Altered Expression of Retinol Metabolism-Related Genes in an ANIT-Induced Cholestasis Rat Model
Abstract
1. Introduction
2. Results
2.1. Biochemical Characterization of ANIT Rats
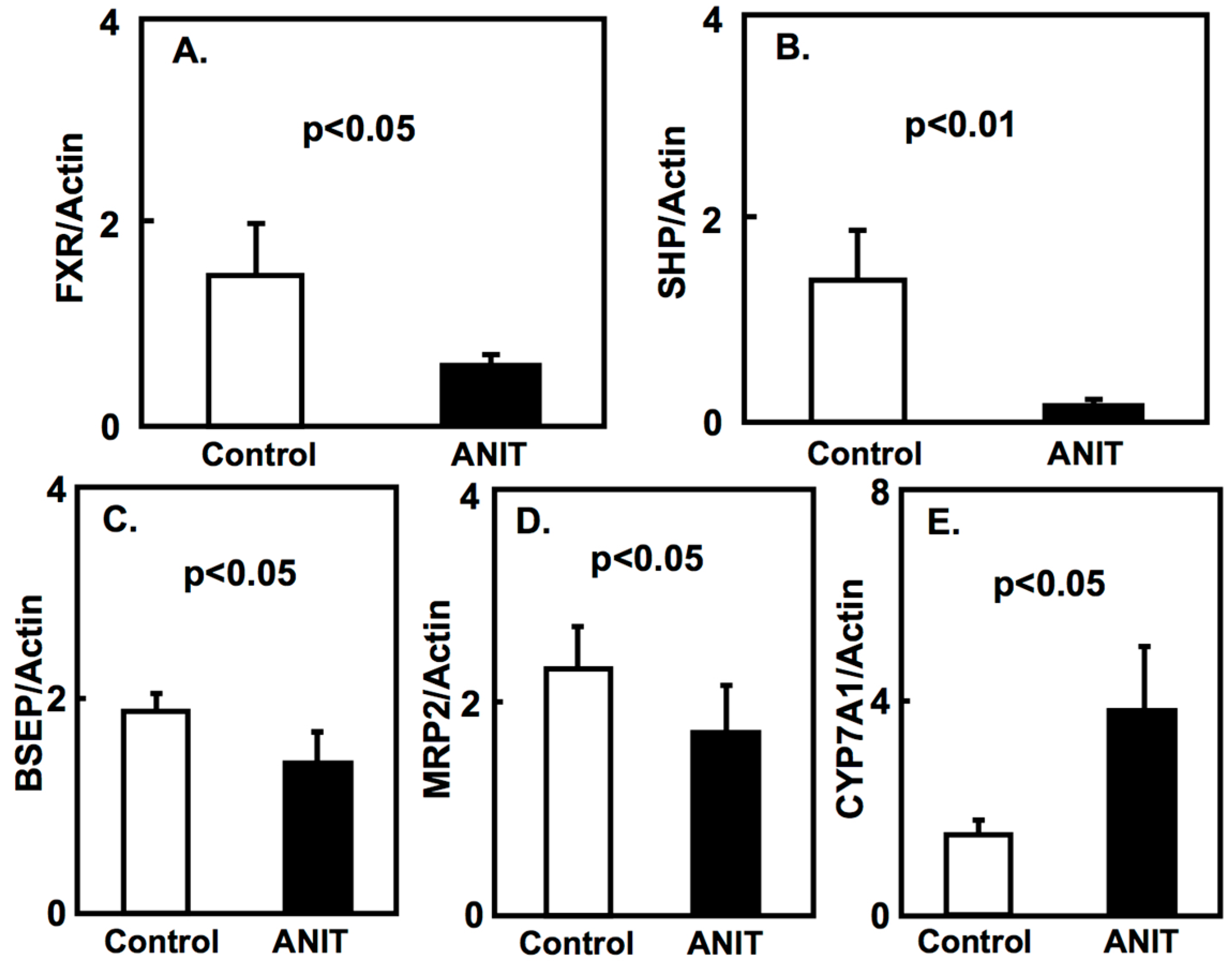
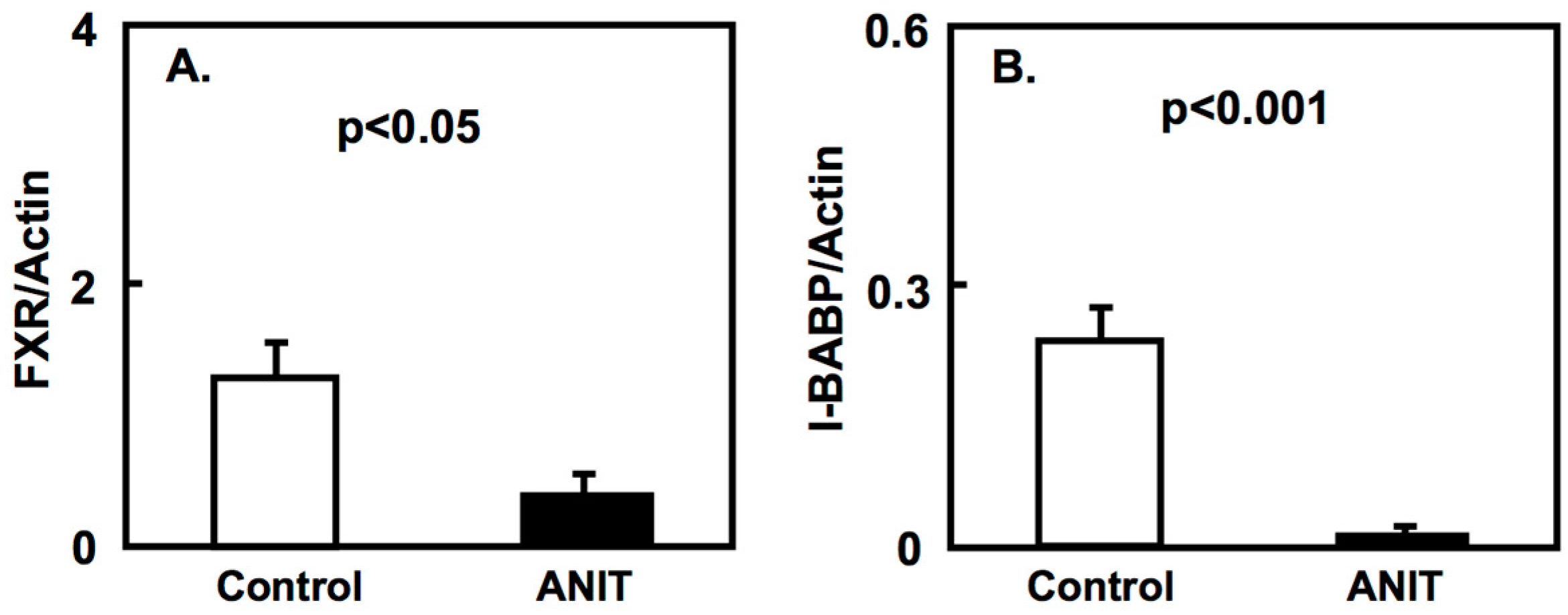
2.2. Gene Expression of Bile Acid-Metabolizing and Transporter Proteins
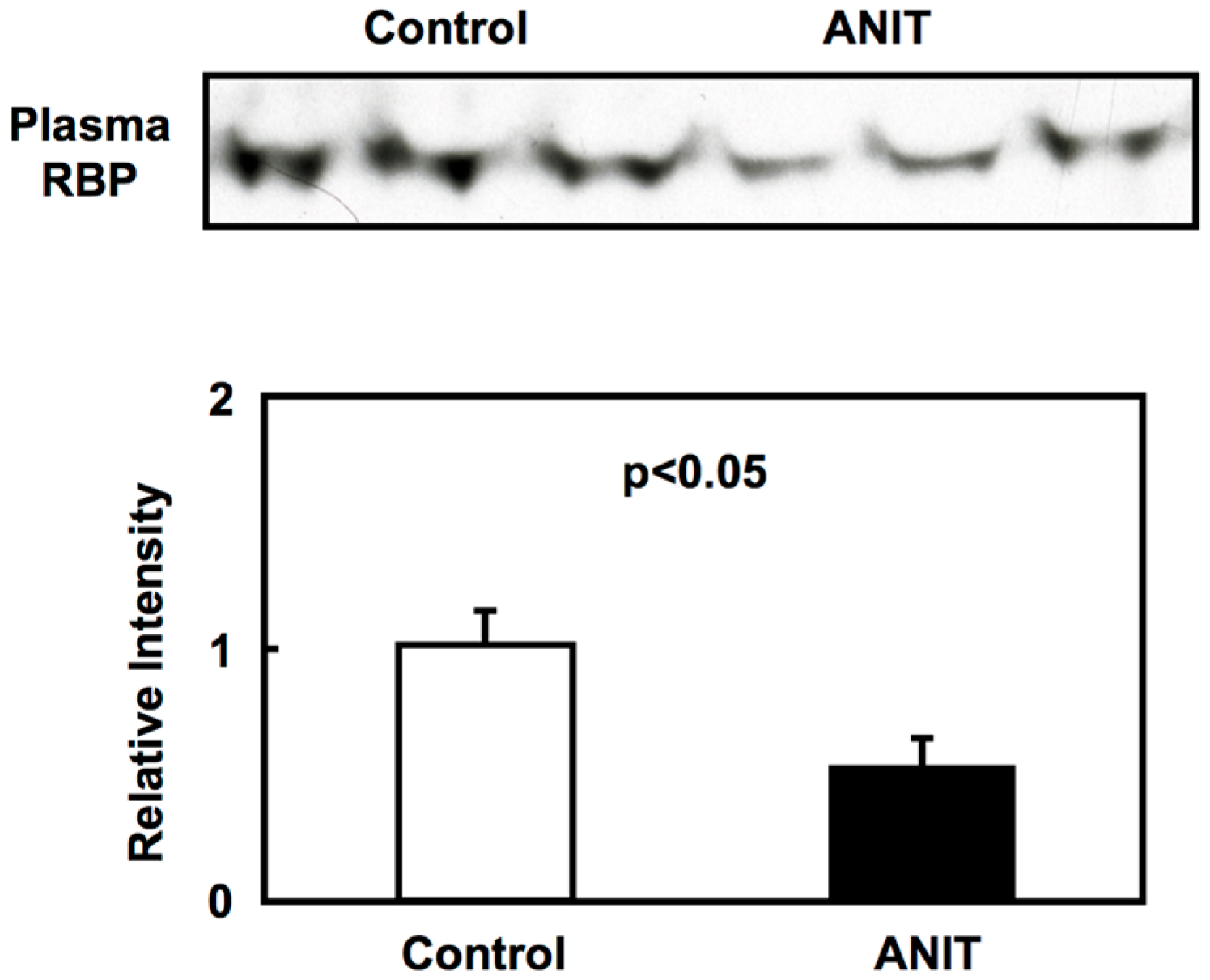
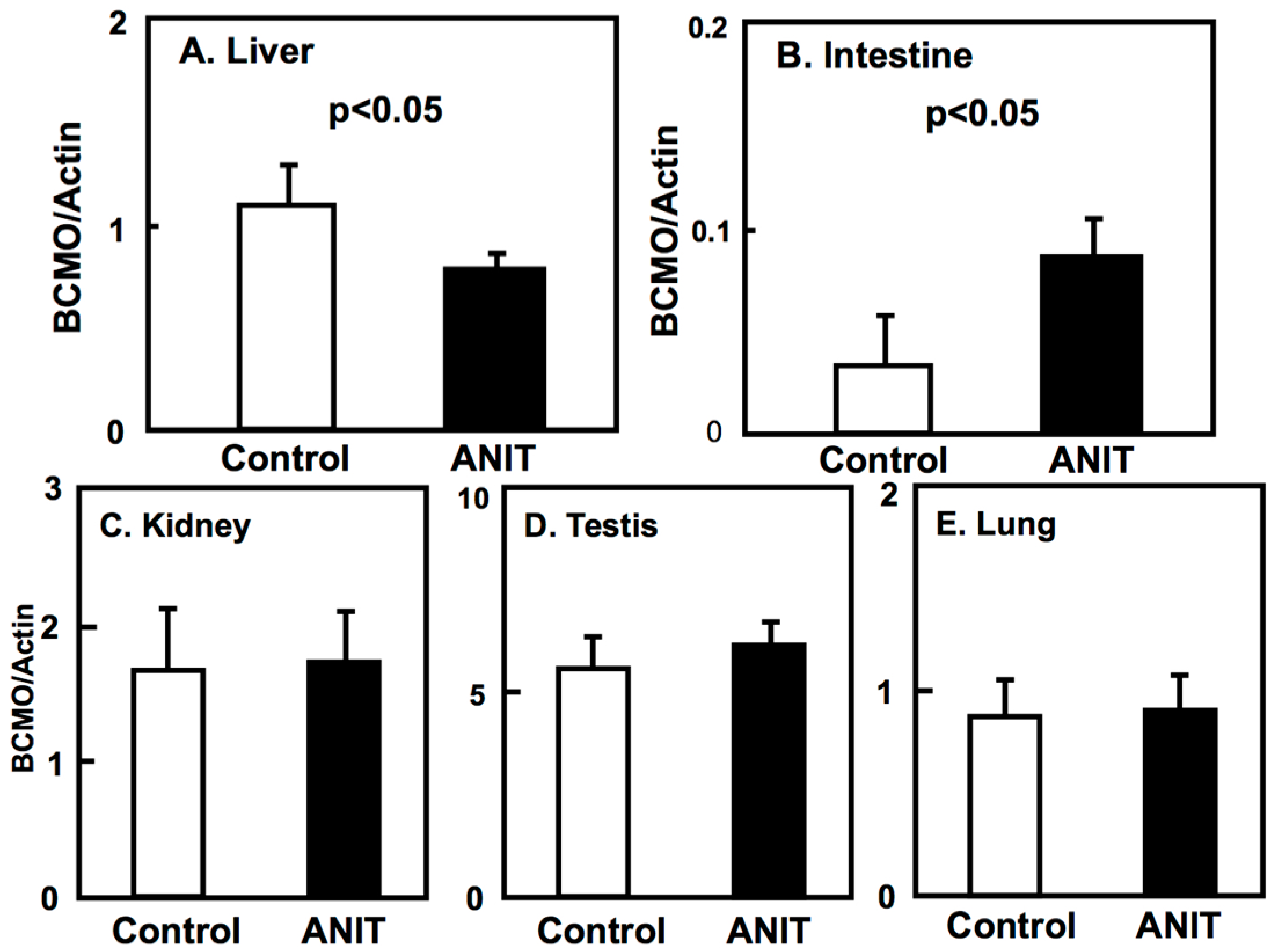
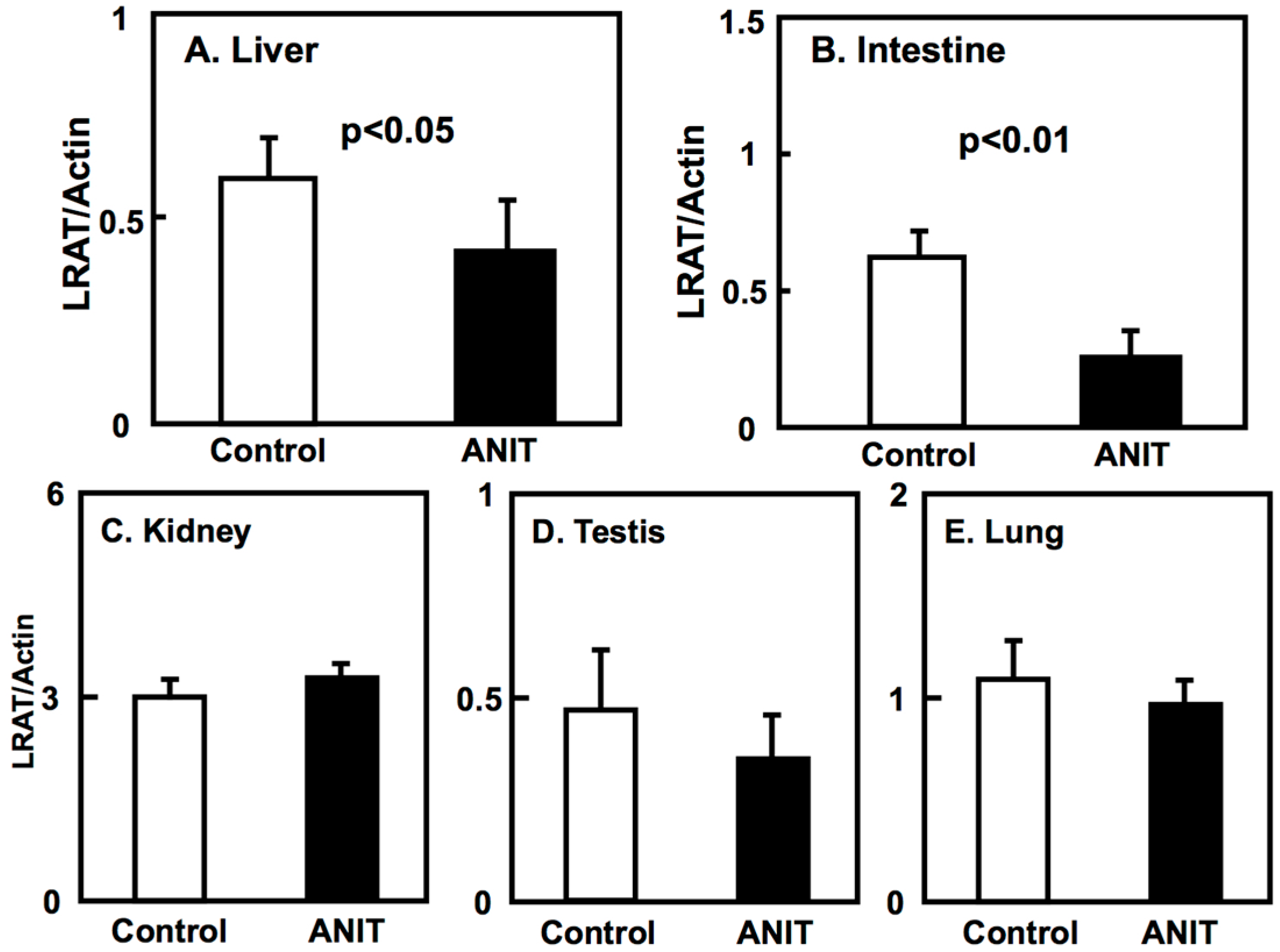
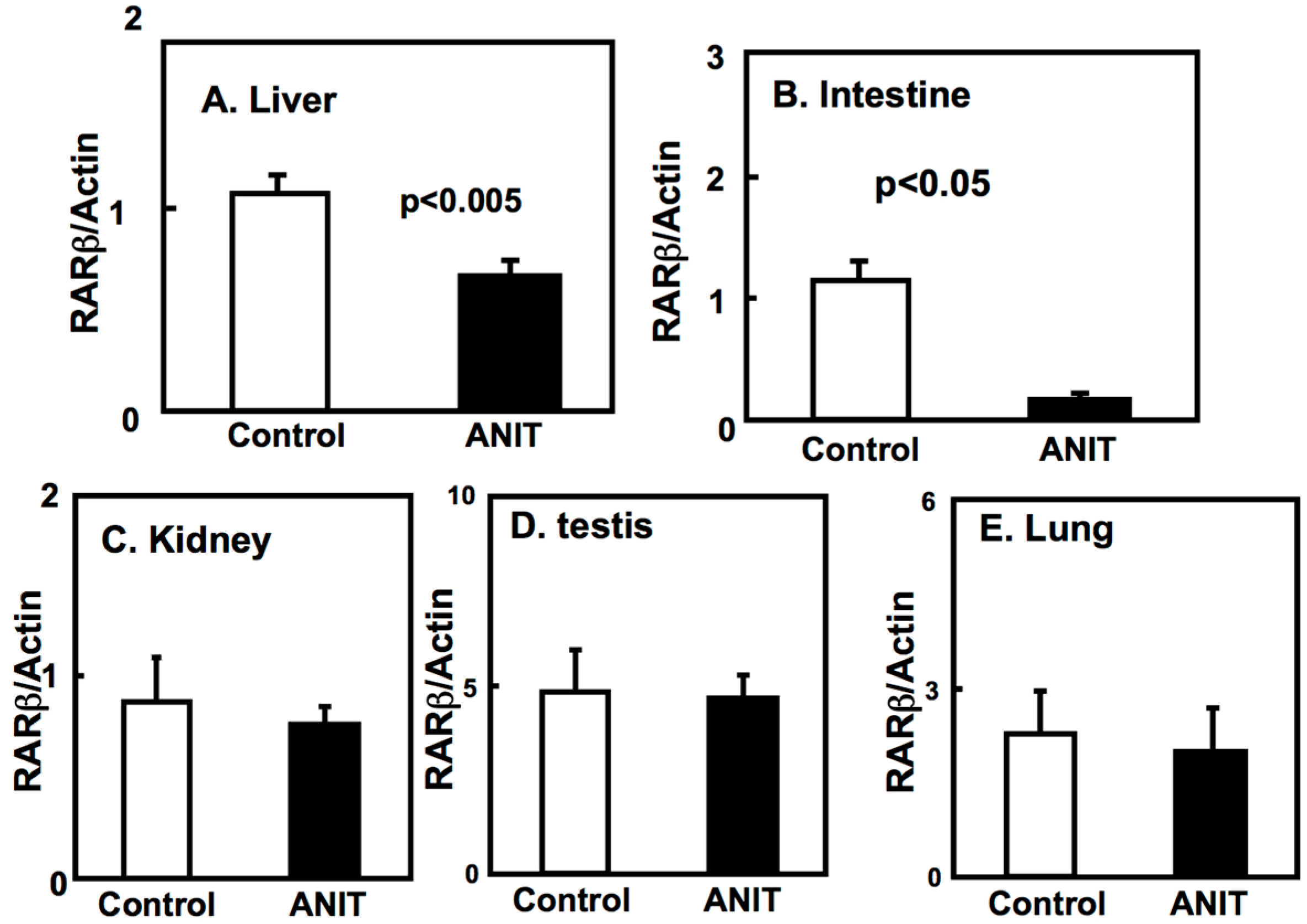
2.3. Gene Expression of BCMO, LRAT, and RARβ in Peripheral Tissues
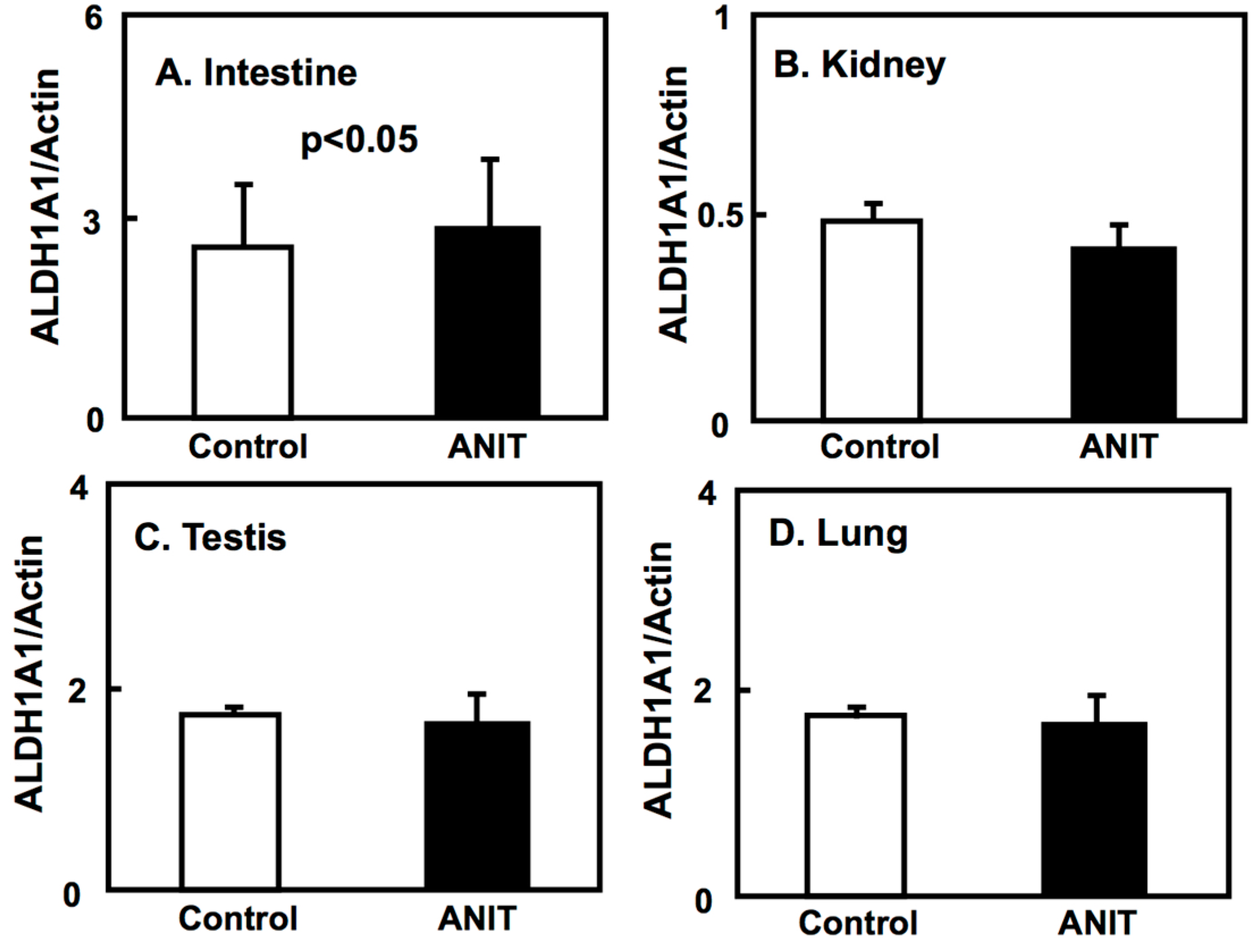
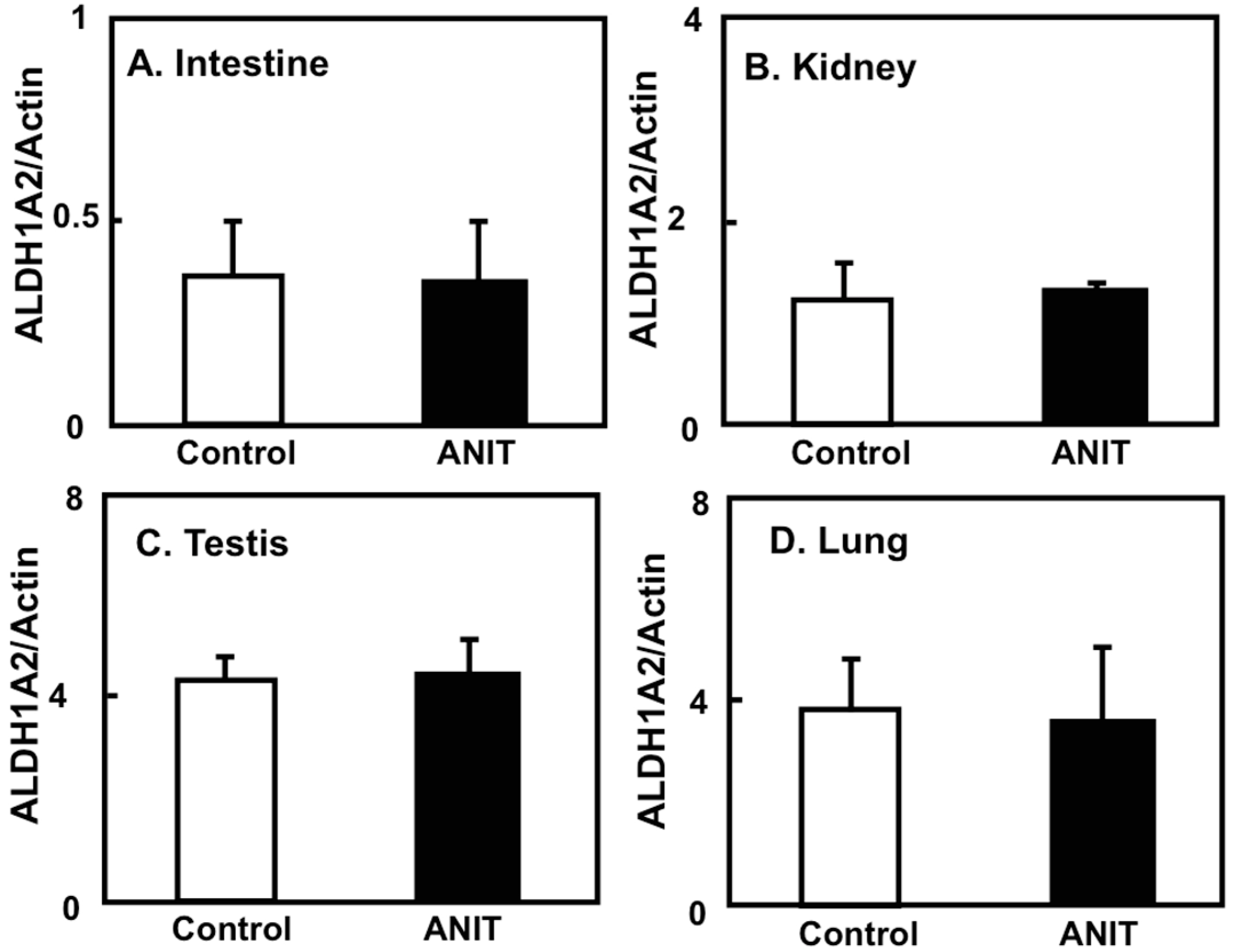
2.4. Gene Expression of CRBPI, ALDH1As, and CYP26A1 in the Liver, and ALDH1As Expression in Peripheral Tissues
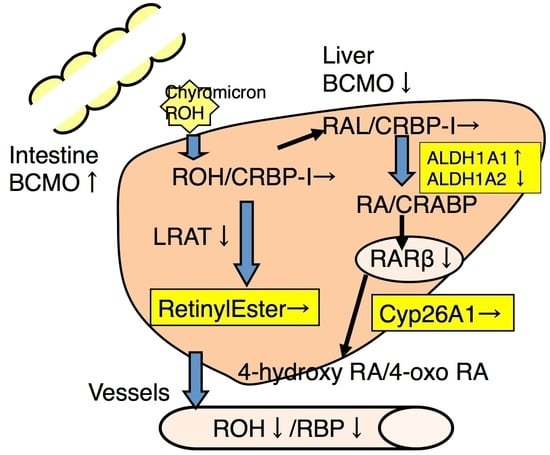
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Biochemical Data Analysis
4.3. Immunoblotting
4.4. Reverse Transcription and Quantitative Real-Time PCR
4.5. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALT | alanine aminotransferase |
| ALDH | aldehyde dehydrogenase |
| ANIT | α-naphthyl isothiocyanate |
| BCMO | β-carotene 15,15′ monooxygenase |
| BSEP | bile salt export pump |
| CRBP | cellular retinol binding protein |
| FXR | farnesoid X receptor |
| I-BABP | intestinal bile-acid binding protein |
| LRAT | lecithin-retinol acyltransferase |
| MRP2 | multidrug resistance protein-2 |
| NAFLD | non-alcoholic fatty liver disease |
| RARE | retinoic acid response element |
| RAR | retinoic acid receptor |
| RXR | retinoid X receptor |
| RBP | retinol binding protein |
| CYP7A1 | 7α-hydroxylase |
| SHP | small heterodimer partner |
References
- Barber, T.; Esteban-Pretel, G.; Marín, M.P.; Timoneda, J. Vitamin A deficiency: An overview. In Vitamin A and Carotenoids: Chemistry, Analysis, Function and Effects; Preedy, V.R., Ed.; RSC Publishing: Cambridge, UK, 2012; pp. 396–416. ISBN 978-1849733687. [Google Scholar]
- Biesalski, H.K.; Chichili, G.R.; Frank, J.; von Lintig, J.; Nohr, D. Conversion of β-carotene to retinal pigment. Vitam. Horm. 2007, 75, 117–130. [Google Scholar] [PubMed]
- Zollner, G.; Trauner, M. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br. J. Pharmacol. 2009, 156, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Loomes, K.M.; Emerick, K.M. Pediatric cholestatic liver disease with genetic etiology. In Pediatric Gastrointestinal and Liver Disease, 4th ed.; Wyllie, R., Hyams, J.S., Kay, M., Eds.; Elsevier Saunders: Philadelphia, PA, USA, 2011; pp. 752–766. ISBN 978-1437707748. [Google Scholar]
- Feranchak, A.P.; Frederick, J.; Suchy, F.J.; Sokol, R.J. Medical and nutritional management of cholestasis in infants and children. In Liver Disease in Children, 4th ed.; Frederick, J., Suchy, F.J., Sokol, R.J., Balistreri, W.F., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 111–139. ISBN 978-1107013797. [Google Scholar]
- Elias, E. Jaundice and cholestasis. In Sherlock’s Diseases of the Liver and Biliary System, 12th ed.; Dooley, J.S., Lok, A., Burroughs, A.K., Heathcote, J., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2011; pp. 234–256. ISBN 978-1405134897. [Google Scholar]
- Saeed, A.; Hoekstra, M.; Hoeke, M.O.; Heegsma, J.; Faber, K.N. The interrelationship between bile acid and vitamin A homeostasis. Biochim. Biophys. Acta 2017, 1862, 496–512. [Google Scholar] [CrossRef] [PubMed]
- Chazouillères, O.; Housset, C. Intrahepatic Cholestasis. In Textbook of Hepatology: From Basic Science Clinical Practice, 3rd ed.; Juan Rodés, J., Benhamou, J.-P., Blei, A.T., Reichen, J., Mario Rizzetto, M., Eds.; Blackwell Publishing Ltd.: Malden, MA, USA, 2007; pp. 1481–1500. ISBN 978-1405127417. [Google Scholar]
- Liu, Y.; Meyer, C.; Xu, C.; Weng, H.; Hellerbrand, C.; ten Dijke, P.; Dooley, S. Animal models of chronic liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G449–G468. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.A.; Dahm, L.J. Neutrophil- and glutathione-mediated hepatotoxicity of α-naphthylisothiocyanate. Drug. Metab. Rev. 1997, 29, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a nuclear receptor for bile acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.T.; Makishima, M.; Repa, J.J.; Schoonjans, K.; Kerr, T.A.; Auwerx, J.; Mangelsdorf, D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 2000, 6, 507–515. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.; Chen, X.; Huang, W. FXR. In Nuclear Receptors; Bunce, C.M., Campbell, M.J., Eds.; Springer: New York, NY, USA, 2010; pp. 307–326. ISBN 978-9048133024. [Google Scholar]
- Laudet, V.; Gronemeyer, H. FXR. In The Nuclear Receptor FactsBook; Laudet, V., Gronemeyer, H., Eds.; Academic Press: London, UK, 2002; pp. 199–203. ISBN 978-0124377356. [Google Scholar]
- Chen, H.; Huang, X.; Min, J.; Li, W.; Zhang, R.; Zhao, W.; Liu, C.; Yi, L.; Mi, S.; Wang, N.; et al. Geniposidic acid protected against ANIT-induced hepatotoxity and acute intrahepatic cholestasis, due to Fxr-mediated regulation of Bsep and Mrp2. J. Ethnopharmacol. 2016, 179, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Ou, Q.Q.; Qian, X.H.; Li, D.Y.; Zhang, Y.X.; Pei, X.N.; Chen, J.W.; Yu, L. Yinzhihuang attenuates ANIT-induced intrahepatic cholestasis in rats through upregulation of Mrp2 and Bsep expressions. Pediatr. Res. 2016, 79, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.Y.; Ai, G.; Zhang, X.J.; Xu, H.J.; Huang, Z.M. Investigations of the total flavonoids extracted from flowers of Abelmoschus manihot (L.) Medic against α-naphthylisothiocyanate- induced cholestatic liver injury in rats. J. Ethnopharmacol. 2015, 172, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Chen, X.L.; Wang, C.Y.; Liu, Q.; Sun, H.J.; Sun, P.Y.; Huo, X.K.; Liu, Z.H.; Yao, J.H.; Liu, K.X. Alisol B 23-acetate protects against ANIT-induced hepatotoxity and cholestasis, due to FXR-mediated regulation of transporters and enzymes involved in bile acid homeostasis. Toxicol. Appl. Pharmacol. 2015, 283, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.J.; Aleksunes, L.M.; Tanaka, Y.; Goedken, M.J.; Klaassen, C.D. Compensatory induction of liver efflux transporters in response to ANIT-induced liver injury is impaired in FXR-null mice. Toxicol. Sci. 2009, 110, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Freund, C.; Gotthardt, D.N. Vitamin A deficiency in chronic cholestatic liver disease: Is vitamin A therapy beneficial? Liver Int. 2017, 37, 1752–1758. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.R.; Angulo, P.; Petterson, T.; Lindor, K.D. Fat-soluble vitamin levels in patients with primary biliary cirrhosis. Am. J. Gastroenterol. 2001, 96, 2745–2750. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Mennone, A.; Boyer, J.L.; Cai, S.Y. Combination of retinoic acid and ursodeoxycholic acid attenuates liver injury in bile duct-ligated rats and human hepatic cells. Hepatology 2011, 53, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Assis, D.N.; Abdelghany, O.; Cai, S.Y.; Gossard, A.A.; Eaton, J.E.; Keach, J.C.; Deng, Y.; Setchell, K.D.; Ciarleglio, M.; Lindor, K.D.; et al. Combination Therapy of all-trans retinoic acid with ursodeoxycholic acid in patients with primary sclerosing cholangitis: A human pilot study. J. Clin. Gastroenterol. 2017, 51, e11–e16. [Google Scholar] [CrossRef] [PubMed]
- Lietz, G.; Lange, J.; Rimbach, G. Molecular and dietary regulation of β, β-carotene 15,15′-monooxygenase 1 (BCMO1). Arch. Biochem. Biophys. 2010, 502, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lobo, G.P.; Amengual, J.; Palczewski, G.; Babino, D.; von Lintig, J. Mammalian carotenoid-oxygenases: Key players for carotenoid function and homeostasis. Biochim. Biophys. Acta 2012, 1821, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Zhu, C.L.; Inoue, A.; Tamai, H. Molecular cloning of the rat β-carotene 15,15′-monooxygenase gene and its regulation by retinoic acid. Eur. J. Nutr. 2006, 45, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Miyazaki, H.; Takaya, R.; Tamai, H. Expression of β-carotene 15,15′-monooxygenase gene and retinol status in type 2 diabetic Goto-Kakizaki rats. BioFactors 2008, 33, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Matsumura, H.; Nakakura, H.; Ashida, A.; Tamai, H. Expression of β-carotene 15,15′- monooxygenase gene and retinol status in rats with puromycin aminonucleoside- induced nephrosis. BioFactors 2008, 33, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Miyazaki, H.; Fukunishi, S.; Takaya, R.; Yoden, A.; Higuchi, K.; Tamai, H. Altered expression of both β-carotene 15,15′ monooxygenase and lecithin:retinol acyltransferase in obese Zucker rats. J. Nutr. Sci. Vitaminol. 2011, 57, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, H.; Takitani, K.; Koh, M.; Inoue, A.; Kishi, K.; Tamai, H. Retinol status and expression of retinol-related proteins in methionine-choline deficient rats. J. Nutr. Sci. Vitaminol. 2014, 60, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Inoue, K.; Koh, M.; Miyazaki, H.; Inoue, A.; Kishi, K.; Tamai, H. Altered retinol status and expression of retinol-related proteins in streptozotocin-induced type 1 diabetic model rats. J. Clin. Biochem. Nutr. 2015, 56, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Takitani, K.; Miyazaki, H.; Koh, M.; Kishi, K.; Inoue, A.; Tamai, H. Dehydroepiandrosterone alters retinol status and expression of the β-carotene 15, 15′- monooxygenase and lecithin: Retinol acyltransferase genes. J. Nutr. Sci. Vitaminol. 2016, 62, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Ray, W.J.; Bain, G.; Yao, M.; Gottlieb, D.I. CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J. Biol. Chem. 1997, 272, 18702–18708. [Google Scholar] [CrossRef] [PubMed]
- White, J.A.; Beckett-Jones, B.; Guo, Y.D.; Dilworth, F.J.; Bonasoro, J.; Jones, G.; Petkovich, M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J. Biol. Chem. 1997, 272, 18538–18541. [Google Scholar] [CrossRef] [PubMed]
- Loudig, O.; Babichuk, C.; White, J.; Abu-Abed, S.; Mueller, C.; Petkovich, M. Cytochrome P450RAI (CYP26) promoter: A distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 2000, 14, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Husmann, M.; Hoffmann, B.; Stump, D.G.; Chytil, F.; Pfahl, M. A retinoic acid response element from the rat CRBPI promoter is activated by an RAR/RXR heterodimer. Biochem. Biophys. Res. Commun. 1992, 187, 1558–1564. [Google Scholar] [CrossRef]
- de Thé, H.; Vivanco-Ruiz, M.M.; Tiollais, P.; Stunnenberg, H.; Dejean, A. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature 1990, 343, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.C.; Zolfaghari, R. Regulation of hepatic retinol metabolism: Perspectives from studies on vitamin A. status. J. Nutr. 2004, 134, 269S–275S. [Google Scholar] [CrossRef] [PubMed]
- Zolfaghari, R.; Ross, A.C. An essential set of basic DNA response elements is required for receptor- dependent transcription of the lecithin:retinol acyltransferase (Lrat) gene. Arch. Biochem. Biophys. 2009, 489, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sandell, L.L.; Trainor, P.A.; Koentgen, F.; Duester, G. Alcohol and aldehyde dehydrogenases: Retinoid metabolic effects in mouse knockout models. Biochim. Biophys. Acta 2012, 1821, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Duester, G.; Mic, F.A.; Molotkov, A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem. Biol. Interact. 2003, 143–144, 201–210. [Google Scholar] [CrossRef]
- Petrosino, J.M.; Disilvestro, D.; Ziouzenkova, O. Aldehyde dehydrogenase 1A1: Friend or foe to female metabolism? Nutrients 2014, 6, 950–973. [Google Scholar] [CrossRef] [PubMed]
- Modica, S.; Gadaleta, R.M.; Moschetta, A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl. Recept. Signal. 2010, 8, e005. [Google Scholar] [CrossRef] [PubMed]
- Jonker, J.W.; Liddle, C.; Downes, M. FXR and PXR: Potential therapeutic targets in cholestasis. J. Steroid Biochem. Mol. Biol. 2012, 130, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.Y.; Mennone, A.; Soroka, C.J.; Boyer, J.L. All-trans-retinoic acid improves cholestasis in α-naphthylisothiocyanate-treated rats and Mdr2-/- mice. J. Pharmacol. Exp. Ther. 2014, 349, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Wang, G.; Qu, J.; Wang, X.; Li, K. 9-cis-retinoic acid elevates MRP3 expression by inhibiting sumoylation of RXRα to alleviate cholestatic liver injury. Biochem. Biophys. Res. Commun. 2018, 503, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Ohata, M.; Lin, M.; Satre, M.; Tsukamoto, H. Diminished retinoic acid signaling in hepatic stellate cells in cholestatic liver fibrosis. Am. J. Physiol. 1997, 272, G589–G596. [Google Scholar] [CrossRef] [PubMed]
- Denson, L.A.; Bohan, A.; Held, M.A.; Boyer, J.L. Organ-specific alterations in RARα:RXRα abundance regulate rat Mrp2 (Abcc2) expression in obstructive cholestasis. Gastroenterology 2002, 123, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.; Barshack, I.; Onaca, N.; Goldberg, I.; Berkovich, Z.; Melzer, E.; Jonas, A.; Reifen, R. Vitamin A deficiency associated with enhanced proliferation of bile duct epithelial cells in the rat. Isr. Med. Assoc. J. 2010, 12, 82–86. [Google Scholar] [PubMed]
- Bieri, J.G.; Tolliver, T.J.; Catignani, G.L. Simultaneous determination of α-tocopherol and retinol in plasma or red cells by high pressure liquid chromatography. Am. J. Clin. Nutr. 1979, 32, 2143–2149. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Control | ANIT | |
|---|---|---|
| (n = 5) | (n = 5) | |
| Total cholesterol (mg/dL) | 66.8 ± 3.2 | 218.8 ± 18.1 *** |
| Total Bilirubin (mg/dL) | 0.04 ± 0.01 | 0.53 ± 0.3 * |
| ALT (U/L) | 35.8 ± 3.3 | 376.2 ± 180.4 * |
| Retinol (plasma) (µg/dL) | 50.8 ± 4.6 | 35.6 ± 3.4 *** |
| Retinol (liver) (µg/mg protein) | 0.17 ± 0.02 | 0.15 ± 0.02 |
| Gene (Accession Number) | Forward | Reverse | Length (bp) |
|---|---|---|---|
| ALDH1A1 (BC061526) | 5′-ATGGTCTAGCAGCAGGAGT-3′ | 5′-CCAGACATCTTGAATCCACCGAA-3′ | 142 |
| ALDH1A2 (NM_053896) | 5′-TTGCCTCACAACAAGTGAGC-3′ | 5-ACAAAATGGGGTTCATTGGA-3' | 125 |
| BCMO (NM_053648) | 5′-CAAGTCCTCCTTAAAGTGGT-3′ | 5′-AATAAACCATGCAGGTCCA-3′ | 225 |
| 5′-CATCATCTCTACAGATCCCCAAAAGC-3′-FITC | |||
| LC-5′-GCCCTTTTTACTTATTCTGGATGCG-3′ | |||
| BSEP (NM_031760) | 5′-GCCATTGTGCGAGATCCTAAA-3′ | 5′-TGCAGGTCCGACCCTCTCT-3′ | 118 |
| β-actin (V01217 J00691) | 5′-CCTGTATGC CTC TGG TCG TA-3′ | 5′-CCATCTCTTGCTCGAAGTCT-3′ | 260 |
| 5′-CGGGACCTGACAGACTACCTCATG-3′-FITC | |||
| LC-5′-AGATCCTGACCGAGCGTGGCTAC-3′ | |||
| CYP7A1 (NM_012942) | 5′-GCTTTACAGAGTGCTGGCCAA-3′ | 5′-CTGTCTAGTACCGGCAGGTCATT-3′ | 92 |
| CYP26A1 (DQ266888) | 5′-GTGCCAGTGATTGCTGAAGA-3′ | 5'-AGAGAAGAGATTGCGGGTCA-3′ | 213 |
| FXR (U18374) | 5′-GTGACAAAGAAGCCGCGAAT-3′ | 5′-GCAGGTGAGCGCGTTGTAAT-3′ | 114 |
| I-BABP (L22788) | 5′-CACTATGGCCTTCACCGGCAAATA-3′ | 5-ACCCTCCATCTTCACGGTTGCCTT-3′ | 268 |
| LRAT (AF255060) | 5′-GGAACAACTGCGAACACT-3′ | 5′-ACACTAATCCCAAGACAGC-3′ | 141 |
| MRP2 (NM_012833) | 5′-CTGGTGTGGATTCCCTTGG-3′ | 5′-CAAAACCAGGAGCCATGTGC-3′ | 252 |
| RARβ (NM_031529) | 5′-ATACCCCAGAGCAAGACACC-3′ | 5′-AGCAGATGGCACTGAGAAGA-3′ | 170 |
| SHP (D86839) | 5′-CTCGGTTTGCATACAGTGTTTGAC-3′ | 5′-GCATATTGGCCTGGAGGTTTT-3′ | 75 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takitani, K.; Kishi, K.; Miyazaki, H.; Koh, M.; Tamaki, H.; Inoue, A.; Tamai, H. Altered Expression of Retinol Metabolism-Related Genes in an ANIT-Induced Cholestasis Rat Model. Int. J. Mol. Sci. 2018, 19, 3337. https://doi.org/10.3390/ijms19113337
Takitani K, Kishi K, Miyazaki H, Koh M, Tamaki H, Inoue A, Tamai H. Altered Expression of Retinol Metabolism-Related Genes in an ANIT-Induced Cholestasis Rat Model. International Journal of Molecular Sciences. 2018; 19(11):3337. https://doi.org/10.3390/ijms19113337
Chicago/Turabian StyleTakitani, Kimitaka, Kanta Kishi, Hiroshi Miyazaki, Maki Koh, Hirofumi Tamaki, Akiko Inoue, and Hiroshi Tamai. 2018. "Altered Expression of Retinol Metabolism-Related Genes in an ANIT-Induced Cholestasis Rat Model" International Journal of Molecular Sciences 19, no. 11: 3337. https://doi.org/10.3390/ijms19113337
APA StyleTakitani, K., Kishi, K., Miyazaki, H., Koh, M., Tamaki, H., Inoue, A., & Tamai, H. (2018). Altered Expression of Retinol Metabolism-Related Genes in an ANIT-Induced Cholestasis Rat Model. International Journal of Molecular Sciences, 19(11), 3337. https://doi.org/10.3390/ijms19113337