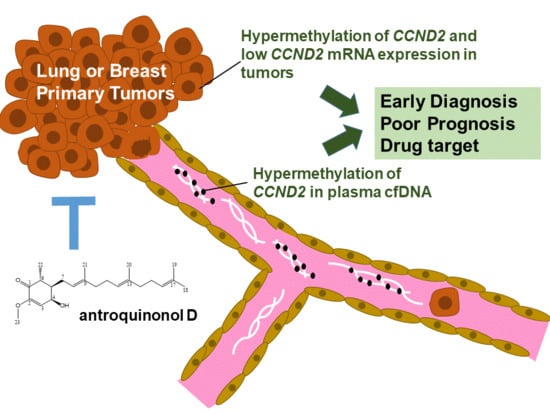
Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target
Abstract
1. Introduction
2. Results
2.1. CCND2 is a Common Target in Lung and Breast Cancer
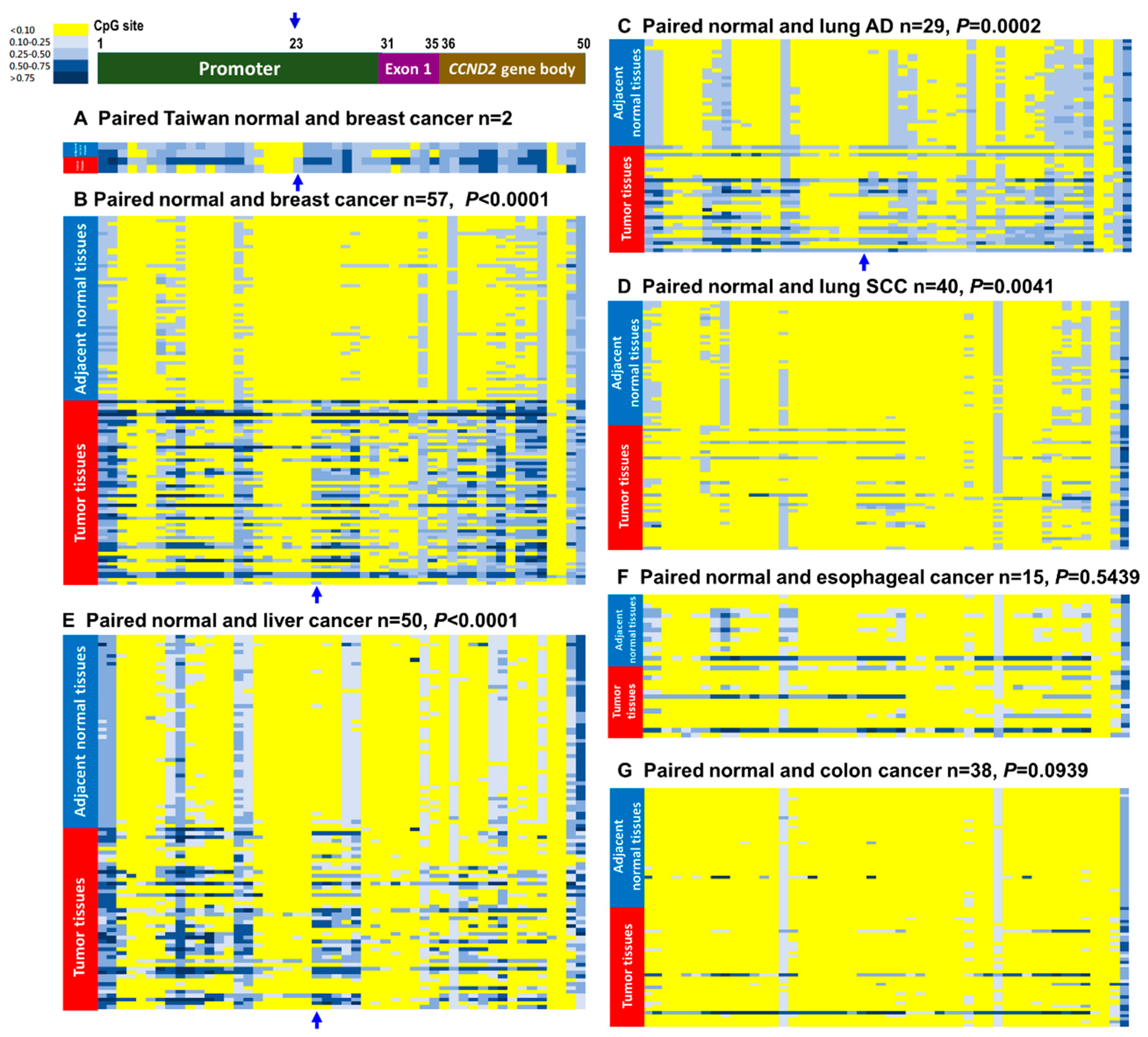
2.2. CCND2 Reveals Promoter Hypermethylation and mRNA Downregulation in Lung Cancer and Breast Cancer
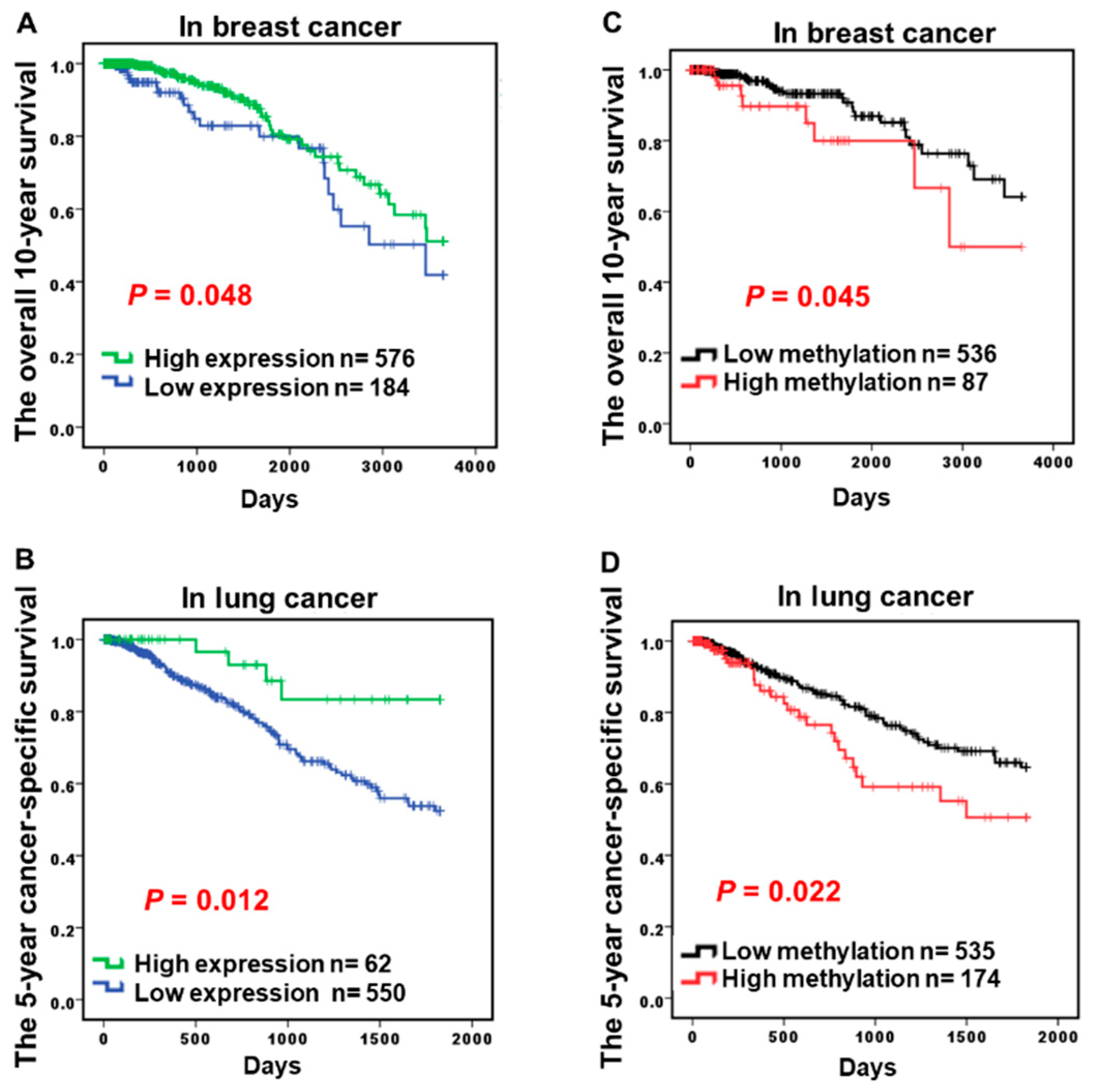
2.3. Hypermethylation of CCND2 is an Independent Poor Prognostic Factor
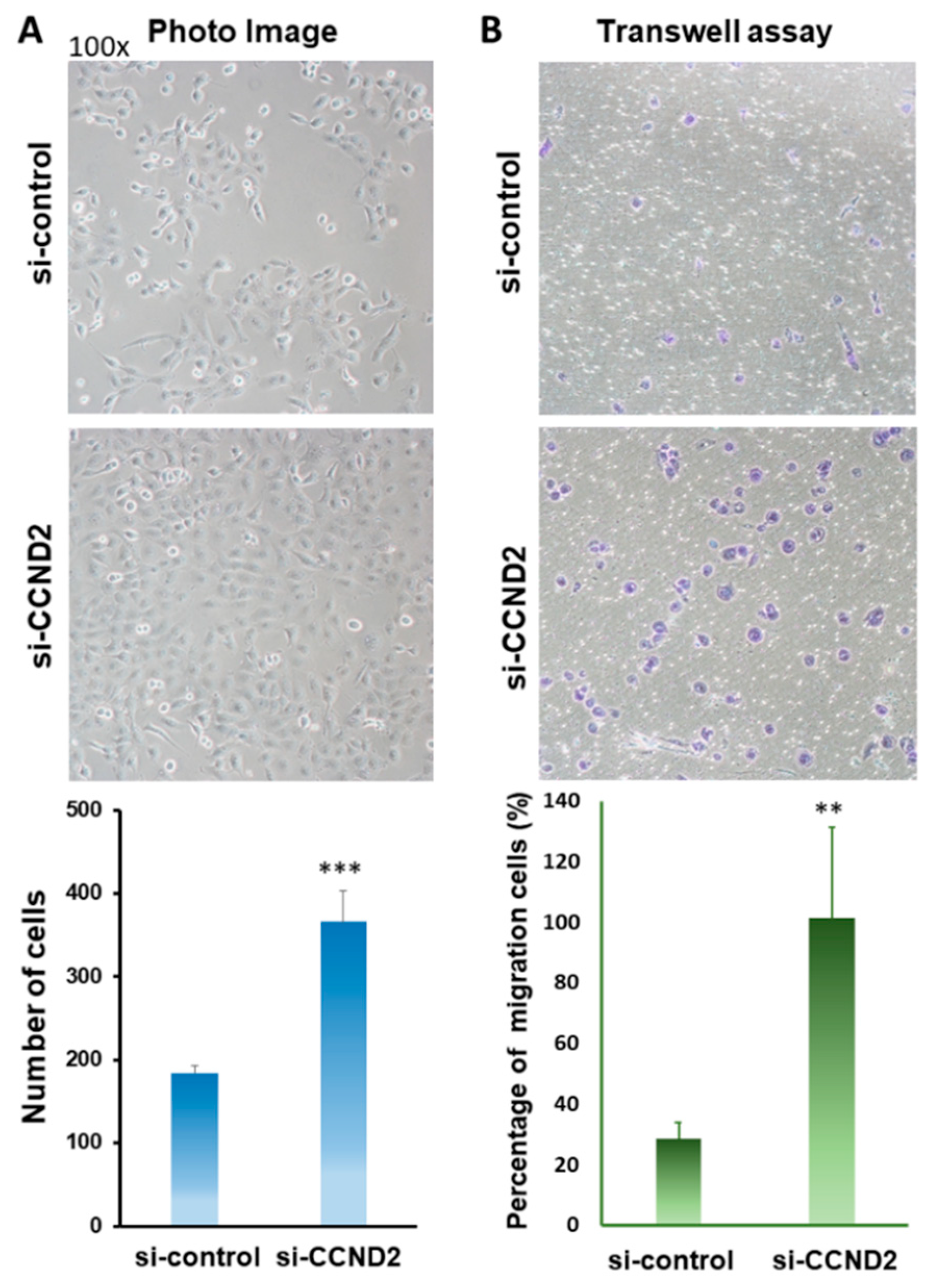
2.4. CCND2 is Involved in the Inhibition of Lung Cancer Cell Growth and Cell Migration
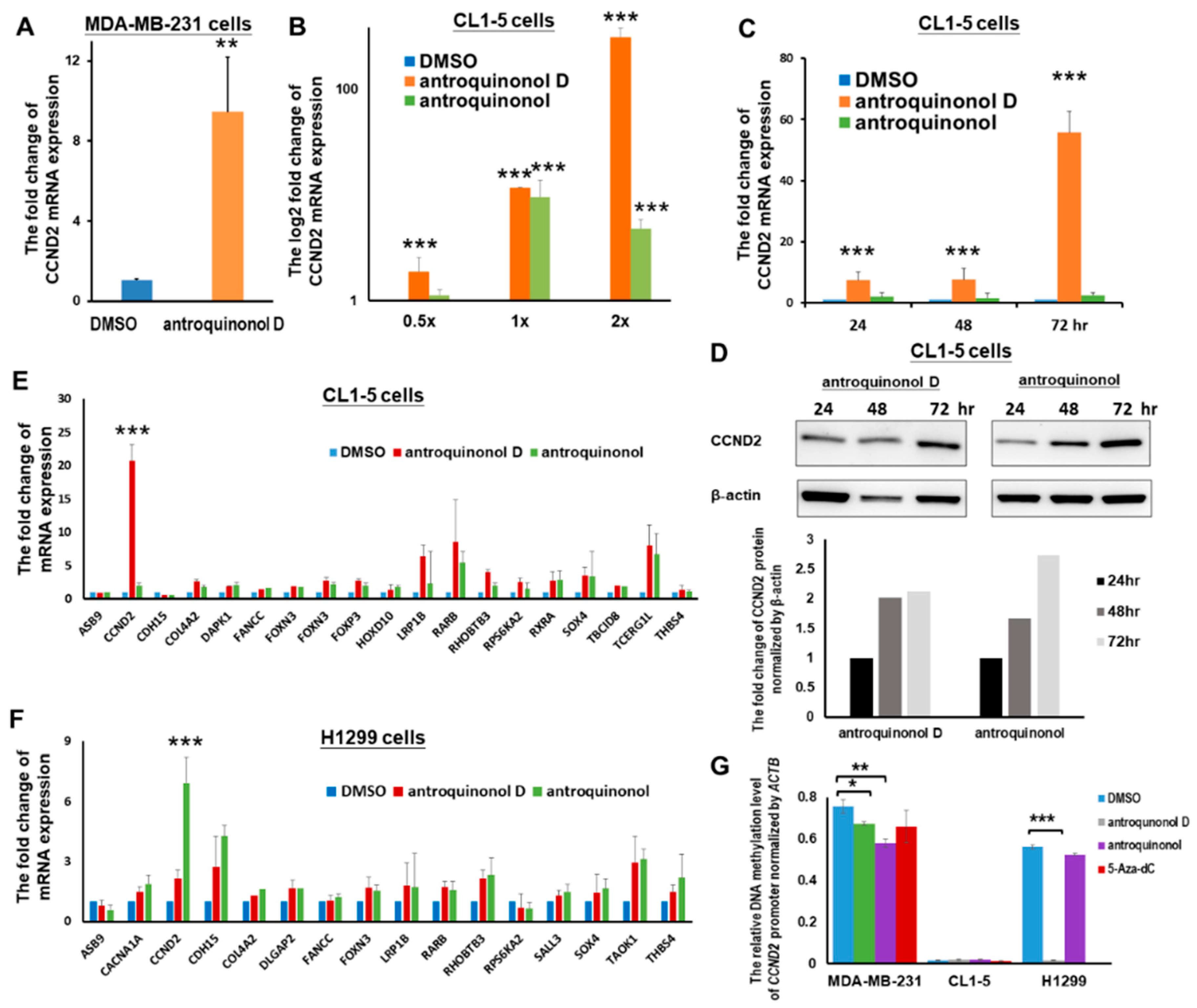
2.5. The Candidate CCND2 Stimulator Antroquinonol D Induces CCND2 mRNA and Protein Expression
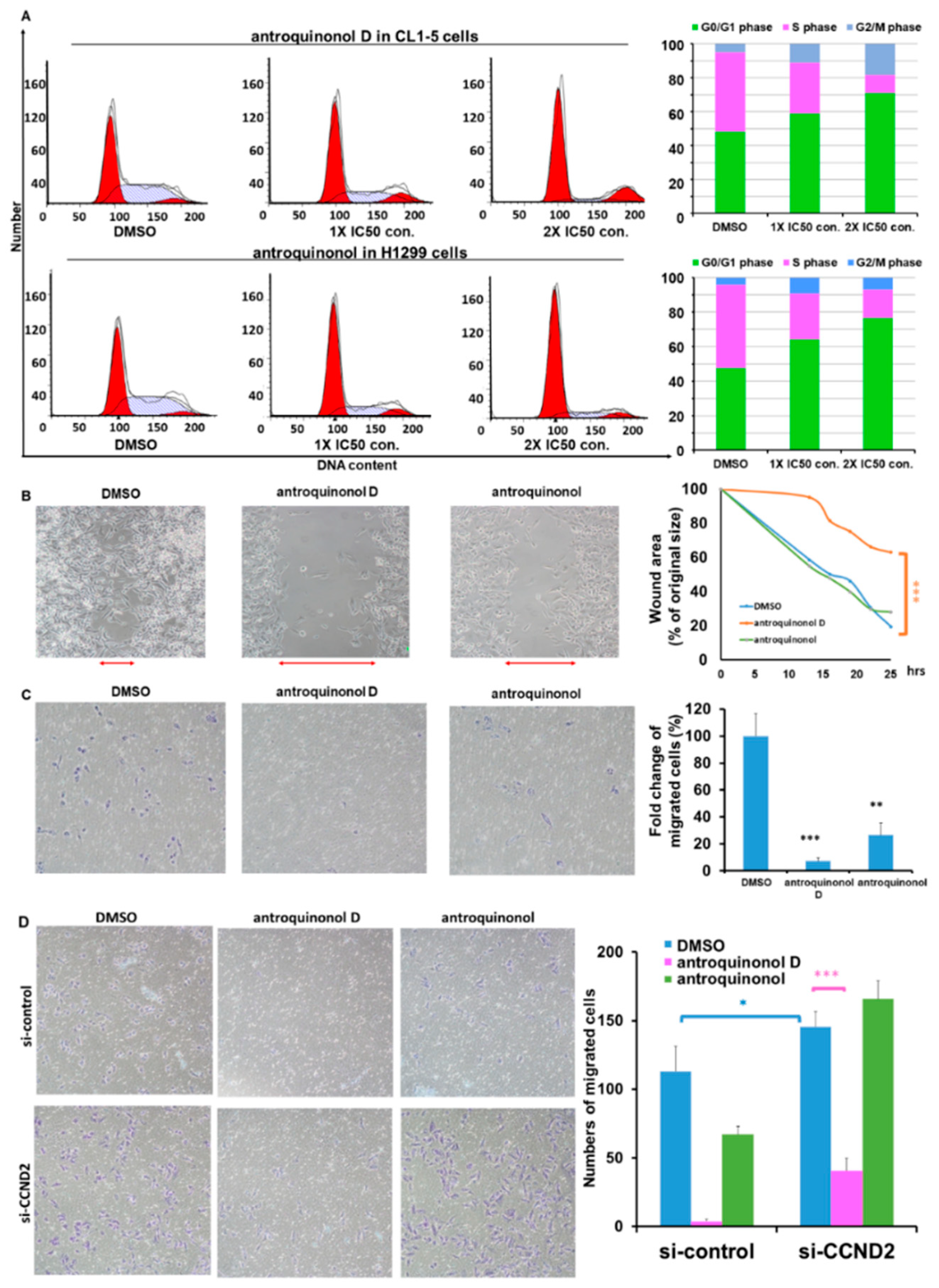
2.6. Antroquinonol and Antroquinonol D Inhibit Growth and Induce Cell Cycle Arrest in the G1 Phase in a Dose-Dependent Manner in Lung Cancer Cell Lines
2.7. Antroquinonol D Inhibits Cell Proliferation and Migration Ability through CCND2 Induction
3. Discussion
4. Materials and Methods
4.1. Patients and Plasma and Tissue Collection
4.2. Genomic DNA, Circulating Cell-Free DNA and RNA Extraction
4.3. DNA Methylation Assays
4.4. Real-Time RT–PCR
4.5. The Cancer Genome Atlas Data Processing and Candidate Gene Selection
4.6. Statistical Analyses
4.7. Antroquinonol and Antroquinonol D
4.8. Cell. Lines and Drug Treatments
4.9. Sulphorhodamine B Assay
4.10. Wound-Healing Assays for Migration Analysis
4.11. Transwell Assays for Migration Analysis
4.12. Cell. Cycle Distribution Assay
4.13. Cell. Lyses and Immunoblotting Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| antroquinonol D | 3-demethoxyl antroquinonol |
| CCND2 | cyclin D2 |
| cfDNA | circulating cell-free DNA |
| HER2 | growth factor receptor 2 |
| NSCLC | non-small cell lung cancer |
| TCGA | The Cancer Genome Atlas |
| TMU | Taipei Medical University |
| TNBC | triple-negative breast cancer |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute. 2017. Available online: http://www.cdc.gov/uscs (accessed on 31 December 2017).
- Gadgeel, S.M. Personalized Therapy of Non-small Cell Lung Cancer (NSCLC). Adv. Exp. Med. Boil. 2016, 890, 203–222. [Google Scholar]
- Sacco, J.; Al-Akhrass, H.; Wilson, C.M. Challenges and strategies in precision medicine for non-small cell lung cancer. Curr. Pharm. Des. 2016, 22, 4374–4385. [Google Scholar] [CrossRef] [PubMed]
- Nixon, N.A.; Hannouf, M.B.; Verma, S. A review of the value of human epidermal growth factor receptor 2 (HER2)-targeted therapies in breast cancer. Eur. J. Cancer 2018, 89, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Lortet-Tieulent, J.; Soerjomataram, I.; Ferlay, J.; Rutherford, M.; Weiderpass, E.; Bray, F. International trends in lung cancer incidence by histological subtype: Adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer 2014, 84, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Chalela, R.; Curull, V.; Enriquez, C.; Pijuan, L.; Bellosillo, B.; Gea, J. Lung adenocarcinoma: From molecular basis to genome-guided therapy and immunotherapy. J. Thorac. Dis. 2017, 9, 2142–2158. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Djamgoz, M.B.A. Triple negative breast cancer: Emerging therapeutic modalities and novel combination therapies. Cancer Treat. Rev. 2018, 62, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T.; Lee, Y.M.; Huang, R.S. The impact of the Cancer Genome Atlas on lung cancer. Transl. Res. J. Lab. Clin. Med. 2015, 166, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Malumbres, M.; Barbacid, M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim. Biophys. Acta 2002, 1602, 73–87. [Google Scholar] [CrossRef]
- Kwapisz, D. Cyclin-dependent kinase 4/6 inhibitors in breast cancer: Palbociclib, ribociclib, and abemaciclib. Breast Cancer Res. Treat. 2017, 166, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Evron, E.; Umbricht, C.B.; Korz, D.; Raman, V.; Loeb, D.M.; Niranjan, B.; Buluwela, L.; Weitzman, S.A.; Marks, J.; Sukumar, S. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001, 61, 2782–2787. [Google Scholar] [PubMed]
- Evron, E.; Dooley, W.C.; Umbricht, C.B.; Rosenthal, D.; Sacchi, N.; Gabrielson, E.; Soito, A.B.; Hung, D.T.; Ljung, B.; Davidson, N.E.; et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet 2001, 357, 1335–1336. [Google Scholar] [CrossRef]
- Salskov, A.; Hawes, S.E.; Stern, J.E.; Feng, Q.; Jordan, C.D.; Wiens, L.; Rasey, J.; Lu, H.; Kiviat, N.B.; Vesselle, H. Hypermethylation of CCND2 May Reflect a Smoking-Induced Precancerous Change in the Lung. J. Oncol. 2011, 2011, 950140. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Grau, L.; Puerta, P.; Gimenez, L.; Venditti, J.; Quadrelli, S.; Sanchez-Carbayo, M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J. Transl. Med. 2010, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.H.; Lee, H.J.; Kim, B.H.; Cho, N.Y.; Kang, G.H. DNA methylation profile during multistage progression of pulmonary adenocarcinomas. Virchows Arch. Int. J. Pathol. 2011, 459, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, M.; Akahira, J.; Ito, K.; Niikura, H.; Moriya, T.; Okamura, K.; Sasano, H.; Yaegashi, N. Promoter methylation status of the Cyclin D2 gene is associated with poor prognosis in human epithelial ovarian cancer. Cancer Sci. 2007, 98, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, M.; Iizuka, N.; Moribe, T.; Miura, T.; Kimura, N.; Tamatsukuri, S.; Ishitsuka, H.; Fujita, Y.; Hamamoto, Y.; Tsunedomi, R.; et al. Methylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinoma. Clin. Chim. Acta Int. J. Clin. Chem. 2010, 411, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Lee, T.H.; Hsu, C.H.; Chang, Y.J.; Chang, M.S.; Wang, Y.C.; Ho, Y.S.; Wen, W.C.; Lin, R.K. Antroquinonol D, isolated from Antrodia camphorata, with DNA demethylation and anticancer potential. J. Agric. Food Chem. 2014, 62, 5625–5635. [Google Scholar] [CrossRef] [PubMed]
- Geethangili, M.; Tzeng, Y.M. Review of Pharmacological Effects of Antrodia camphorata and Its Bioactive Compounds. Evid. Based Complement. Altern. Med. 2011, 2011, 212641. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.C.; Chiang, P.C.; Lu, P.H.; Kuo, M.T.; Wen, W.C.; Chen, P.; Guh, J.H. Antroquinonol, a natural ubiquinone derivative, induces a cross talk between apoptosis, autophagy and senescence in human pancreatic carcinoma cells. J. Nutr. Biochem. 2012, 23, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.T.; Lee, T.H.; Cheng, C.H.; Chen, K.C.; Chen, Y.C.; Lin, C.W. Antroquinonol from Antrodia Camphorata suppresses breast tumor migration/invasion through inhibiting ERK-AP-1- and AKT-NF-kappaB-dependent MMP-9 and epithelial-mesenchymal transition expressions. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Boil. Res. Assoc. 2015, 78, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.B.; Yuan, T.C.; Liou, J.W.; Yang, C.J.; Sung, P.J.; Weng, C.F. Antroquinonol inhibits NSCLC proliferation by altering PI3K/mTOR proteins and miRNA expression profiles. Mutat. Res. 2011, 707, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.L.; Wang, J.L.; Lee, C.C.; Cheng, H.Y.; Wen, W.C.; Cheng, H.H.; Chen, M.C. Antroquinonol blocks Ras and Rho signaling via the inhibition of protein isoprenyltransferase activity in cancer cells. Biomed. Pharmacother. Biomed. Pharmacother. 2014, 68, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.C.; Lin, S.C.; Pan, S.L.; Kuo, C.H.; Tsai, I.L.; Kuo, M.T.; Wen, W.C.; Chen, P.; Guh, J.H. Antroquinonol displays anticancer potential against human hepatocellular carcinoma cells: A crucial role of AMPK and mTOR pathways. Biochem. Pharmacol. 2010, 79, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Villaume, M.T.; Sella, E.; Saul, G.; Borzilleri, R.M.; Fargnoli, J.; Johnston, K.A.; Zhang, H.; Fereshteh, M.P.; Dhar, T.G.; Baran, P.S. Antroquinonol A: Scalable Synthesis and Preclinical Biology of a Phase 2 Drug Candidate. ACS Cent. Sci. 2016, 2, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Ho, C.L.; Kao, W.Y.; Chen, Y.M. A phase I multicenter study of antroquinonol in patients with metastatic non-small-cell lung cancer who have received at least two prior systemic treatment regimens, including one platinum-based chemotherapy regimen. Mol. Clin. Oncol. 2015, 3, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Sulake, R.S.; Jiang, Y.F.; Lin, H.H.; Chen, C. Total synthesis of (+/−)-antroquinonol d. J. Org. Chem. 2014, 79, 10820–10828. [Google Scholar] [CrossRef] [PubMed]
- Sulake, R.S.; Chen, C. Total synthesis of (+)-antroquinonol and (+)-antroquinonol D. Org. Lett. 2015, 17, 1138–1141. [Google Scholar] [CrossRef] [PubMed]
- McCullough, L.E.; Chen, J.; Cho, Y.H.; Khankari, N.K.; Bradshaw, P.T.; White, A.J.; Teitelbaum, S.L.; Terry, M.B.; Neugut, A.I.; Hibshoosh, H.; et al. Modification of the association between recreational physical activity and survival after breast cancer by promoter methylation in breast cancer-related genes. Breast Cancer Res. 2017, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.K.; Kohlschmidt, J.; Schwind, S.; Nicolet, D.; Blachly, J.S.; Orwick, S.; Shah, C.; Bainazar, M.; Kroll, K.W.; Walker, C.J.; et al. Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia 2017, 31, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Hoglund, M.; Johansson, B.; Pedersen-Bjergaard, J.; Marynen, P.; Mitelman, F. Molecular characterization of 12p abnormalities in hematologic malignancies: Deletion of KIP1, rearrangement of TEL, and amplification of CCND2. Blood 1996, 87, 324–330. [Google Scholar] [PubMed]
- Buschges, R.; Weber, R.G.; Actor, B.; Lichter, P.; Collins, V.P.; Reifenberger, G. Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas. Brain Pathol. 1999, 9, 435–442; discussion 432–433. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U.; Berg-Ribbe, I.; Wingen, L.U.; Brakensiek, K.; Becker, T.; Klempnauer, J.; Schlegelberger, B.; Kreipe, H.; Flemming, P. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin. Cancer Res. 2005, 11, 3654–3660. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Q.; Wang, Q.; Li, J.; Sipeky, C.; Xia, J.; Gao, P.; Hu, Y.; Zhang, H.; Yang, X.; et al. Genetic association analysis of the RTK/ERK pathway with aggressive prostate cancer highlights the potential role of CCND2 in disease progression. Sci. Rep. 2017, 7, 4538. [Google Scholar] [CrossRef] [PubMed]
- Oshimo, Y.; Nakayama, H.; Ito, R.; Kitadai, Y.; Yoshida, K.; Chayama, K.; Yasui, W. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int. J. Oncol. 2003, 23, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.K.; Hsieh, Y.S.; Lin, P.; Hsu, H.S.; Chen, C.Y.; Tang, Y.A.; Lee, C.F.; Wang, Y.C. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J. Clin. Investig. 2010, 120, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.T.; Bai, B.; Zhu, J.; Di, C.X.; Li, X.; Zhou, W.C. Epigenetic actions of environmental factors and promising drugs for cancer therapy. Oncol. Lett. 2018, 15, 2049–2056. [Google Scholar] [CrossRef] [PubMed]
- Clements, E.G.; Mohammad, H.P.; Leadem, B.R.; Easwaran, H.; Cai, Y.; Van Neste, L.; Baylin, S.B. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012, 40, 4334–4346. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.H.; Sieber, S.A. Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep. 2016, 33, 681–708. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Chen, Y.; Zhou, J.; Zhao, H.; Zhang, H.; Wang, G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int. J. Infect. Dis. 2018, 67, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Zhou, M.Q.; Tian, W.; Meng, K.X.; He, H.F. Effect of Age on Breast Cancer Patient Prognoses: A Population-Based Study Using the SEER 18 Database. PLoS ONE 2016, 11, e0165409. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.; Chia, Y.L.; Plevritis, S. The effect of age, race, tumor size, tumor grade, and disease stage on invasive ductal breast cancer survival in the U.S. SEER database. Breast Cancer Res. Treat. 2005, 89, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gold, K.A.; Lin, H.Y.; Swisher, S.G.; Xing, Y.; Lee, J.J.; Kim, E.S.; William, W.N., Jr. Relationship between tumor size and survival in non-small-cell lung cancer (NSCLC): An analysis of the surveillance, epidemiology, and end results (SEER) registry. J. Thorac. Oncol. 2015, 10, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Lee, C.K.; Tsou, W.L.; Liu, S.Y.; Kuo, M.T.; Wen, W.C. A new cytotoxic agent from solid-state fermented mycelium of Antrodia camphorata. Planta Med. 2007, 73, 1412–1415. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Breast Cancer | Lung Cancer | Liver Cancer | Esophageal Cancer | Gastric Cancer | Colon Cancer | |||
|---|---|---|---|---|---|---|---|---|---|
| All 1 | TNBC 2 | All | AD 3 | SCC 4 | |||||
| Hypermethylation on CCND2 gene (paired) | 31/57 (54.4%) | 3/6 (60.0%) | 22/69 (31.9%) | 14/29 (48.3%) | 8/40 (20.0%) | 26/50 (52.0%) | 2/15 (13.3%) | 1/2 (50.0%) | 3/38 (7.9%) |
| Low CCND2 mRNA expressions 5 (paired) | 62/73 *** (84.9%) | 7/7 *** (100%) | 73/90 (81.1%) | 45/49 *** (91.8%) | 28/51 (54.9%) | 21/50 (42.0%) | 7/13 (53.9%) | 22/32 (68.6%) | 13/41 (31.7%) |
| % of methylation and low CCND2 | 43/49 (63.3%) | - | 6/23 (26.1%) | 5/16 (31.3%) | 1/7 (14.3%) | 6/41 (14.6%) | - | - | 2/11 (18.2%) |
| Concordance 6 between methylation & mRNA expression | p = 0.002 ** | - | p = 0.015 * | p = 0.029 * | p > 0.05 | p > 0.05 | - | - | p > 0.05 |
| Prognosis 7 | p = 0.048 * | p = 0.012 * | p = 0.011 * | p > 0.05 | p > 0.05 | p > 0.05 | |||
| Lung 2 | 5-Year Overall Survival 1 | |||||
| Cancer | Univariate Analysis | Multivariate Analysis | ||||
| Variable | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| Gender | 0.982 | 0.761–1.268 | 0.890 | 0.883 | 0.633–1.232 | 0.465 |
| Race | 1.088 | 0.767–1.543 | 0.635 | 1.158 | 0.742–1.805 | 0.519 |
| Age | 1.005 | 0.994–1.016 | 0.380 | 1.005 | 0.992–1.018 | 0.474 |
| Tumor type | 1.001 | 0.779–1.287 | 0.993 | 0.742 | 0.507–1.084 | 0.123 |
| Stage *** | 1.466 | 1.287–1.669 | <0.001 *** | 1.477 | 1.247–1.751 | <0.001 *** |
| CCND2(RNA) 3 | 0.742 | 0.600–0.919 | 0.006 ** | 0.688 | 0.501–0.946 | 0.021 * |
| CCND2(methyl) | 1.386 | 0.982–1.957 | 0.063 | 1.544 | 1.022–2.333 | 0.039 * |
| Breast 4 | 10-Year Overall Survival 1 | |||||
| Cancer | Univariate Analysis | Multivariate Analysis | ||||
| Variable | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| Race | 0.902 | 0.568–1.433 | 0.662 | 1.949 | 0.781–4.860 | 0.153 |
| Age | 2.055 | 1.366–3.091 | 0.001 *** | 0.725 | 0.218–2.415 | 0.600 |
| Tumor type | 1.391 | 1.028–1.882 | 0.033 * | 1.455 | 0.620–3.414 | 0.388 |
| Stage | 1.685 | 1.287–2.208 | <0.001 *** | 3.525 | 1.700–7.313 | <0.001 *** |
| Menopause | 1.256 | 0.904–1.746 | 0.174 | 1.757 | 0.904–3.415 | 0.096 |
| CCND2(RNA) 4 | 0.594 | 0.353–1.000 | 0.050 * | 0.173 | 0.064–0.467 | 0.001 *** |
| CCND2(methyl) | 2.207 | 0.998–4.881 | 0.051 | 4.693 | 1.466–15.017 | 0.009 ** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, C.-S.; Wang, S.-C.; Yen, Y.-T.; Lee, T.-H.; Wen, W.-C.; Lin, R.-K. Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. Int. J. Mol. Sci. 2018, 19, 3096. https://doi.org/10.3390/ijms19103096
Hung C-S, Wang S-C, Yen Y-T, Lee T-H, Wen W-C, Lin R-K. Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. International Journal of Molecular Sciences. 2018; 19(10):3096. https://doi.org/10.3390/ijms19103096
Chicago/Turabian StyleHung, Chin-Sheng, Sheng-Chao Wang, Yi-Ting Yen, Tzong-Huei Lee, Wu-Che Wen, and Ruo-Kai Lin. 2018. "Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target" International Journal of Molecular Sciences 19, no. 10: 3096. https://doi.org/10.3390/ijms19103096
APA StyleHung, C.-S., Wang, S.-C., Yen, Y.-T., Lee, T.-H., Wen, W.-C., & Lin, R.-K. (2018). Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. International Journal of Molecular Sciences, 19(10), 3096. https://doi.org/10.3390/ijms19103096