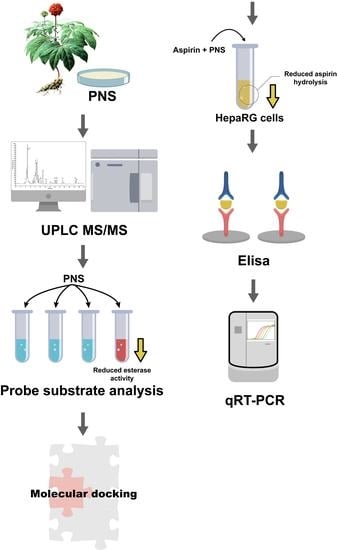
Effects of Panax Notoginseng Saponins on Esterases Responsible for Aspirin Hydrolysis In Vitro
Abstract
1. Introduction
2. Results
2.1. UPLC/LTQ-Orbitrap MS/MS Analysis
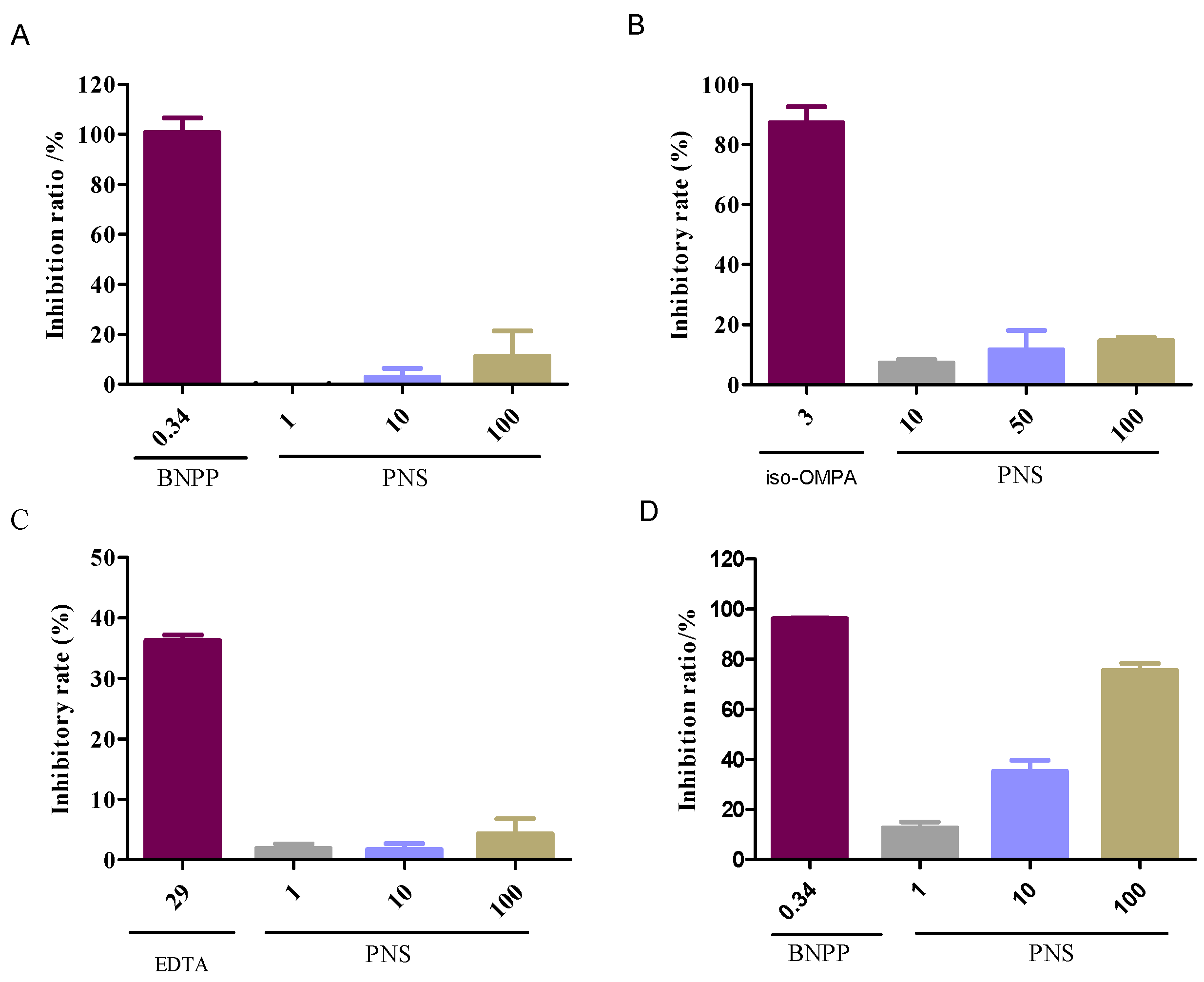
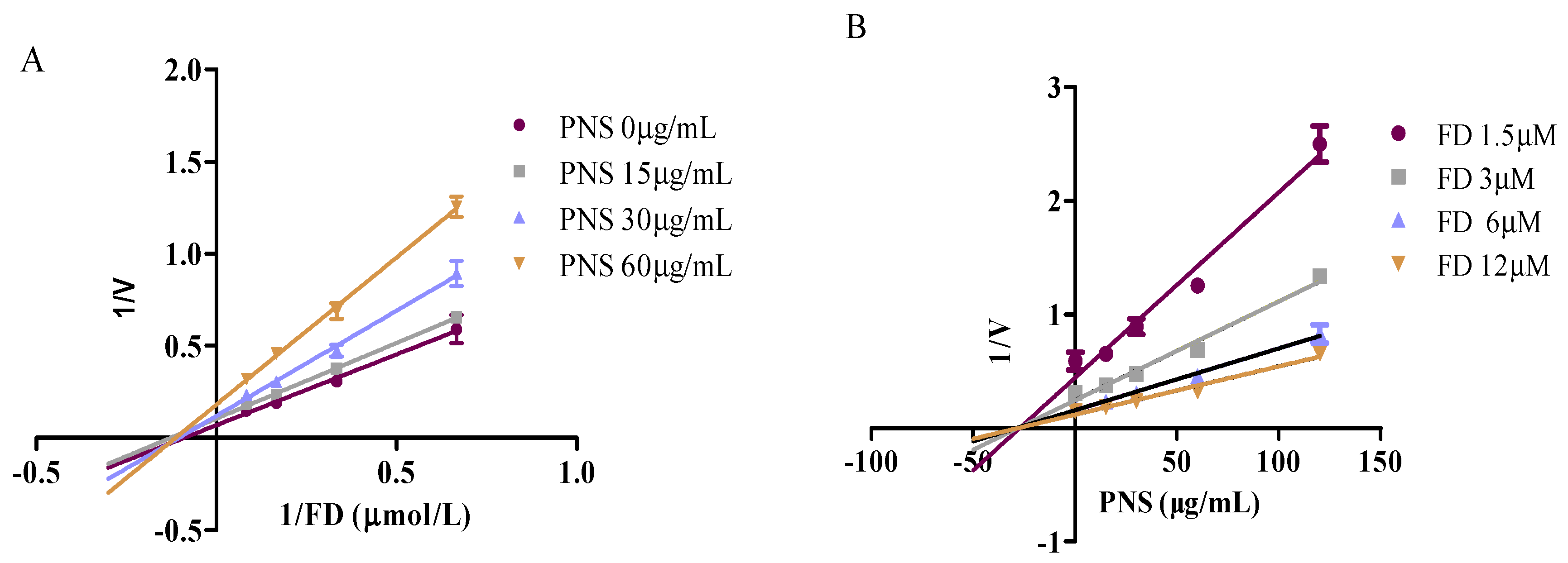
2.2. Enzyme-Specific Probe Substrate Analysis
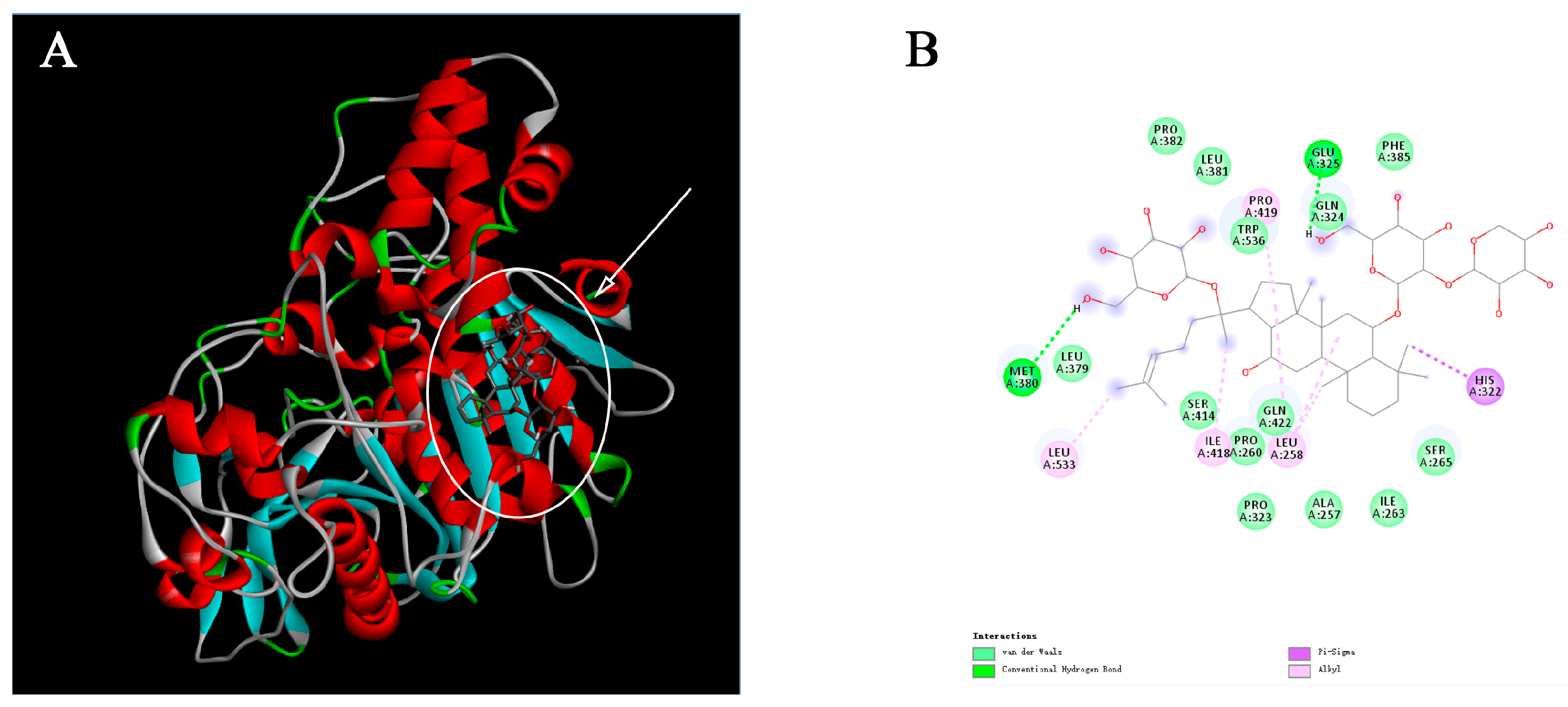
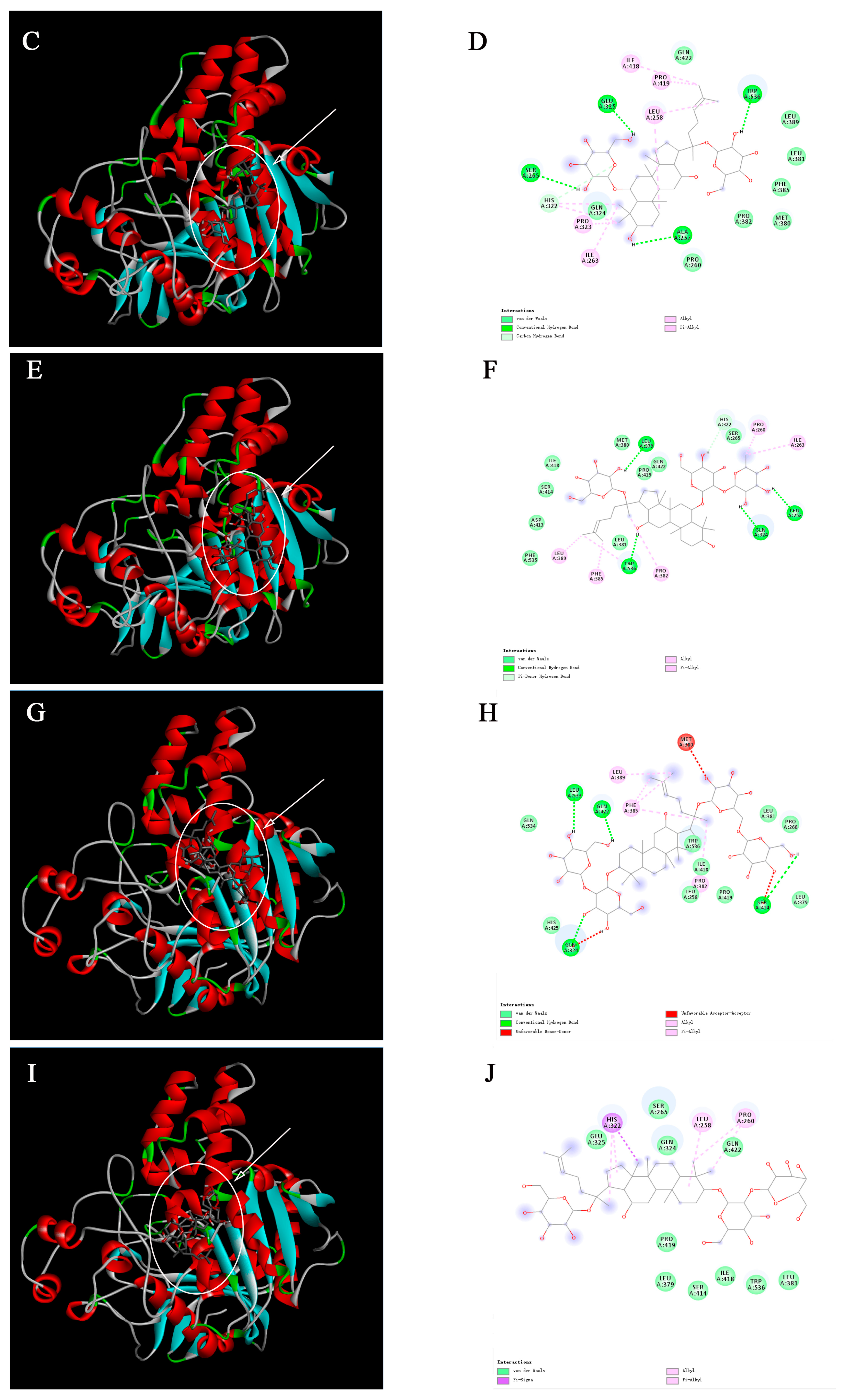
2.3. Molecular Docking Analysis
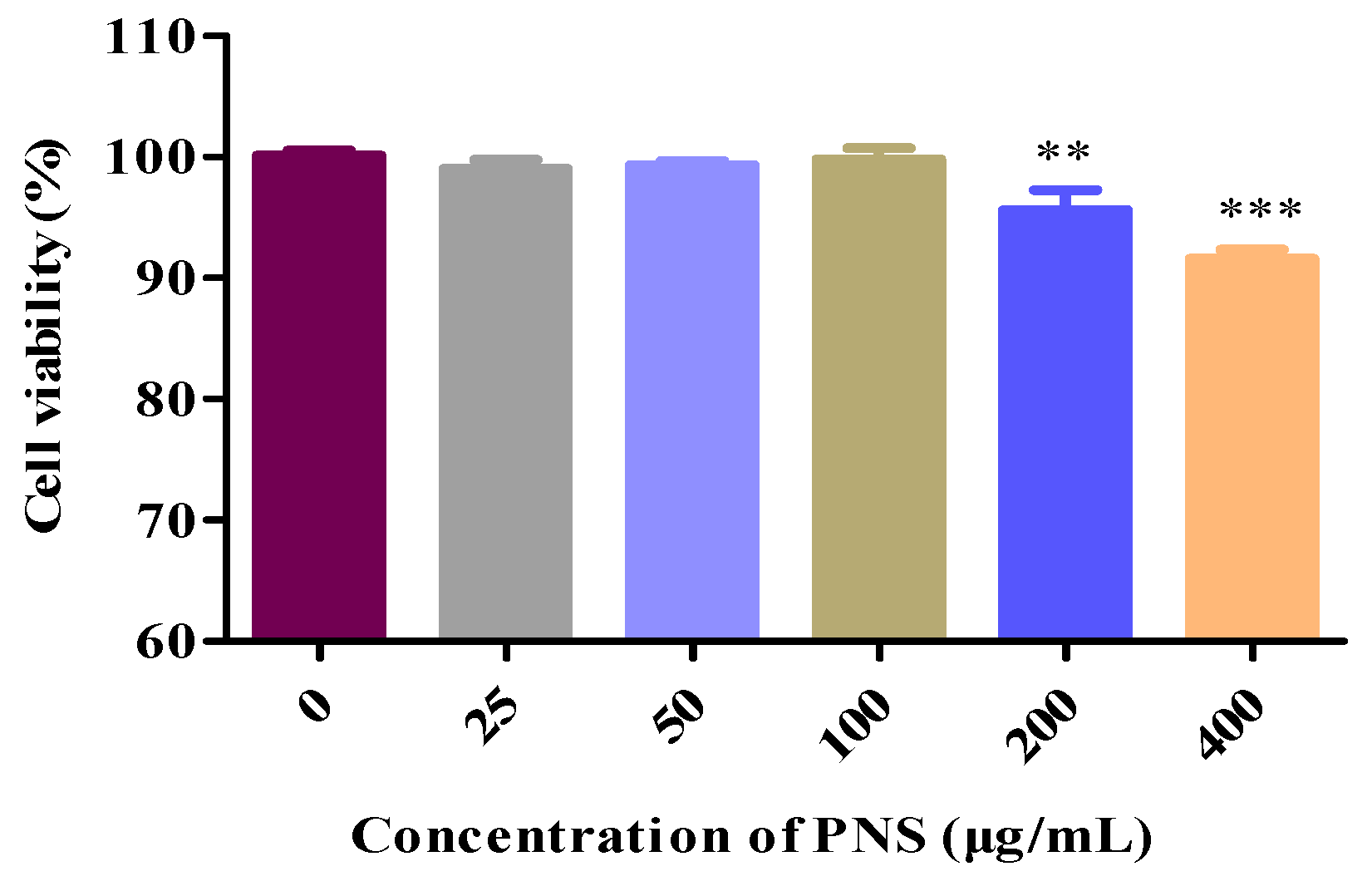
2.4. Cytotoxicology of PNS in HepaRG Cells
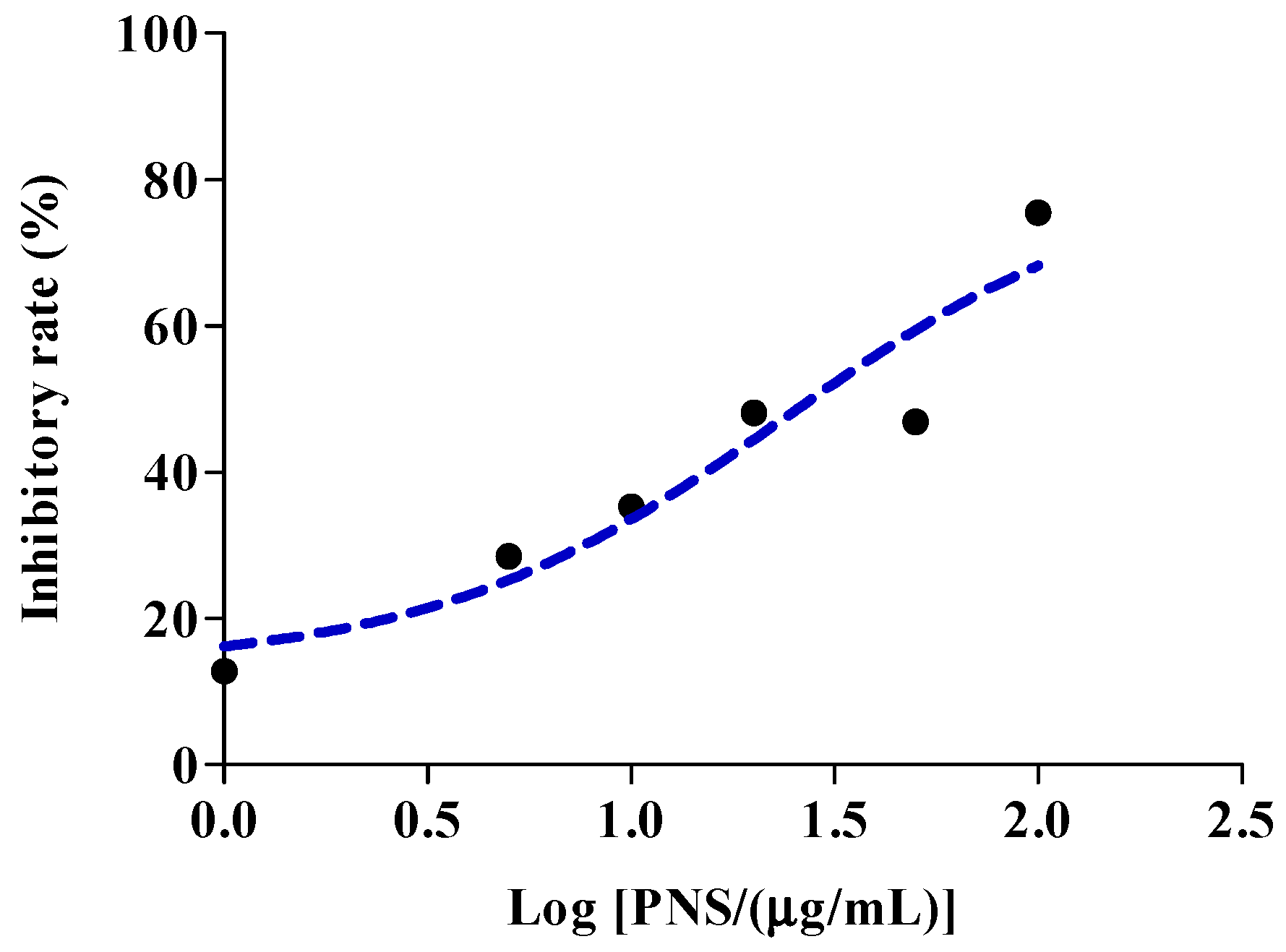
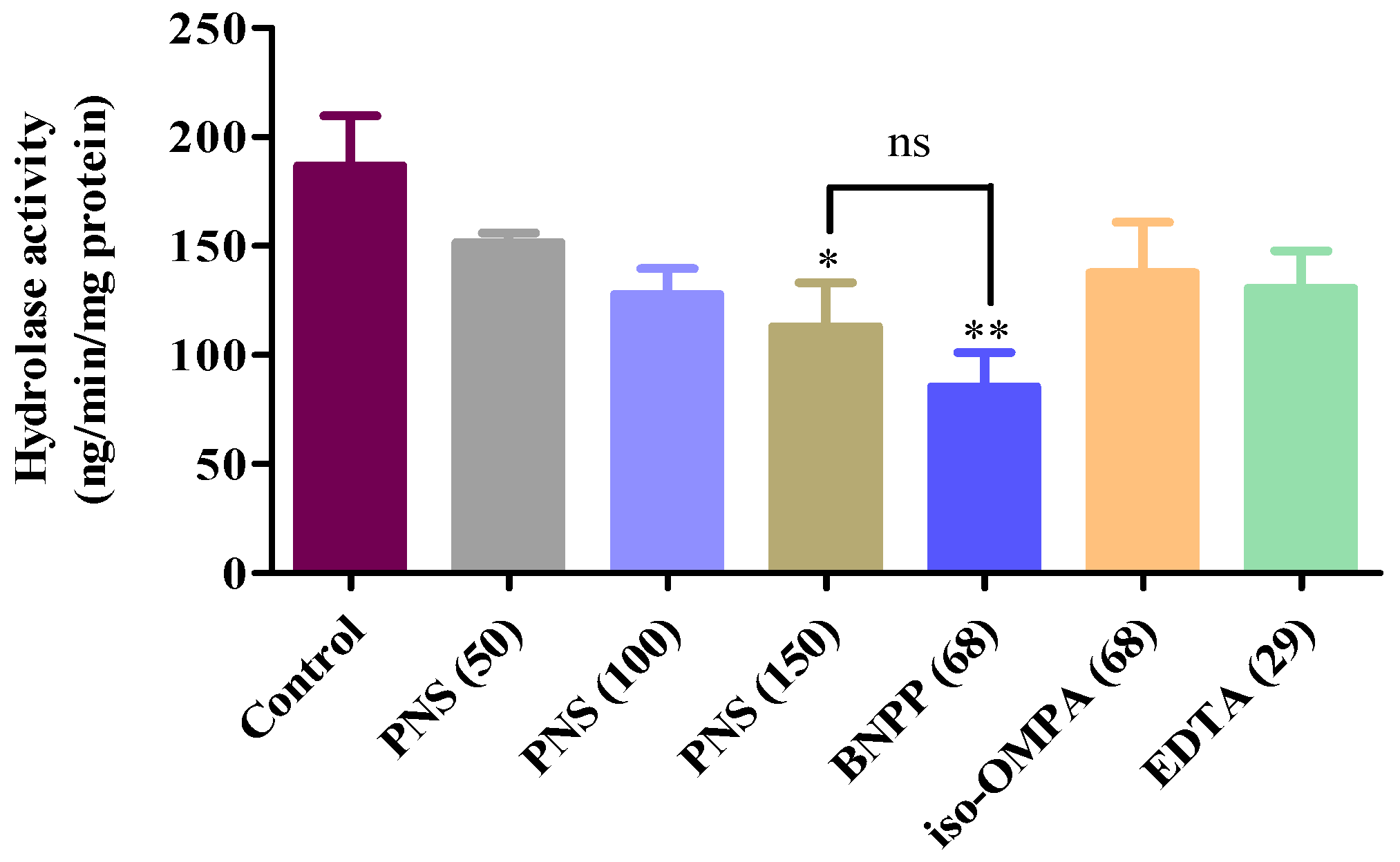
2.5. Aspirin Hydrolysis after PNS Treatment
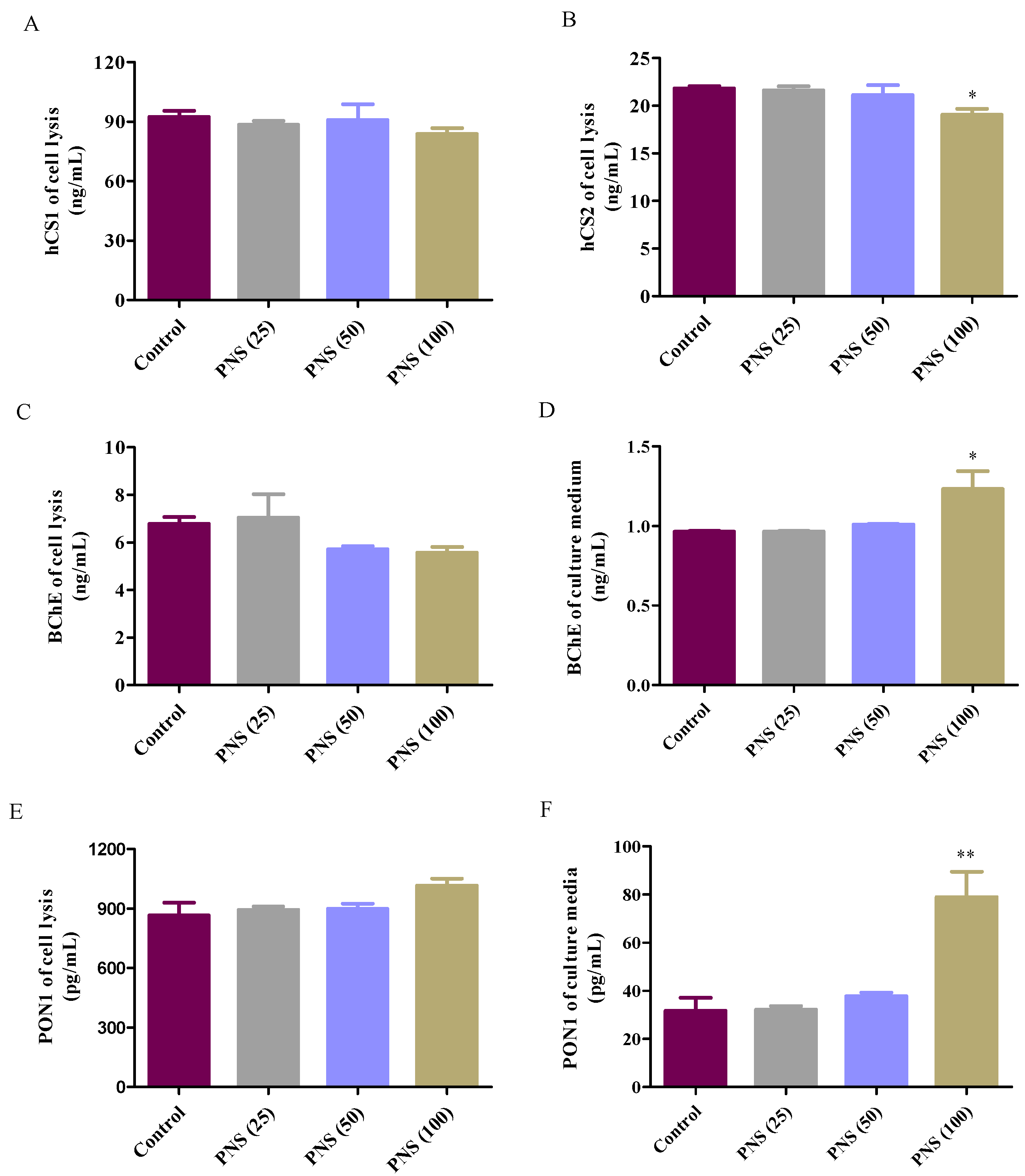
2.6. ELISA Analysis for Esterases
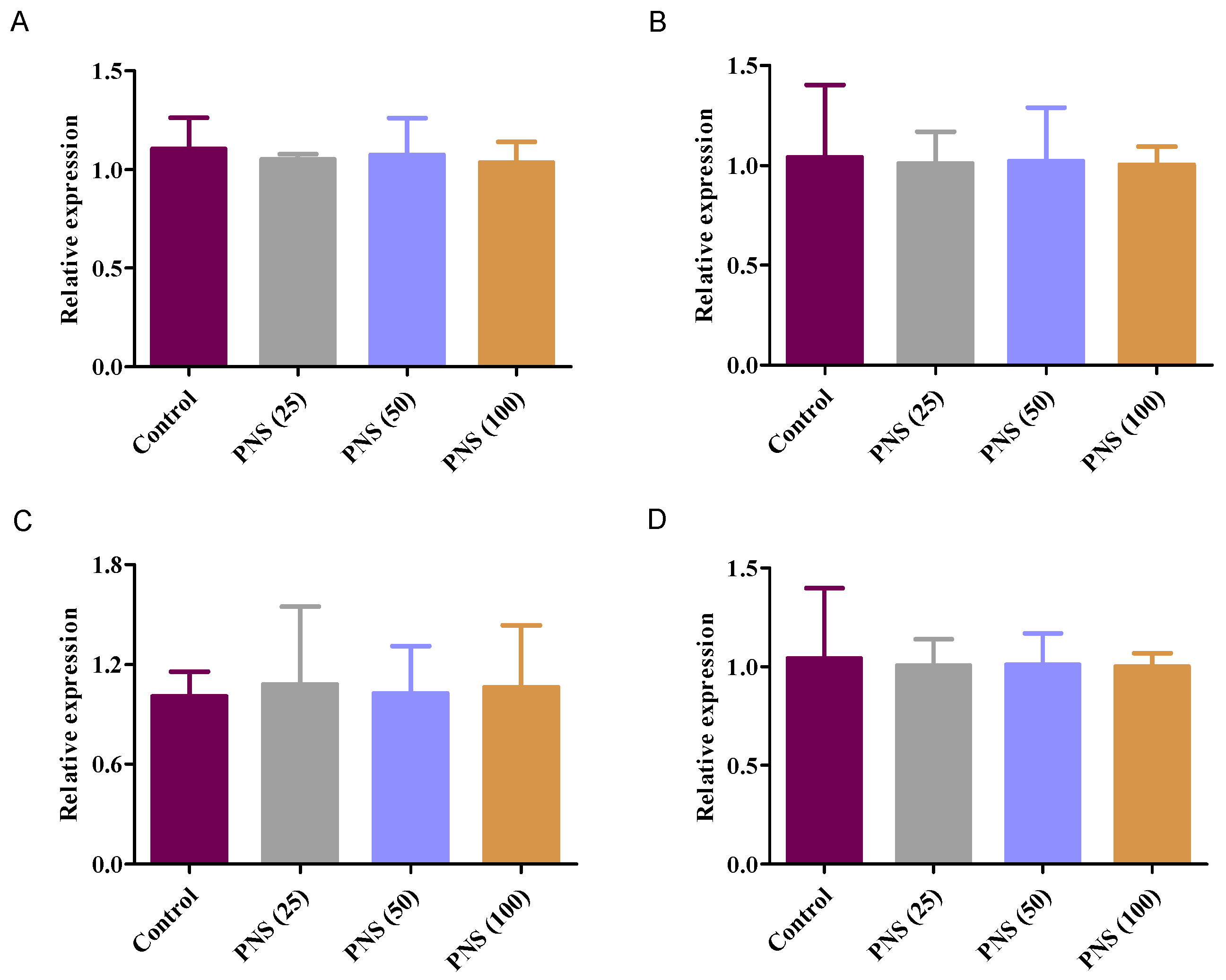
2.7. qRT-PCR Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. UPLC/LTQ-Orbitrap MS/MS Analysis
4.3. Enzyme-Specific Probe Substrate Analysis
4.3.1. Enzyme Inhibition Experiments
4.3.2. Inhibitory Kinetics Evaluation and In Vitro-In Vivo Extrapolation
4.4. Molecular Docking Analysis
4.5. Cell Culture
4.6. Cell Viability Assay
4.7. Hydrolysis Experiments
4.8. ELISA Analysis for Esterases
4.9. qRT-PCR Analysis
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Choi, Y.H.; Chin, Y.; Kim, Y.G. Herb-drug interactions: Focus on metabolic enzymes and transporters. Arch. Pharm. Res. 2011, 34, 1843–1863. [Google Scholar] [CrossRef] [PubMed]
- Brantley, S.J.; Argikar, A.A.; Lin, Y.S.; Nagar, S.; Paine, M.F. Herb-drug interactions: Challenges and opportunities for improved predictions. Drug Metab. Dispos. 2013, 12, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Q.; Ren, T.; Zhang, Y.; Lam, C.W.K.; Chow, M.S.S.; Zuo, Z. Non-linear pharmacokinetics of piperine and its herb-drug interactions with docetaxel in Sprague-Dawley rats. J. Pharm. Biomed. Anal. 2016, 128, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Picking, D.; Lamm, A.; McKenzie, J.; Hartley, S.; Watson, C.; Williams, L.; Lowe, H.; Delgoda, R. Significant inhibitory impact of dibenzyl trisulfide and extracts of Petiveria alliacea on the activities of major drug-metabolizing enzymes in vitro: An assessment of the potential for medicinal plant-drug interactions. Fitoterapia 2016, 111, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Aninat, C.; Piton, A.; Glaise, D.; Le Charpentier, T.; Langouet, S.; Morel, F.; Guguen-Guillouzo, C.; Guillouzo, A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006, 34, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Kanebratt, K.P.; Andersson, T.B. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab. Dispos. 2008, 36, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.N.; Ye, L.; Nakamoto, K.; Subileau, E.; Steen, D.; Zhong, X. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab. Dispos. 2010, 38, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.B.; Kanebratt, K.P.; Kenna, J.G. The HepaRG cell line: A unique in vitro tool for understanding drug metabolism and toxicology in human. Expert. Opin. Drug. Metab. Toxicol. 2012, 8, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhu, L.; Zou, Y.; Liu, W.; Zhang, X.; Wei, X.; Hu, B.; Chen, J. Panax notoginseng saponins promotes stroke recovery by influencing expression of Nogo-A, NgR and p75NGF, in vitro and in vivo. Biol. Pharm. Bull. 2014, 37, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Lee, S.M.; Wang, J.D.; Wang, N.; He, C.W.; Wang, Y.T.; Kang, J.X. Panax notoginseng reduces atherosclerotic lesions in apoE-deficient mice and inhibits TNF-α-Induced endothelial adhesion molecule expression and monocyte adhesion. J. Agric. Food Chem. 2009, 57, 6692–6697. [Google Scholar] [CrossRef] [PubMed]
- Levy, G. Clinical pharmacokinetics of aspirin. Pediatrics 1978, 62, 867–872. [Google Scholar] [PubMed]
- Levy, G. Comparative pharmacokinetics of aspirin and acetaminophen. Arch. Intern. Med. 1981, 141, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Williams, F.M.; Mutch, E.M.; Nicholson, E.; Wynne, H.; Wright, P.; Lambert, D.; Rawlins, M.D. Human liver and plasma aspirin esterase. J. Pharm. Pharmacol. 1989, 41, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Morikawa, M.; Tsuboi, M.; Ito, Y.; Sugiura, M. Comparative study of human intestinal and hepatic esterases as related to enzymatic properties and hydrolizing activity forester-type drugs. Jpn. J. Pharmacol. 1980, 30, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Mukundan, M.; Yang, J.; Charpentier, N.; LeCluyse, E.L.; Black, C.; Yang, D.; Shi, D.; Yan, B. Antiplatelet agents aspirin and clopidogrel are hydrolyzed by distinct carboxylesterases, and clopidogrel is transesterificated in the presence of ethyl alcohol. J. Pharmacol. Exp. Ther. 2006, 319, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Marathe, G.K.; Hartiala, J.; Hazen, S.L.; Allayee, H.; Tang, W.H.; McIntyre, T.M. Aspirin hydrolysis in plasma is a variable function of butyrylcholinesterase and PAF acetylhydrolase 1b2. J. Biol. Chem. 2013, 288, 11940–11948. [Google Scholar] [CrossRef] [PubMed]
- Santanam, N.; Parthasarathy, S. Aspirin is a substrate for paraoxonase-like activity: Implications in atherosclerosis. Atherosclerosis 2007, 191, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Jaichander, P.; Selvarajan, K.; Garelnabi, M.; Parthasarathy, S. Induction of paraoxonase 1 and apolipoprotein A-I gene expression by aspirin. J. Lipid. Res. 2008, 49, 2142–2148. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.X.; Wu, Y.L.; Yang, B.; Zhu, B.C.; Hu, S.N.; Lu, Y.; Zhao, B.; Du, S.Y. Inhibitory influence of Panax notoginseng saponins on aspirin hydrolysis in human intestinal Caco-2 cells. Molecules 2018, 23, 455. [Google Scholar] [CrossRef]
- Li, X.; Wang, G.; Xiong, Y.; Sun, J.; Hao, H.; Zou, L.; Zheng, Y.; Yan, B.; Xia, C.; Liu, G. Population pharmacokinetic and pharmacodynamic evaluation of Xuesaitong Injection, a typical multiple constituent tradition Chinese medicine in patients with cerebral ischemia. Chin. J. Clin. Pharmacol. Ther. 2007, 12, 1183–1184. [Google Scholar]
- Zou, L.W.; Dou, T.Y.; Wang, P.; Lei, W.; Weng, Z.M.; Hou, J.; Wang, D.D.; Fan, Y.M.; Zhang, W.D.; Ge, G.B.; et al. Structure-activity relationships of pentacyclic triterpenoids as potent and selective inhibitors against human carboxylesterase 1. Front. Pharmacol. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Mai, Z.P.; Zhou, K.; Ge, G.B.; Wang, C.; Huo, X.K.; Dong, P.P.; Deng, S.; Zhang, B.J.; Zhang, H.L.; Huang, S.S.; et al. Protostane triterpenoids from the rhizome of Alisma orientale exhibit inhibitory effects on human carboxylesterase 2. J. Nat. Prod. 2015, 78, 2372–2380. [Google Scholar] [CrossRef] [PubMed]
- Jamila, N.; Khairuddean, M.; Yeong, K.K.; Osman, H.; Murugaiyah, V. Cholinesterase inhibitory triterpenoids from the bark of Garcinia hombroniana. J. Enzyme Inhib. Med. Chem. 2015, 30, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.H.; Pang, H.H.; Du, S.Y.; Lu, Y.; Zhang, L.; Wu, H.C.; Guo, S.; Wang, M.; Zhang, Q. Effect of Panax notoginseng saponins on the pharmacokinetics of aspirin in rats. J. Chromatogr. B 2017, 1040, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Wienkers, L.C.; Heath, T.G. Predicting in vivo drug interactions from in vitro drug design data. Nat. Rev. Drug Discov. 2005, 4, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, N.; Guo, Q.; Ji, H.; Zhao, D.; Xie, S.; Li, X.; Qiu, Z.; Han, D.; Chen, X.; et al. Inhibitory effects of wogonin on catalytic activity of cytochrome P450 enzyme in human liver microsomes. Eur. J. Drug Metab. Pharmacokinet. 2011, 36, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, N.; Richert, L.; Coassolo, P.; Lavé, T. Qualitative and quantitative assessment of drug-drug interaction potential in man, based on Ki, IC50 and inhibitor concentration. Curr. Drug Metab. 2004, 5, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.G.; Santos, R.N.D.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- López-Vallejo, F.; Caulfield, T.; Martínez-Mayorga, K.; Giulianotti, M.A.; Houghten, R.A.; Nefzi, A.; Nefzi, A.; Houghten, R.A.; Medina-Franco, J.L. Integrating virtual screening and combinatorial chemistry for accelerated drug discovery. Comb. Chem. High Scr. 2011, 14, 475–487. [Google Scholar] [CrossRef]
- Huang, S.Y.; Zou, X. Advances and challenges in protein-ligand docking. Int. J. Mol. Sci. 2010, 11, 3016–3034. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.F.; Yang, J.; Du, F.J.; Gao, X.M.; Ma, X.T.; Huang, Y.H.; Xu, F.; Niu, W.; Wang, F.; Mao, Y.; et al. Absorption and disposition of ginsenosides after oral administration of Panax notoginseng extract to rats. Drug Metab. Dispos. 2009, 37, 2290–2298. [Google Scholar] [CrossRef] [PubMed]
- Hermann, R.; von Richter, O. Clinical evidence of herbal drugs as perpetrators of pharmacokinetic drug interactions. Planta Med. 2012, 78, 1458–1477. [Google Scholar] [CrossRef] [PubMed]
- Durrington, P.N.; Mackness, B.; Mackness, M.I. Paraoxonase and atherosclerosis. Arterioscl. Throm. Vas. 2001, 21, 473–480. [Google Scholar] [CrossRef]
- Blatter-Garin, M.C.; Kalix, B.; De Pree, S.; James, R.W. Aspirin use is associated with higher serum concentrations of the anti-oxidant enzyme, paraoxonase-1. Diabetologia 2003, 46, 594–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Jin, Q.; Hou, J.; Feng, L.; Li, N.; Li, S.Y.; Zhou, Q.; Zou, L.W.; Ge, G.B.; Wang, J.G.; et al. Highly sensitive and selective detection of human carboxylesterase 1 activity by liquid chromatography with fluorescence detection. J. Chromatogr. B 2016, 1008, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Williams, E.T.; Bourgea, J.; Wong, Y.N.; Patten, C.J. Characterization of recombinant human carboxylesterases: fluorescein diacetate as a probe substrate for human carboxylesterase 2. Drug Metab. Dispos. 2011, 39, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Gulcin, I.; Scozzafava, A.; Supuran, C.T.; Koksal, Z.; Turkan, F.; Cetinkaya, S.; Bingöl, Z.; Huyut, Z.; Alwasel, S.H. Rosmarinic acid inhibits some metabolic enzymes including glutathione S-transferase, lactoperoxidase, acetylcholinesterase, butyrylcholinesterase and carbonic anhydrase isoenzymes. J. Enzyme Inhib. Med. Chem. 2016, 31, 1698–1702. [Google Scholar] [CrossRef] [PubMed]
- Furlong, C.E.; Richter, R.J.; Seidel, S.L.; Motulsky, A.G. Role of genetic polymorphism of human plasma paraoxonase/arylesterase in hydrolysis of the insecticide metabolites chlorpyrifos oxon and paraoxon. Am. J. Hum. Genet. 1988, 43, 230–238. [Google Scholar] [PubMed]
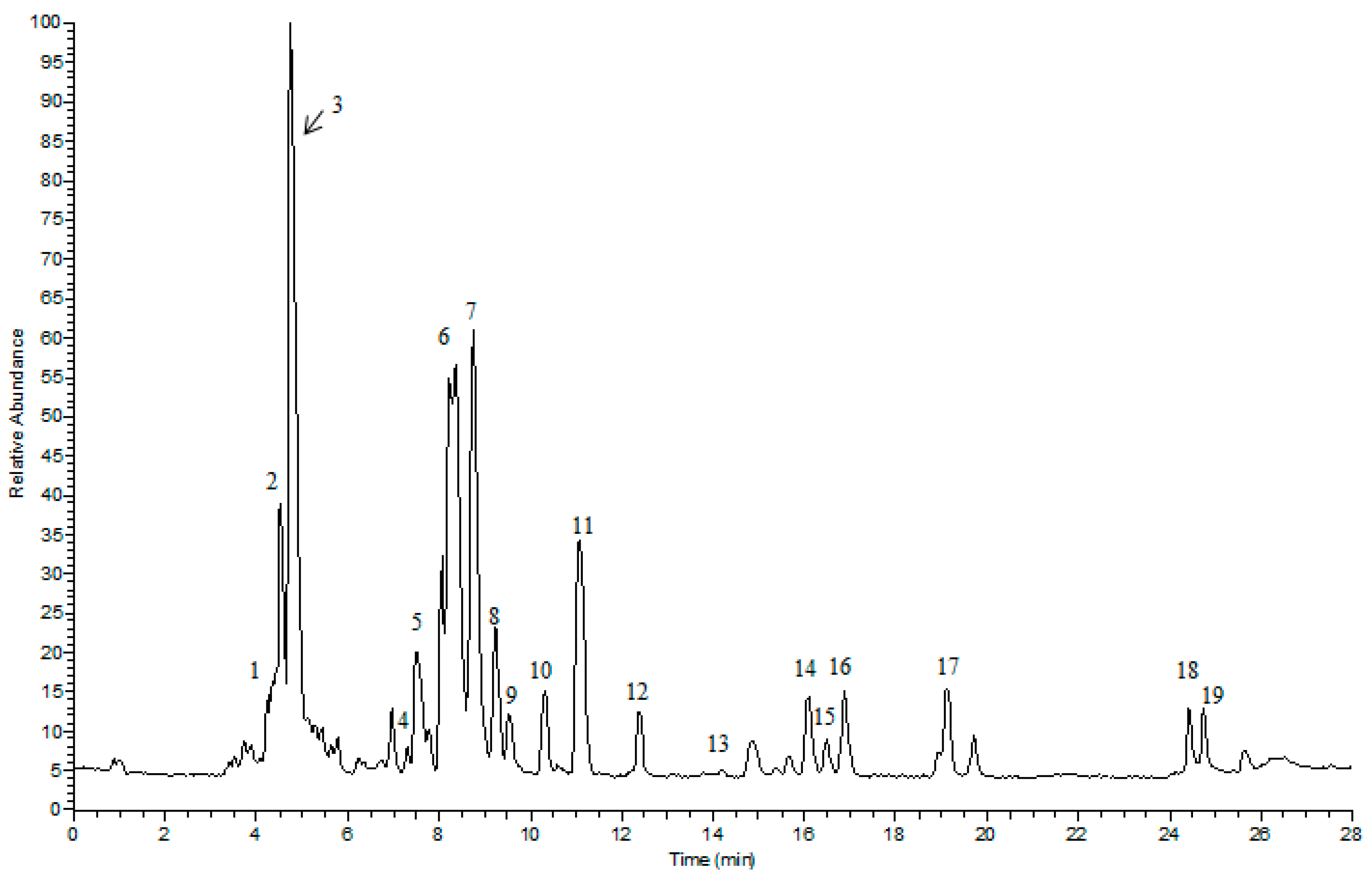
| No. | TR (min) | Theoretical Mass (m/z) | Experimental Mass (m/z) | Mass Error (ppm) | MS2 Fragment Ions | Formula | Identification |
|---|---|---|---|---|---|---|---|
| 1 | 4.42 | 1007.5421 | 1007.5417 | −0.40 | 799(100), 637(20.49), 475(1.24) | C48H82O19 | notoginsenoside R3/R6 |
| 2 | 4.48 | 977.5315 | 977.5316 | 0.10 | 799(100), 637(4.13), 475(3.7) | C47H80O18 | notoginsenoside R1 |
| 3 | 4.73 | 845.4891 | 845.4893 | 0.24 | 637(34.70), 475(32.28) | C42H72O14 | ginsenoside Rg1 |
| 4 | 7.30 | 845.4891 | 845.4890 | −0.12 | 637(10.01), 475(6.54) | C42H72O14 | ginsenoside F11 |
| 5 | 8.02 | 815.4787 | 815.4785 | −0.25 | 637(52.99), 475(100), 391(1.53) | C41H70O13 | notoginsenoside R2 or F3 |
| 6 | 8.30 | 1107.5946 | 1107.5994 | 4.33 | 945(100), 783(22.44), 621(16.01), 459(3.33) | C54H92O23 | ginsenoside Rb1 |
| 7 | 8.62 | 829.4944 | 829.4941 | −0.36 | 637(26.69), 475(100), 391(3.34) | C42H72O13 | ginsenoside Rg2 |
| 8 | 9.24 | 683.4365 | 683.4363 | −0.29 | 945(100), 783(35.03) | C53H90O22 | ginsenoside Rb2 or Rb3 |
| 9 | 9.55 | 1123.5895 | 1123.5892 | 0.27 | 637(100), 475(16.41) | C36H62O9 | ginsenoside Rh1 |
| 10 | 10.31 | 683.4365 | 683.4362 | −0.44 | 637(100), 475(45.43) | C36H62O9 | ginsenoside F1 |
| 11 | 11.08 | 991.5472 | 991.5465 | −0.71 | 783(70.63), 675(32.46), 475(3.7) | C48H82O18 | ginsenoside Re |
| 12 | 12.36 | 991.5472 | 991.5465 | −0.71 | 783(100), 621(7.47), 459(5.62), 375(0.86) | C48H82O18 | ginsenoside Rd |
| 13 | 13.76 | 961.5367 | 961.5376 | 0.94 | 783(0.68), 621(8.38) | C47H80O17 | notoginsenoside Fd |
| 14 | 16.09 | 665.4259 | 665.4281 | 3.31 | - | C36H60O8 | ginsenoside Rh4 |
| 15 | 16.47 | 829.4947 | 829.4944 | −0.36 | 715(100), 621(9.06), 459(18.43), 375(9.79) | C42H72O13 | ginsenoside Rg3 |
| 16 | 16.88 | 665.4259 | 665.4261 | 0.30 | - | C36H60O8 | ginsenoside Rk3 |
| 17 | 19.11 | 829.4947 | 829.4944 | −0.36 | 715(100), 621(9.06), 459(18.43), 375(9.79) | C42H72O13 | ginsenoside F2 |
| 18 | 24.17 | 667.4416 | 667.4431 | 2.25 | - | C36H62O8 | ginsenoside Rh2 |
| 19 | 24.37 | 811.4838 | 811.4843 | 0.62 | 603(100) | C42H70O12 | ginsenoside RK1 |
| Rank | Compound Name | Affinity (kcal/mol) | Rank | Compound Name | Affinity (kcal/mol) |
|---|---|---|---|---|---|
| 1 | ginsenoside Rg2 | −8.8 | 12 | ginsenoside F2 | −6.9 |
| 2 | ginsenoside Rh1 | −8.7 | 12 | notoginsenoside F3 | −6.9 |
| 3 | ginsenoside Rh4 | −8.6 | 14 | ginsenoside Fe | −6.8 |
| 4 | ginsenoside F1 | −8.3 | 14 | ginsenoside Rg3 | −6.8 |
| 5 | ginsenoside R2 | −8.1 | 16 | notoginsenoside R1 | −6.7 |
| 6 | notoginsenoside Fd | −7.7 | 16 | ginsenoside Rb3 | −6.7 |
| 7 | ginsenoside Re | −7.4 | 18 | ginsenoside Rb1 | −6.5 |
| 8 | notoginsenoside R6 | −7.3 | 19 | ginsenoside Rb2 | −6.1 |
| 9 | ginsenoside RK3 | −7.2 | 20 | ginsenoside F11 | −6.0 |
| 10 | ginsenoside RK1 | −7.1 | 20 | notoginsenoside R3 | −6.0 |
| 10 | ginsenoside Rg1 | −7.1 | 22 | ginsenoside Rd | −5.5 |
| Gene | Forward Sequences 5′–3′ | Reverse Sequences 5′–3′ |
|---|---|---|
| hCE1 | GGGCACTGTGATTGATGGGA | CCTTCGGAGAGTGGATAGCTC |
| hCE2 | TCAGAAGTTTCTTTGGGGGCA | GCAGGTATTGCTCCTCCTGG |
| BChE | GCTGGCCTGTCTTCAAAAGC | TCCTGCTTTCCACTCCCATTC |
| PON1 | AAGTTCGAGTGGTGGCAGAA | TGGCATCCAACCCAAAGGTC |
| GAPDH | CTCCTCCACCTTTGACGCTG | TCCTCTTGTGCTCTTGCTGG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Wu, Y.; Liu, S.; Hu, S.; Zhao, B.; Li, P.; Du, S. Effects of Panax Notoginseng Saponins on Esterases Responsible for Aspirin Hydrolysis In Vitro. Int. J. Mol. Sci. 2018, 19, 3144. https://doi.org/10.3390/ijms19103144
Sun Z, Wu Y, Liu S, Hu S, Zhao B, Li P, Du S. Effects of Panax Notoginseng Saponins on Esterases Responsible for Aspirin Hydrolysis In Vitro. International Journal of Molecular Sciences. 2018; 19(10):3144. https://doi.org/10.3390/ijms19103144
Chicago/Turabian StyleSun, Zongxi, Yali Wu, Song Liu, Shaonan Hu, Bo Zhao, Pengyue Li, and Shouying Du. 2018. "Effects of Panax Notoginseng Saponins on Esterases Responsible for Aspirin Hydrolysis In Vitro" International Journal of Molecular Sciences 19, no. 10: 3144. https://doi.org/10.3390/ijms19103144
APA StyleSun, Z., Wu, Y., Liu, S., Hu, S., Zhao, B., Li, P., & Du, S. (2018). Effects of Panax Notoginseng Saponins on Esterases Responsible for Aspirin Hydrolysis In Vitro. International Journal of Molecular Sciences, 19(10), 3144. https://doi.org/10.3390/ijms19103144