specific methods |
| Comprehensive tumor profile | False-positive and false-negative results |
| Minimal pain and risk | Personnel microenvironment may influence the released of cfDNA amount |
| Serial assessments | Not-standardized cfDNA/ctDNA concentration as a cancer biomarker |
| Directly able to assess for specific mutations | |
| Present in many biological fluids | |
| Potential to evaluate prognosis, recurrence, response to therapy | |
| Detection of minimal residual disease | |
| Assessment of cancer high-risk populations | |
| Enabling of early cancer diagnosis | |
| Evaluation of tumor heterogeneity |
| Technique | Limit of Detection | Type of Alteration Detection | Advantages | Disadvantages |
|---|---|---|---|---|
| PCR based approaches (COLD-PCR, PNAs-LNA, ARMS, etc.) | 0.1–1% | SNV, indels | Low cost; Easy to perform | Low sensitivity; A limited number of studied genes at a time; Genes need to be pre-determined |
| Digital PCR (ddPCR and BEAMing) | 0.05% or less | SNV, indels, CNV | High sensitivity and specificity; Reasonable cost; Easy to perform | A limited number of studied genes at a time; Genes need to be pre-determined |
| NGS (Deep sequencing, TAM-seq, Safe-Seqs, CAPP-Seq, cSMART, digital sequencing) | 0.01–2% | SNV, indels, CNV, rearrangements | Allows more genes to be analyzed at a time | Wide range of sensitivity depending on the NGS platform used (PCR amplicon strategies are more sensitive and less expensive than whole genome or exome sequencing); Higher cost |
| SERS-nanotags | 0.01% | SNV | Reduced susceptibility to photobleaching; Bandwidths are significantly narrower | Raman signal deterioration upon prolonged laser illumination |
| UltraSEEK | 0.01% | SNV, indels | Low cost and low DNA input | Lower sensitivity |
| References | Patients | Controls | Origin of the Study | Source of cfDNA/ctDNA | Abnormalities | Methodology | Target | Clinical Relevance |
|---|---|---|---|---|---|---|---|---|
| Leon et al. [25] | 9 | 55 | USA | Serum | Concentration | Diagnostic | ||
| Anker et al. [82] | 14 | Switzerland | Plasma | Mutation | PCR | KRAS | Diagnostic | |
| de Kok et al. [83] | 14 | - | Netherland | Serum | Mutation | KRAS | Diagnostic | |
| Kopreski et al. [84] | 31 | 28 | USA | Plasma Serum | Mutation | PCR | KRAS | Diagnostic |
| Hibi et al. [85] | 44 | - | USA | Serum | Mutation | PCR | KRAS TP53 | Diagnostic |
| Lecomte et al. [86] | 58 | - | France | Plasma | Mutation Methylation | MS-PCR qPCR | KRAS p16 | Prognostic |
| Ryan et al. [87] | 94 | - | Ireland | Serum | Mutation | PCR sequencing | KRAS2 | Diagnostic Prognostic |
| Wang et al. [27] | 104 | 50 | Taiwan | Serum | Mutation Concentration | PCR-SSCP | APC KRAS TP53 | Diagnostic |
| Leung et al. [88] | 49 | 41 | Hong Kong | Serum | Methylation | MethyLight | APC hMLH1 HLTF | Diagnostic |
| Lindforss et al. [89] | 25 | - | Sweden | Plasma | Mutation | PCR | KRAS | Prognostic |
| Bazan et al. [90] | 66 | - | Italy | Plasma | Mutation Methylation | PCR | KRAS TP53 p16INK4a | Prognostic |
| Flamini et al. [91] | 75 | 75 | Italy | Serum | Concentration | qPCR | Diagnostic | |
| Frattini et al. [92] | 70 | 20 | Italy | Plasma | Concentration | qPCR | Diagnostic Prognostic | |
| Trevisiol et al. [93] | 86 | Italy | Serum | Mutation | qPCR | KRAS2 | Diagnostic Prognostic | |
| Wallner et al. [94] | 38 | 20 | Germany | Serum | Methylation | MS-PCR | HPP1/TPEF HLTF hMLH1 | Prediction |
| Boni et al. [95] | 67 | 67 | Italy | Plasma | Concentration | qPCR | Diagnostic | |
| Nakayama et al. [96] | 94 | - | Japan | Serum | Methylation | MS-PCR | p16 | Diagnostic |
| Diehl et al. [97] | 18 | - | USA | Plasma | Concentration | qPCR | Diagnostic Prognostic | |
| Frattini et al. [98] | 70 | 20 | Italy | Plasma | Concentration Mutation Methylation | qPCR MS-PCR ME-PCR | KRAS p16INK4a | Diagnostic Predictive |
| Lofton-Day et al. [99] | 133 | 179 | Germany | Plasma | Methylation | MS-PCR | TMEFF2 NGFR SEPT9 | Diagnostic |
| Schwarzenbach et al. [100] | 55 | 20 | Germany | Serum | Concentration | qPCR | Diagnostic | |
| Su et al. [39] | 20 | - | USA | Serum Plasma Urine | Concentration Mutation | PCR | KRAS | Diagnostic |
| Zitt et al. [101] | 26 | - | Austria | Plasma | Concentration | qPCR | Prognostic Predictive | |
| DeVos et al. [102] | 97 | 172 | Germany | Plasma | Methylation | MS-PCR | SEPT9 | Diagnostic |
| Herbst et al. [103] | 106 | - | Germany | Serum | Methylation | MethyLight | HLTF HPP1/TPEF | Prognostic |
| Agostini et al. [104] | 67 | 35 | Italy | Plasma | Concentration Dna Integrity | qPCR | Predictive | |
| Herbst et al. [103] | 106 | - | Germany | Plasma | Methylation | MS-PCR | HPP1/TPEF HLTF NEUROG1 | Diagnostic |
| Diaz et al. [105] | 28 | - | USA | Serum | Mutation | PCR | KRAS | Diagnostic Predictive |
| Morgan et al. [106] | 71 | - | UK | Plasma Serum | Mutation | qPCR | KRAS | Diagnostic |
| Phillip et al. [107] | 311 | - | Germany | Serum | Methylation | MS-PCR | HLTF HPP1 | Prognostic |
| Spindler et al. [108] | 108 | - | Denmark | Plasma | Concentration Mutation | qPCR | KRAS | Prognostic Predictive |
| Bai et al. [109] | 106 | - | China | Plasma | Mutation | PCR | KRAS | Diagnostic Prognostic |
| Cassinotti et al. [110] | 223 | - | Italy | Plasma | Concentration | qPCR | Prognostic | |
| Lee et al. [111] | 101 | 96 | Korea | Plasma | Methylation | PCR | Septin9 | Diagnostic |
| Spindler et al. [112] | 211 | - | Denmark | Plasma | Mutation | qPCR | KRAS BRAF | Diagnostic Predictive Prognostic |
| Sun et al. [113] | 34 | 10 | China | Plasma | Concentration Methylation Mutation | qPCR MS-PCR PCR-RFLP | MGMT KRAS | Diagnostic Predictive |
| Bettegowda et al. [21] | 24 | - | USA | Plasma | Concentration Mutation | PCR | KRAS | Diagnosis |
| Kuo et al. [16] | 52 | - | Taiwan | Plasma | Mutation | PCR | KRAS | Predictive |
| Lin et al. [114] | 133 | - | Taiwan | Plasma | Mutation | qPCR | 74 genes | Prognostic |
| Mohan et al. [115] | 10 | - | Austria | Plasma | Mutation | WGS | KRAS BRAF PIK3CA EGFR | Diagnostic Predictive |
| Perrone et al. [116] | 170 | - | Italy | Plasma | Mutation Concentration | ME-PCR, qPCR | KRAS | Diagnostic |
| Spindler et al. [117] | 108 | - | Denmark | Plasma | Mutation Concentration | PCR | KRAS BRAF | Predictive |
| Spindler et al. [118] | 100 | 100 | Denmark | Plasma | Concentration Mutation | PCR | KRAS | Diagnostic Predicitve |
| Tham et al. [119] | 150 | - | Singapore | Serum | Methylation | MS-PCR | TAC1 Septin9 NELL1 | Prognostic |
| Thierry et al. [23] | 106 | 29 | France | Plasma | Mutation | qPCR | KRAS BRAF | Diagnostic Predictive |
| Xu et al. [120] | 242 | - | China | Plasma | Mutation | PCR | KRAS | Prognostic |
| Carpinetti et al. [121] | 4 | - | Brazil | Plasma | Chromosomal Rearrangements | SOLiD | Predictive | |
| Lin et al. [122] | 353 | - | Taiwan | Plasma | Methylation | Methylation array | >450,000 CpG sites | Diagnostic |
| Reinert et al. [123] | 118 | - | Denmark | Plasma | Concentration | ddPCR | Diagnostic | |
| Sefrioui et al. [124] | 34 | - | France | Plasma | Mutation Concentration | dPCR | KRAS | Diagnostic Prognostic |
| Siravegna et al. [125] | 100 | - | Italy | Plasma | Mutation | PCR | KRAS | Prognostic Predictive |
| Spindler et al. [126] | 229 | 100 | Denmark | Plasma | Mutation Concentration | qPCR | KRAS | Diagnostic Prognostic |
| Liu et al. [127] | 165 | - | Singapore | Serum | Methylation | MS-PCR | SST | Prognostic |
| Matthaios et al. [128] | 155 | - | Greece | Plasma | Methylation | MS-PCR | APC RASSF1A | Prognostic |
| El Messaoudi et al. [129] | 97 | - | Francie | Plasma | Mutation Concentration | qPCR | KRAS BRAF | Diagnostic Prognostic |
| Tie et al. [130] | 230 | - | Australia | Plasma | Mutation | PCR | APC TP53 KRAS | Prognostic |
| Agah et al. [131] | 74 | - | Iran | Plasma | Concentration | qPCR | Diagnostic | |
| Bhangu et al. [132] | 30 | 17 | Austria | Plasma | Concentration | qPCR | Diagnostic | |
| Herbst et al. [133] | 467 | - | Germany | Plasma | Methylation | MS-PCR | HPP1 | Predictive Prognostic |
| Kloten et al. [134] | 50 | 8 | Germany | Plasma | Concentration | qPCR mutation | KRAS | Diagnostic |
| Li et al. [135] | 80 | 35 | USA | Serum Plasma | Concentration Cnvs | WGS | Diagnostic Prognostic | |
| Pereira et al. [136] | 128 | - | USA | Plasma | Mutation | sequencing | Diagnostic | |
| Yamauchi et al. [137] | 21 | - | Japan | Plasma | Mutation | sequencing | Predictive | |
| Liu et al. [138] | 27 | - | USA | Plasma | Methylation | Infinium HM450 array | Diagnostic | |
| Takayama et al. [139] | 85 | - | Japan | Plasma | Concentration Mutation | dPCR | KRAS | Diagnostic Predictive |
| Toledo et al. [140] | 1 | - | Spain | Plasma | Whole Exome Sequencing | sequencing | Predicitve | |
| Schou et al. [141] | 123 | - | Denmark | Plasma | Concentration | fluorescence | Diagnostic | |
| Sclafani et al. [142] | 51 | - | Clinical Trial | Plasma | Mutation | ddPCR | KRAS BRAF | Diagnostic Predictive |
| Boysen et al. [143] | 273 | 94 | Denmark Norway Sweden | Plasma | Concentration | ddPCR DFA | Diagnostic | |
| Myint et al. [144] | 131 | 37 | UK | Plasma Stool | Concentration Mutation | qPCR | KRAS BRAF | Diagnostic |
| Demuth et al. [145] | 28 | - | Denmark | Plasma | Mutation | ddPCR | KRAS | Prognostic |
| Rokni et al. [146] | 50 | - | Iran | Plasma | Methylation | High methylation resolution PCR | BMP3 | Prognostic |
| Fu et al. [147] | 98 CRC 101 adenomas 76 nCRC | 253 | China | Plasma | Methylation | MS-PCR | SEPT9 | Prognostic |
| Molparia et al. [148] | 24 | 25 | USA | Plasma | Cnvs | sequencing | Diagnostic Prognostic | |
| Gallardo-Gómez et al. [149] | 20 CRC 20 adenomas | 20 | Spain | Serum | Methylation | microarray | Diagnostic | |
| Nunes et al. [150] | 72 | 103 | Portugal | Plasma | Methylation | MS-PCR | APC, FOXA1 MGMT RARβ2 RASSF1A SCGB3A1 SEPT9 SHOX2 SOX17 | Prognostic |
| Song et al. [151] | 150 | - | China | Urine | Concentration | ddPCR | Predictive Prognostic | |
| Yang et al. [152] | 47 | - | China | Plasma | Mutation | sequencing | 37 genes | Diagnostic Prognostic |
| Suehiro et al. [153] | 113 | 25 | Japan | Serum | Methylation | ddPCR | TWIST1 | Diagnostic |
| Sun et al. [154] | 11 | - | China | Plasma | Mutation | sequencing | 85 genes | Prognostic |
| Thomsen et al. [155] | 138 | - | Denmark | Plasma | Concentration Mutation | ddPCR | RAS/RAF | Prognostic |
| Furuki et al. [156] | 22 | - | Japan | Serum | Mutation | sequencing | TP53 KRAS APC PIK3CA BRAF FBXW7 NRAS | Diagnostic Prognostic |
| Klein-Scory et al. [157] | 3 | - | Germany | Plasma | Mutation | BEAMing | BRAF PIK3CA | Predictive |
| Schøler et al. [158] | 45 | - | Denmark | Plasma | Concentration | WGS | Prognostic | |
| Vandeputte et al. [159] | 20 | - | Belgium | Plasma | Concentration | ddPCR | Predictive |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. https://doi.org/10.3390/ijms19113356
Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. International Journal of Molecular Sciences. 2018; 19(11):3356. https://doi.org/10.3390/ijms19113356
Chicago/Turabian StyleVymetalkova, Veronika, Klara Cervena, Linda Bartu, and Pavel Vodicka. 2018. "Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review" International Journal of Molecular Sciences 19, no. 11: 3356. https://doi.org/10.3390/ijms19113356
APA StyleVymetalkova, V., Cervena, K., Bartu, L., & Vodicka, P. (2018). Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. International Journal of Molecular Sciences, 19(11), 3356. https://doi.org/10.3390/ijms19113356






