Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas
Abstract
1. Introduction
2. Results
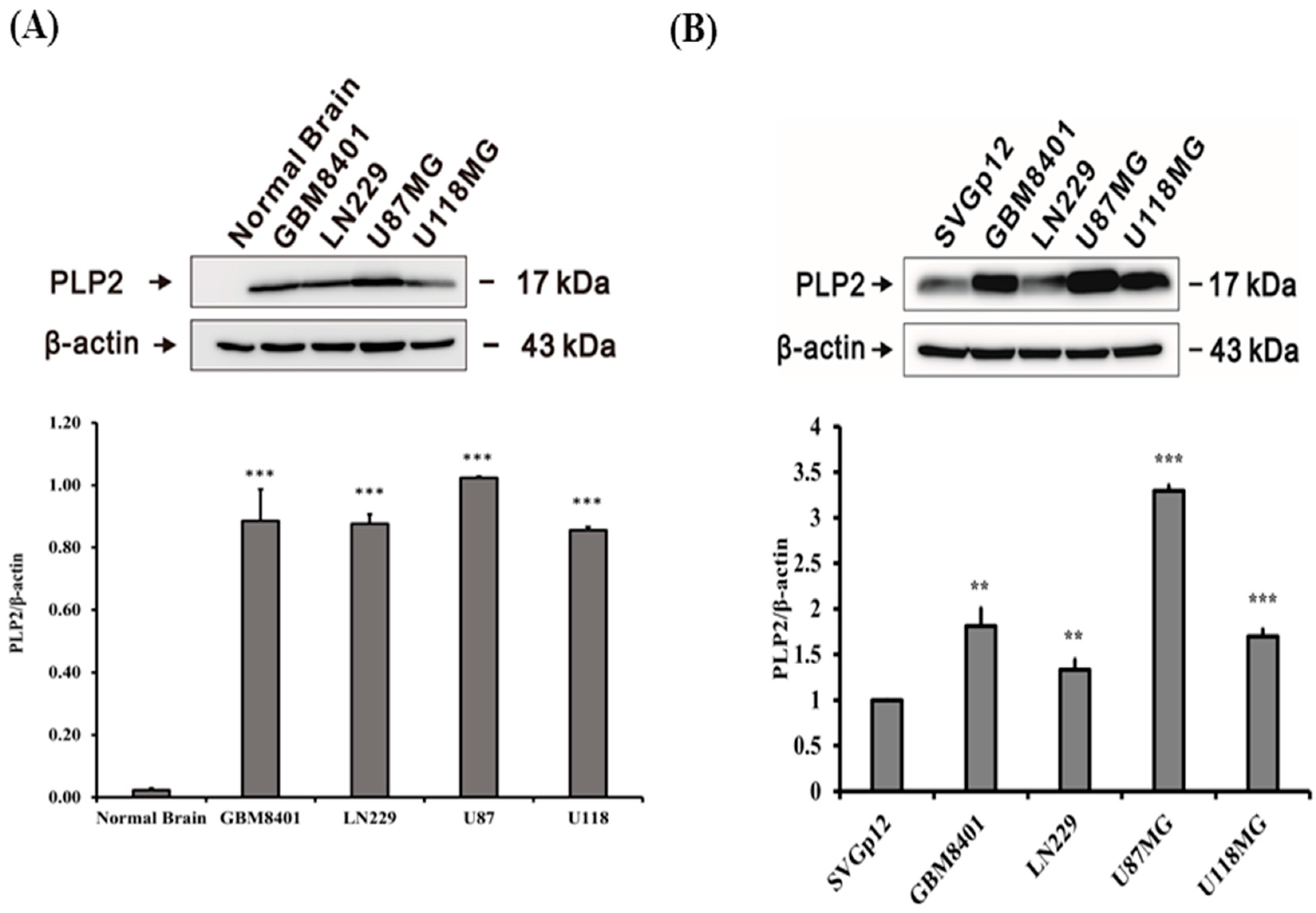
2.1. PLP2 Protein Overexpression in Human Glioma Cell Lines
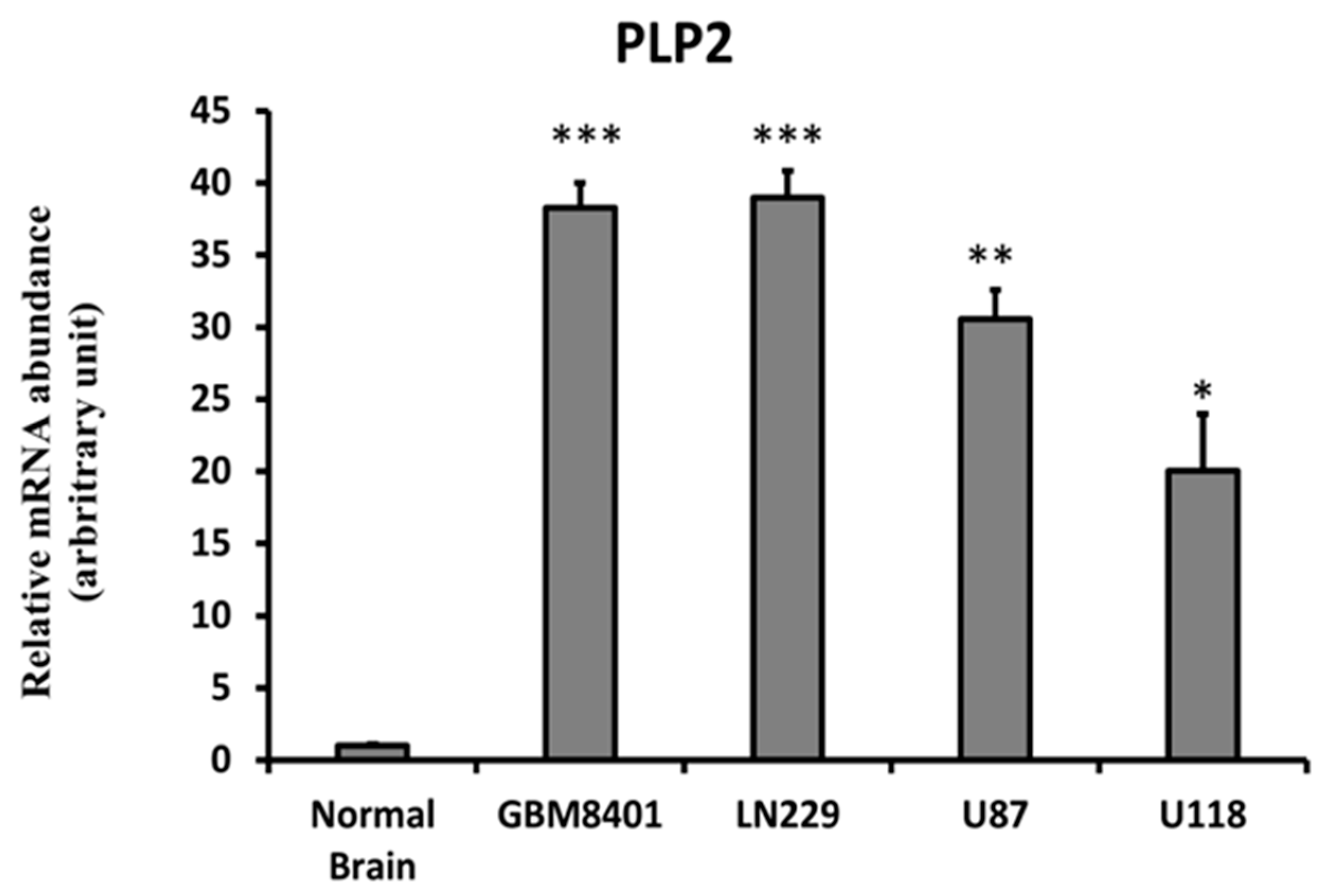
2.2. Higher PLP2 mRNA Expression in Human Glioma Cell Lines than in Normal Brain Tissues
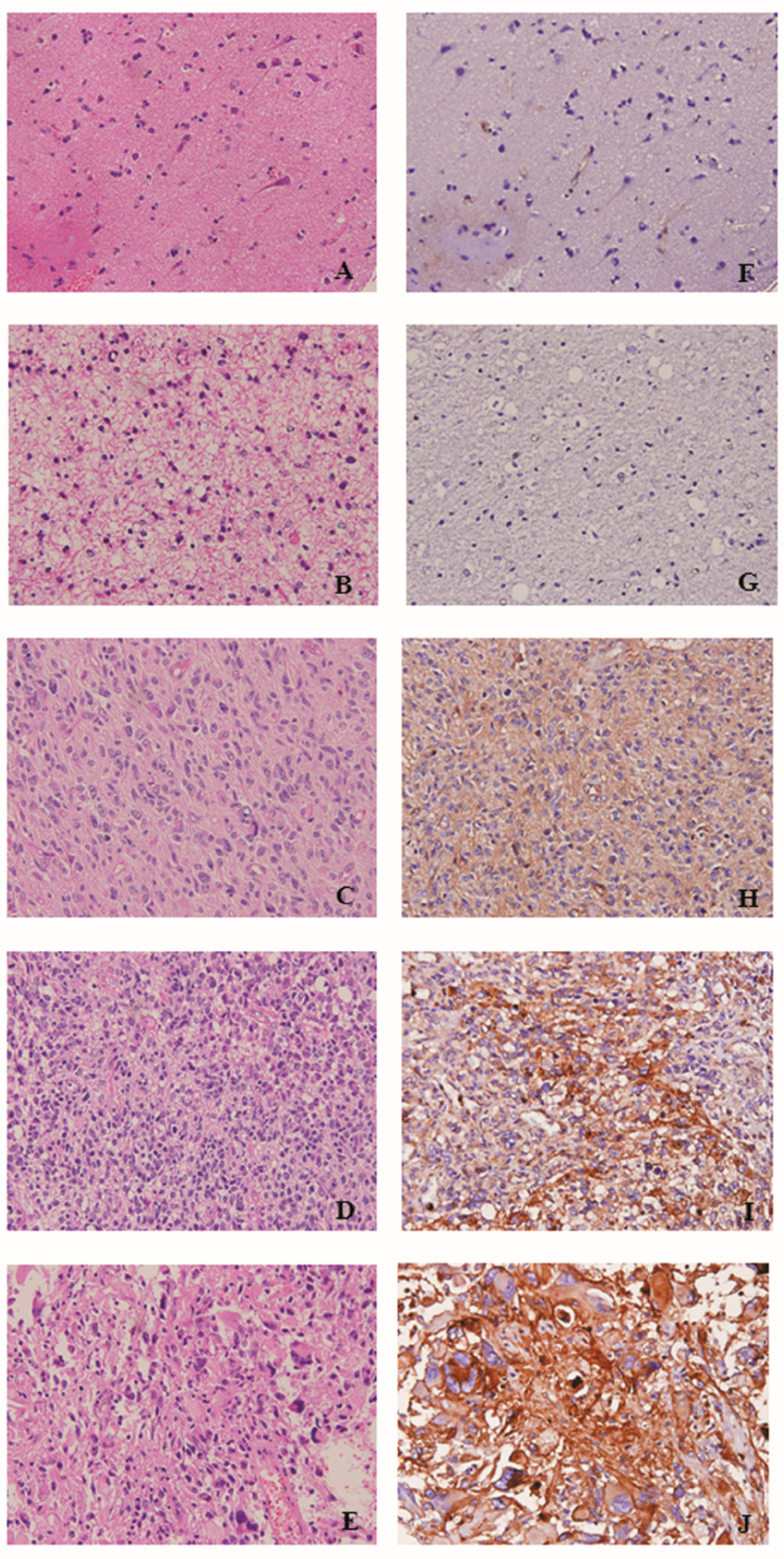
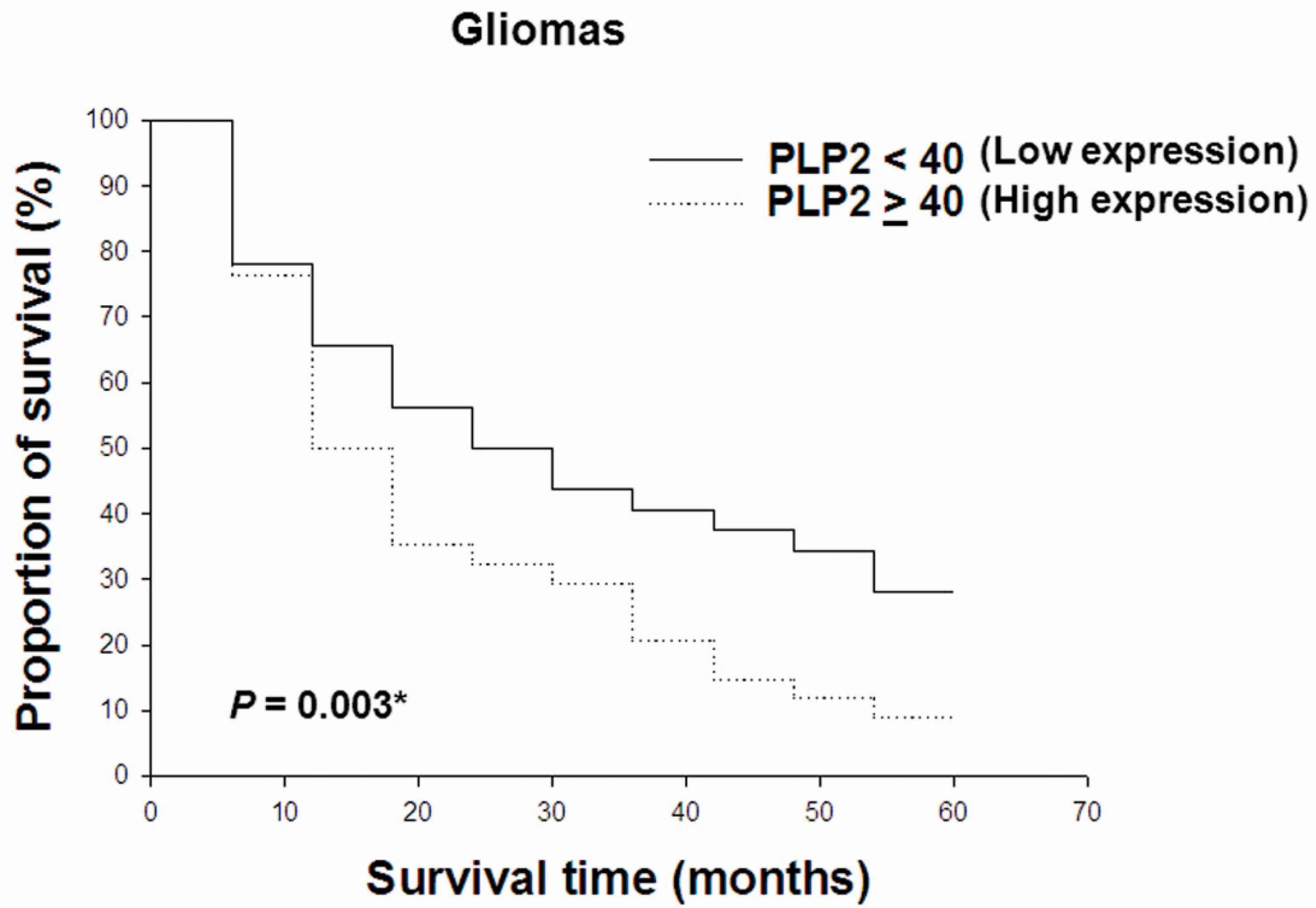
2.3. PLP2 IHC Staining Correlated with WHO Grades and Poor Prognosis in Gliomas
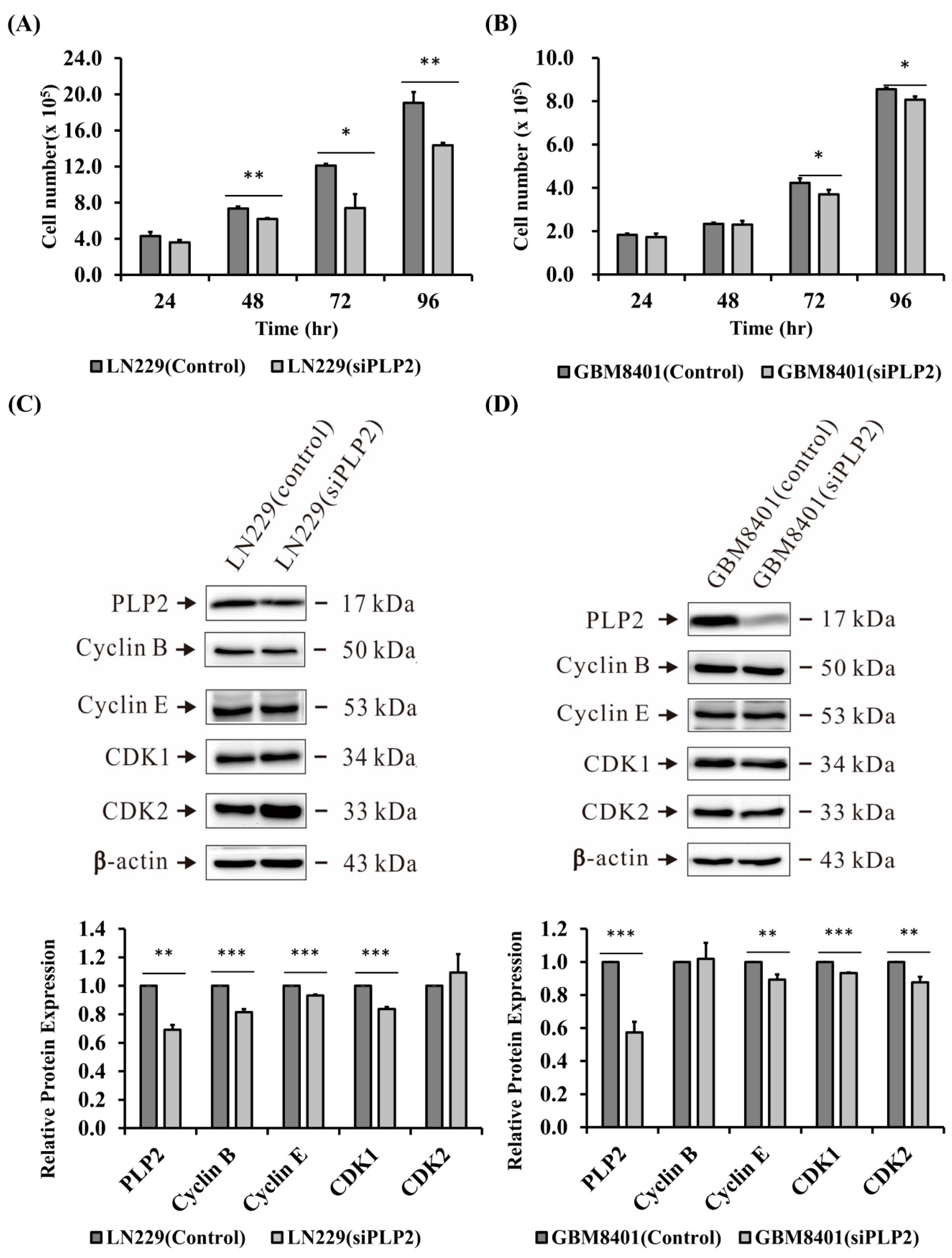
2.4. Suppression of PLP2 Inhibits Glioma Cell Proliferation
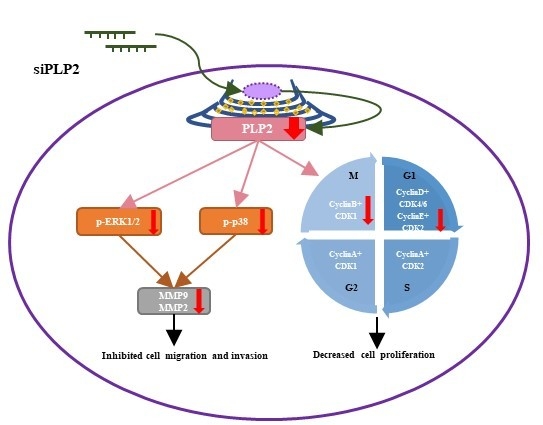
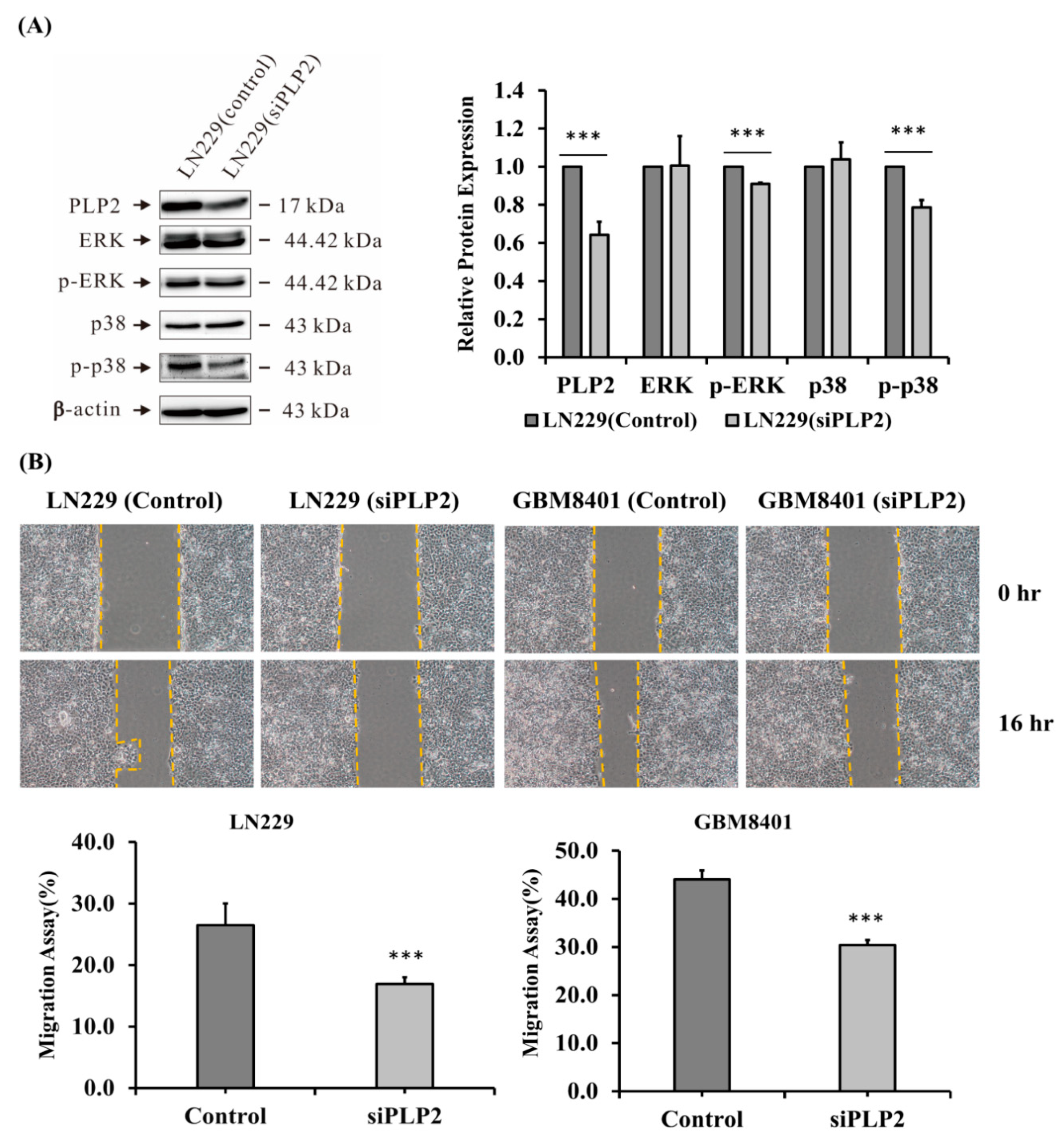
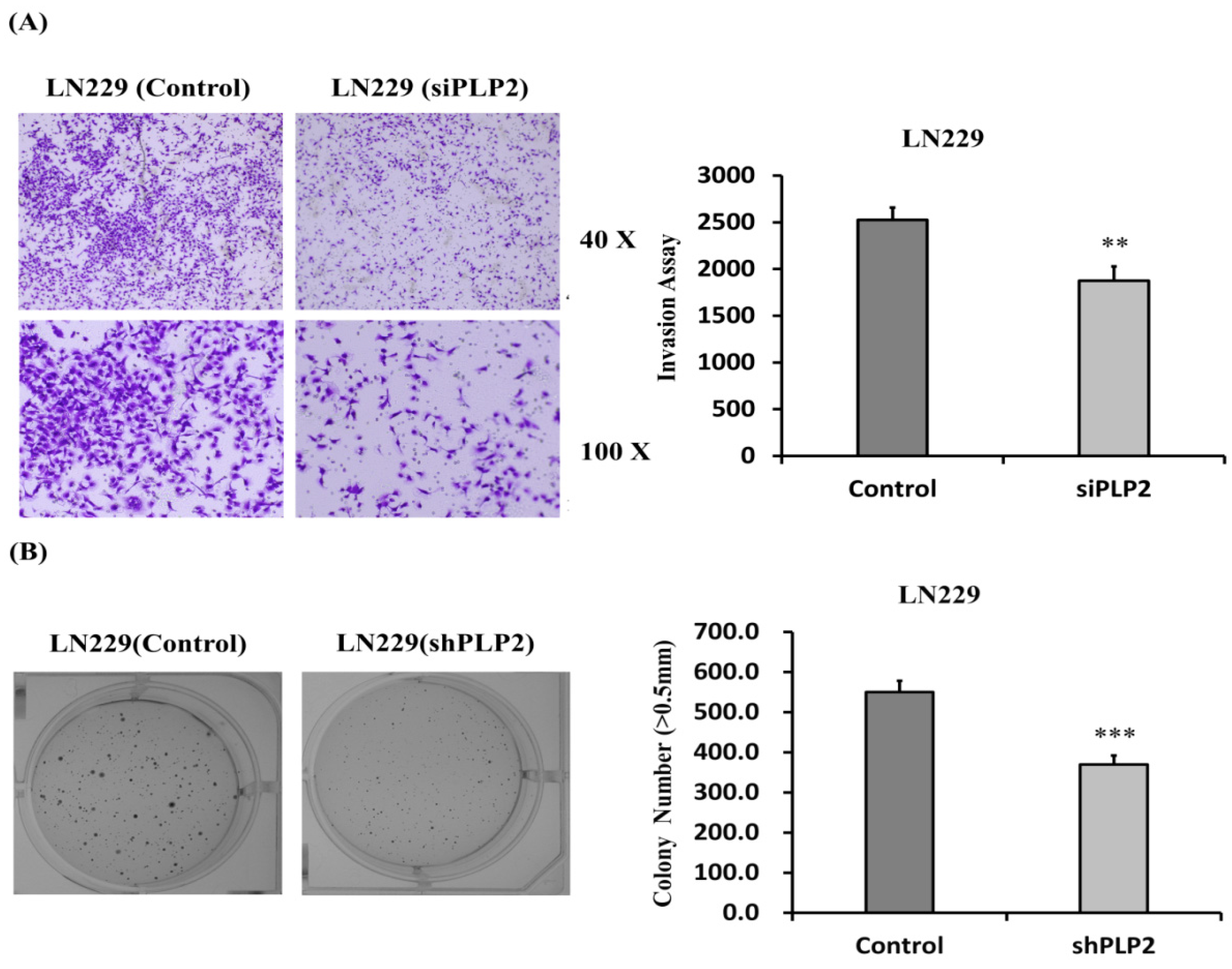
2.5. PLP2 Inhibition Down-Regulates Glioma Cell Migration and Invasion
2.6. Knockdown of PLP2 Expression Inhibits Glioma Tumorigeneity
3. Discussion
4. Materials and Methods
4.1. Human Glioma Cell Lines and Lysate Preparation
4.2. Western Blot Analysis
4.3. RNA Isolation and Real-Time Reverse Transcription-PCR
4.4. Tissue Microarray Slide Preparation and Immunohistochemistry
4.5. Assessment of Immunostain Scores in Gliomas
4.6. Statistical Analysis of PLP2 Expression
4.7. Transfection of siPLP2 and siRNA-A into LN229 and GBM8401 Cell Lines and Cell Proliferation Assay
4.8. Wound Healing Assay
4.9. Cell Invasion Assay
4.10. Soft Agar Colony Formation Assay
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nowosielski, M.; Galldiks, N.; Iglseder, S.; Kickingereder, P.; von Deimling, A.; Bendszus, M.; Wick, W.; Sahm, F. Diagnostic challenges in meningioma. Neuro Oncol. 2017, 19, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Karim, A.B.; Maat, B.; Hatlevoll, R.; Menten, J.; Rutten, E.H.; Thomas, D.G.; Mascarenhas, F.; Horiot, J.C.; Parvinen, L.M.; van Reijn, M.; et al. A randomized trial on dose-response in radiation therapy of low-grade cerebral glioma: European Organization for Research and Treatment of Cancer (EORTC) Study 22844. Int. J. Radiat. Oncol. Biol. Phys. 1996, 36, 549–556. [Google Scholar] [CrossRef]
- Van den Bent, M.J.; Afra, D.; de Witte, O.; Ben Hassel, M.; Schraub, S.; Hoang-Xuan, K.; Malmström, P.O.; Collette, L.; Piérart, M.; Mirimanoff, R.; et al. Long-term efficacy of early versus delayed radiotherapy for low-grade astrocytoma and oligodendroglioma in adults: The EORTC 22845 randomised trial. Lancet 2005, 366, 985–990. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Weistler, O.D.; Cavenee, W.K. World Health Organization Classification of Tumours of the Central Nervous System, 5th ed.; IARC Press: Lyon, France, 2016; pp. 16–122. [Google Scholar]
- Gu, J.; Liu, Y.; Kyritsis, A.P.; Bondy, M.L. Molecular epidemiology of primary brain tumors. Neurotherapeutics 2009, 6, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Lee, H.S.; Lin, C.K.; Chen, A.; Nieh, S.; Ma, H.I. The association of osteopontin and LMX1A expression with World Health Organization grade in meningiomas and gliomas. Histopathology 2012, 61, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Lukas, R.V.; Nicholas, M.K. Update in the treatment of high-grade Gliomas. Neurol. Clin. 2013, 31, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Binello, E.; Green, S.; Germano, I.M. Radiosurgery for high-grade glioma. Surg. Neurol. Int. 2012, 3, S118–S126. [Google Scholar] [PubMed]
- Mandell, L.R.; Walker, R.W.; Steinherz, P.; Fuks, Z. Reduced incidence of the somnolence syndrome in leukemic children with steroid coverage during prophylactic cranial radiation therapy. Results of a pilot study. Cancer 1989, 63, 1975–1978. [Google Scholar] [CrossRef]
- Lee, A.W.; Foo, W.; Chappell, R.; Fowler, J.F.; Sze, W.M.; Poon, Y.F.; Law, S.C.; Ng, S.H.; O, S.K.; Tung, S.Y.; et al. Effect of time, dose, and fractionation on temporal lobe necrosis following radiotherapy for nasopharyngeal carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 1998, 40, 35–42. [Google Scholar] [CrossRef]
- Rajasekhar, A.; George, T.J., Jr. Gemcitabine-induced reversible posterior leukoencephalopathy syndrome: A case report and review of the literature. Oncologist 2007, 12, 1332–1335. [Google Scholar] [CrossRef] [PubMed]
- Onujiogu, N.; Lengyel, E.; Yamada, S.D. Reversible posterior leukoencephalopathy syndrome following intravenous paclitaxel and intraperitoneal cisplatin chemotherapy for fallopian tube cancer. Gynecol. Oncol. 2008, 111, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Sierra del Rio, M.; Rousseau, A.; Soussain, C.; Ricard, D.; Hoang-Xuan, K. Primary CNS lymphoma in immunocompetent patients. Oncologist 2009, 14, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Noronha, V.; Berliner, N.; Ballen, K.K.; Lacy, J.; Kracher, J.; Baehring, J.; Henson, J.W. Treatment-related myelodysplasia/AML in a patient with a history of breast cancer and an oligodendroglioma treated with temozolomide: Case study and review of the literature. Neuro Oncol. 2006, 8, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Metellus, P.; Coulibaly, B.; Colin, C.; de Paula, A.M.; Vasiljevic, A.; Taieb, D.; Barlier, A.; Boisselier, B.; Mokhtari, K.; Wang, X.W.; et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010, 120, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Sadighi, Z.; Slopis, J. Pilocytic astrocytoma: A disease with evolving molecular heterogeneity. J. Child. Neurol. 2013, 28, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Horbinski, C. To BRAF or not to BRAF: Is that even a question anymore? J. Neuropathol. Exp. Neurol. 2013, 72, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, A.B.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Nagahashi Marie, S.K.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Agrawal, N.; Jiao, Y.; Sausen, M.; Wood, L.D.; Hruban, R.H.; Rodriguez, F.J.; Cahill, D.P.; McLendon, R.; Riggins, G.; et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011, 333, 1453–1455. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Butterfield, Y.S.; Morozova, O.; Chittaranjan, S.; Blough, M.D.; An, J.; Birol, I.; Chesnelong, C.; Chiu, R.; Chuah, E.; et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012, 226, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Wellenreuther, R.; Waha, A.; Vogel, Y.; Lenartz, D.; Schramm, J.; Wiestler, O.D.; von Deimling, A. Quantitative analysis of neurofibromatosis type 2 gene transcripts in meningiomas supports the concept of distinct molecular variants. Lab. Investig. 1997, 77, 601–606. [Google Scholar] [PubMed]
- Rouleau, G.A.; Merel, P.; Lutchman, M.; Sanson, M.; Zucman, J.; Marineau, C.; Hoang-Xuan, K.; Demczuk, S.; Desmaze, C.; Plougastel, B.; et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 1993, 363, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.R.; Gill, C.M.; Nayyar, N.; D’Andrea, M.R.; Thiede, C.; Juratli, T.A.; Schackert, G.; Borger, D.R.; Santagata, S.; Frosch, M.P.; et al. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J. Neurosurg. 2017, 127, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Erson-Omay, E.Z.; Serin, A.; Yin, J.; Cotney, J.; Ozduman, K.; Avşar, T.; Li, J.; Murray, P.B.; Henegariu, O.; et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 2013, 339, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Abedalthagafi, M.S.; Bi, W.L.; Merrill, P.H.; Gibson, W.J.; Rose, M.F.; Du, Z.; Francis, J.M.; Du, R.; Dunn, I.F.; Ligon, A.H.; et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015, 208, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Solomon, D.A.; Wood, M.D.; Tihan, T.; Bollen, A.W.; Gupta, N.; Phillips, J.J.; Perry, A. Diffuse Midline Gliomas with Histone H3-K27M Mutation: A Series of 47 Cases Assessing the Spectrum of Morphologic Variation and Associated Genetic Alterations. Brain Pathol. 2016, 26, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Network, T.C. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2013, 494, 506. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.W.; Weiss, W.A. Targeting the RTK-PI3K-mTOR axis in malignant glioma: Overcoming resistance. Curr. Top. Microbiol. Immunol. 2010, 347, 279–296. [Google Scholar] [PubMed]
- Fresno Vara, J.A.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, T.; Valle, D. Reduced PLP2 expression increases ER-stress-induced neuronal apoptosis and risk for adverse neurological outcomes after hypoxia ischemia injury. Hum. Mol. Genet. 2015, 24, 7221–7226. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, Y.; Warita, M.; Suzuki, T.; Ozawa, H.; Fukuda, Y.; Funakoshi-Tago, M.; Kasahara, T. Proteolipid protein 2 is associated with melanoma metastasis. Oncol. Rep. 2010, 23, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Longo, A.; Librizzi, M.; Luparello, C. Effect of transfection with PLP2 antisense oligonucleotides on gene expression of cadmium-treated MDA-MB231 breast cancer cells. Anal. Bioanal. Chem. 2013, 405, 1893–1901. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.W.; Pennison, M.J.; Brezovich, I.; Yi, N.; Yang, C.T.; Ramaker, R.; Absher, D.; Myers, R.M.; Kuster, N.; Costa, F.P.; et al. Cancer cell proliferation is inhibited by specific modulation frequencies. Br. J. Cancer 2012, 106, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Shin, H.; Jang, S.W.; Shim, J.J.; Song, I.S.; Son, K.N.; Hwang, J.; Shin, Y.H.; Kim, H.H.; Lee, C.K.; et al. PLP2/A4 interacts with CCR1 and stimulates migration of CCR1-expressing HOS cells. Biochem. Biophys. Res. Commun. 2004, 324, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Miao, M.H.; Ji, X.Q.; Xue, J.; Shao, X.J. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem. Biophys. Res. Commun. 2015, 459, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Hueng, D.Y.; Lin, C.R.; Yang, T.C.; Gao, H.W. Nrf2 Expressions Correlate with WHO Grades in Gliomas and Meningiomas. Int. J. Mol. Sci. 2016, 17, 722. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Chang, J.T.; Andrechek, E.R.; Matsumura, N.; Baba, T.; Yao, G.; Kim, J.W.; Gatza, M.; Murphy, S.; Nevins, J.R. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene 2009, 28, 2796–2805. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Jian, S.; Peng, X.; Liu, Y.; Wang, J.; Zheng, L.; Ou, C.; Wang, Y.; Zeng, W.; Zhou, M. Loss of MiR-664 Expression Enhances Cutaneous Malignant Melanoma Proliferation by Upregulating PLP2. Medicine (Baltimore) 2015, 94, e1327. [Google Scholar] [CrossRef] [PubMed]
- Bradham, C.; McClay, D.R. p38 MAPK in development and cancer. Cell Cycle 2006, 5, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Ge, B.; Gram, H.; Di Padova, F.; Huang, B.; New, L.; Ulevitch, R.J.; Luo, Y.; Han, J. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 2002, 295, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Blenis, J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004, 68, 320–344. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.C.; Shibata, T.; Kleihues, P. The use of the monoclonal antibody Ki-67 in the identification of proliferating cells: Application to surgical neuropathology. Am. J. Surg. Pathol. 1986, 10, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Deckert, M.; Reifenberger, G.; Wechsler, W. Determination of the proliferative potential of human brain tumors using the monoclonal antibody Ki-67. J. Cancer Res. Clin. Oncol. 1989, 115, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Makino, K.; Nakamura, H.; Hide, T.; Yano, S.; Kuroda, J.; Takahashi, Y.; Iyama, K.; Kuratsu, J. Usefulness of immunohistochemical expression analysis of metabolic-related molecules in differentiating between intracranial neoplastic and non-neoplastic lesions. Brain Tumor Pathol. 2013, 30, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Zengotita, M.; Yachnis, A.T. Gliosis versus glioma: Don’t grade until you know. Adv. Anat. Pathol. 2012, 19, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Sahm, F.; Hartmann, C.; Meyermann, R.; von Deimling, A.; Schittenhelm, J. Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy-induced changes. Am. J. Surg. Pathol. 2010, 34, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Camelo-Piragua, S.; Jansen, M.; Ganguly, A.; Kim, J.C.; Louis, D.N.; Nutt, C.L. Mutant IDH1-specific immunohistochemistry distinguishes diffuse astrocytoma from astrocytosis. Acta Neuropathol. 2010, 119, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Lebrun, D.G.; Yang, J.; Zhu, V.F.; Li, M. Deregulated signaling pathways in glioblastoma multiforme: Molecular mechanisms and therapeutic targets. Cancer Investig. 2012, 30, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Han, L.; Dong, Y.; Tan, Y.; Li, Y.; Zhao, M.; Xie, H.; Ju, H.; Wang, H.; Zhao, Y.; et al. EGFRvIII/integrin β3 interaction in hypoxic and vitronectinenriching microenvironment promote GBM progression and metastasis. Oncotarget 2016, 7, 4680–4694. [Google Scholar] [PubMed]
- Stoeckli, E.T.; Landmesser, L.T. Axonin-1, Nr-CAM, and Ng-CAM play different roles in the in vivo guidance of chick commissural neurons. Neuron 1995, 14, 1165–1179. [Google Scholar] [CrossRef]
- Manfioletti, G.; Brancolini, C.; Avanzi, G.; Schneider, C. The protein encoded by a growth arrest-specific gene (gas6) is a new member of the vitamin K-dependent proteins related to protein S, a negative coregulator in the blood coagulation cascade. Mol. Cell Biol. 1993, 13, 4976–4985. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Song, X.; Liu, G.; Li, R.; Xie, J.; Xiao, L.; Du, M.; Zhang, Q.; Xu, X.; Gan, X.; et al. Involvement of TR3/Nur77 translocation to the endoplasmic reticulum in ER stress-induced apoptosis. Exp. Cell Res. 2007, 313, 2833–2844. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.F.; Lin, P.C.; Ho, L.I.; Liu, P.Y.; Wu, W.C.; Chiang, I.P.; Chang, H.W.; Lin, S.Z.; Harn, Y.C.; Harn, H.J.; et al. Overexpression of the orphan receptor Nur77 and its translocation induced by PCH4 may inhibit malignant glioma cell growth and induce cell apoptosis. J. Surg. Oncol. 2011, 103, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Kessler, T.; Sahm, F.; Sadik, A.; Stichel, D.; Hertenstein, A.; Reifenberger, G.; Zacher, A.; Sabel, M.; Tabatabai, G.; Steinbach, J.; et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro Oncol. 2018, 20, 367–379. [Google Scholar] [CrossRef] [PubMed]

| Average Intensity | Average % Tumor | Average Score | Correlation * | |
|---|---|---|---|---|
| Normal Brain Tissue | 0.55 | 10.95 | 10.95 | |
| Classification of Gliomas | ||||
| Pilocytic astrocytoma | 1 | 5 | 5 | No correlation (p = 0.206) |
| Diffuse astrocytoma, IDH-mutant | 0 | 0 | 0 | |
| Diffuse astrocytoma, IDH-WT | 1.31 | 35.38 | 59.23 | |
| Anaplastic astrocytoma, IDH-mutant | 0.67 | 6.67 | 6.67 | |
| Anaplastic astrocytoma, IDH-WT | 1.43 | 36.43 | 82.14 | |
| Glioblastoma, IDH-mutant | 1.2 | 33 | 46 | |
| Glioblastoma, IDH-WT | 1.53 | 41.32 | 79.41 | |
| Oligodendroglioma, NOS | 0.33 | 10 | 10 | No correlation (p = 0.488) |
| Anaplastic oligodendroglioma, NOS | 0.6 | 23 | 23 | |
| Diffuse midline glioma, H3 K27M-mutant | 1.47 | 44.67 | 87.33 | |
| WHO Grades of Gliomas | ||||
| WHO grade I | 1 | 5 | 5 | Positive correlation (p = 0.039 *) |
| WHO grade II | 1.06 | 28.82 | 47.06 | |
| WHO grade III | 1 | 26 | 47.33 | |
| WHO grade IV | 1.48 | 41.48 | 78.52 |
| Variable | Total | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | ||
| Sex | |||||
| Male | 47 | 1 | |||
| Female | 29 | 1.18 (1.10–1.22) | 0.507 | 1.28 (1.24–1.30) | 0.279 |
| Age | |||||
| <50 | 36 | 1 | |||
| ≥50 | 40 | 2.38 (1.59–3.87) | 0.011 # | 2.38 (1.59–3.87) | 0.244 |
| IDH1 R132H | |||||
| Negative | 63 | 1 | |||
| Positive | 13 | 0.46 (0.09–0.79) | 0.065 | 0.46 (0.09–0.80) | 0.069 |
| ATRX | |||||
| Preserve | 39 | 1 | |||
| Loss of expression | 37 | 0.83 (0.76–0.86) | 0.450 | 0.81 (0.74–0.85) | 0.114 |
| H3K27M | |||||
| Negative | 67 | 1 | |||
| Positive | 9 | 1.80 (1.44–2.00) | 0.045 # | 1.68 (1.33–1.88) | 0.029 * |
| MGMT | |||||
| Preserved | 43 | 1 | |||
| Loss of expression | 33 | 1.30 (1.17–1.63) | 0.301 | 1.36 (1.22–1.71) | <0.001 * |
| EGFR | |||||
| Negative | 69 | 1 | |||
| Positive | 7 | 1.89 (1.42–2.16) | 0.053 | 1.77 (1.31–2.03) | 0.268 |
| EGFRvIII | |||||
| Negative | 58 | 1 | |||
| Positive | 18 | 2.26 (2.14–2.49) | 0.001 # | 2.29 (2.14–2.59) | 0.013 * |
| P53 | |||||
| Negative | 36 | 1 | |||
| Overexpression | 40 | 1.59 (1.39–2.14) | 0.066 | 1.43 (1.27–1.87) | 0.050 |
| Neurofilament | |||||
| Negative | 59 | 1 | |||
| Positive | 17 | 1.17 (0.89–1.33) | 0.582 | 1.16 (0.89–1.31) | 0.091 |
| NF1 | |||||
| Negative | 42 | 1 | |||
| Positive | 34 | 1.29 (1.24–1.40) | 0.302 | 1.18 (1.15–1.22) | 0.002 * |
| AxL | |||||
| Negative | 29 | 1 | |||
| Positive | 47 | 2.21 (1.64–4.99) | 0.006 # | 2.07 (1.50–4.67) | <0.001 * |
| p-AxL | |||||
| Negative | 22 | 1 | |||
| Positive | 54 | 0.97 (0.80–1.53) | 0.923 | 0.97 (0.81–1.52) | 0.004 * |
| NUR77 | |||||
| Negative | 27 | 1 | |||
| Positive | 49 | 1.71 (1.35–3.10) | 0.057 | 1.80 (1.42–3.28) | 0.010 * |
| H3Lys27 | |||||
| Preserved | 67 | 1 | |||
| Loss of expression | 9 | 1.59 (1.47–1.84) | 0.021 # | 1.58 (1.43–1.80) | <0.001 * |
| PDGFRA | |||||
| Negative | 5 | 1 | |||
| Positive | 71 | 1.24 (0.41–1.76) | 0.634 | 1.16 (0.30–1.66) | 0.442 |
| Multivariate Analysis | |||
|---|---|---|---|
| Variable | Hazard Ratio | 95% Confidence Interval | p Value |
| Sex (male/female) | 1.71 | 0.73–3.99 | 0.214 |
| Age (<50/>50) | 2.90 | 1.28–6.56 | 0.010 * |
| PLP2 (Negative/Positive) | 1.01 | 0.99–1.03 | 0.030 * |
| IDH1 R132H (Negative/Positive) | 2.69 | 0.87–8.30 | 0.086 |
| ATRX (Preserve/Loss) | 0.28 | 0.12–0.65 | 0.003 * |
| H3K27M (Negative/Positive) | 2.20 | 0.56–8.76 | 0.262 |
| MGMT (Unmethylated/ Methylated) | 0.47 | 0.17–1.29 | 0.142 |
| EGFR (Negative/Positive) | 2.69 | 0.63–11.49 | 0.181 |
| EGFRvIII (Negative/Positive) | 0.90 | 0.39–2.06 | 0.805 |
| P53 (Negative/Overexpression) | 0.79 | 0.36–1.75 | 0.557 |
| Neurofilament (Negative/Positive) | 0.70 | 0.29–1.69 | 0.423 |
| NF1 (Negative/Positive) | 1.01 | 0.40–2.54 | 0.996 |
| AxL (Negative/Positive) | 4.03 | 1.32–12.27 | 0.014 * |
| p-AxL (Negative/Positive) | 2.39 | 0.77–7.42 | 0.130 |
| NUR77 (Negative/Positive) | 0.35 | 0.14–0.85 | 0.021 * |
| H3Lys27 (Negative/Positive) | 0.45 | 0.11–1.94 | 0.284 |
| PDGFRA (Negative/Positive) | 14.078 | 3.27–60.65 | <0.001 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-H.; Hueng, D.-Y.; Tsai, W.-C. Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas. Int. J. Mol. Sci. 2018, 19, 3353. https://doi.org/10.3390/ijms19113353
Chen Y-H, Hueng D-Y, Tsai W-C. Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas. International Journal of Molecular Sciences. 2018; 19(11):3353. https://doi.org/10.3390/ijms19113353
Chicago/Turabian StyleChen, Yi-Hsuan, Dueng-Yuan Hueng, and Wen-Chiuan Tsai. 2018. "Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas" International Journal of Molecular Sciences 19, no. 11: 3353. https://doi.org/10.3390/ijms19113353
APA StyleChen, Y.-H., Hueng, D.-Y., & Tsai, W.-C. (2018). Proteolipid Protein 2 Overexpression Indicates Aggressive Tumor Behavior and Adverse Prognosis in Human Gliomas. International Journal of Molecular Sciences, 19(11), 3353. https://doi.org/10.3390/ijms19113353