Antibacterial Activity and Mechanism of Action of Bovine Lactoferricin Derivatives with Symmetrical Amino Acid Sequences
Abstract
1. Introduction
2. Results
2.1. Design and Sequence Analysis of Peptides
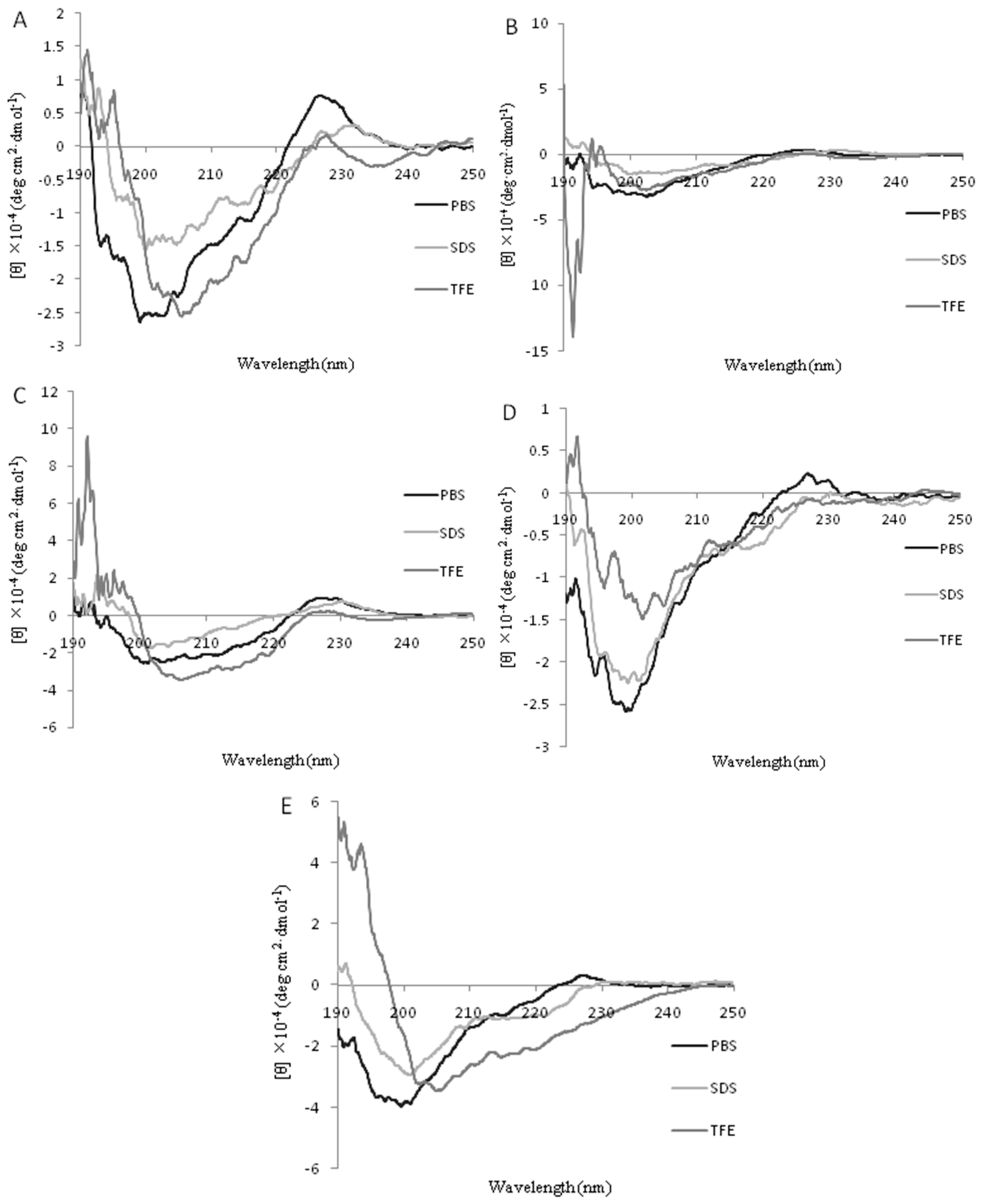
2.2. Circular Dichroism (CD) Spectra
2.3. Antimicrobial Activity
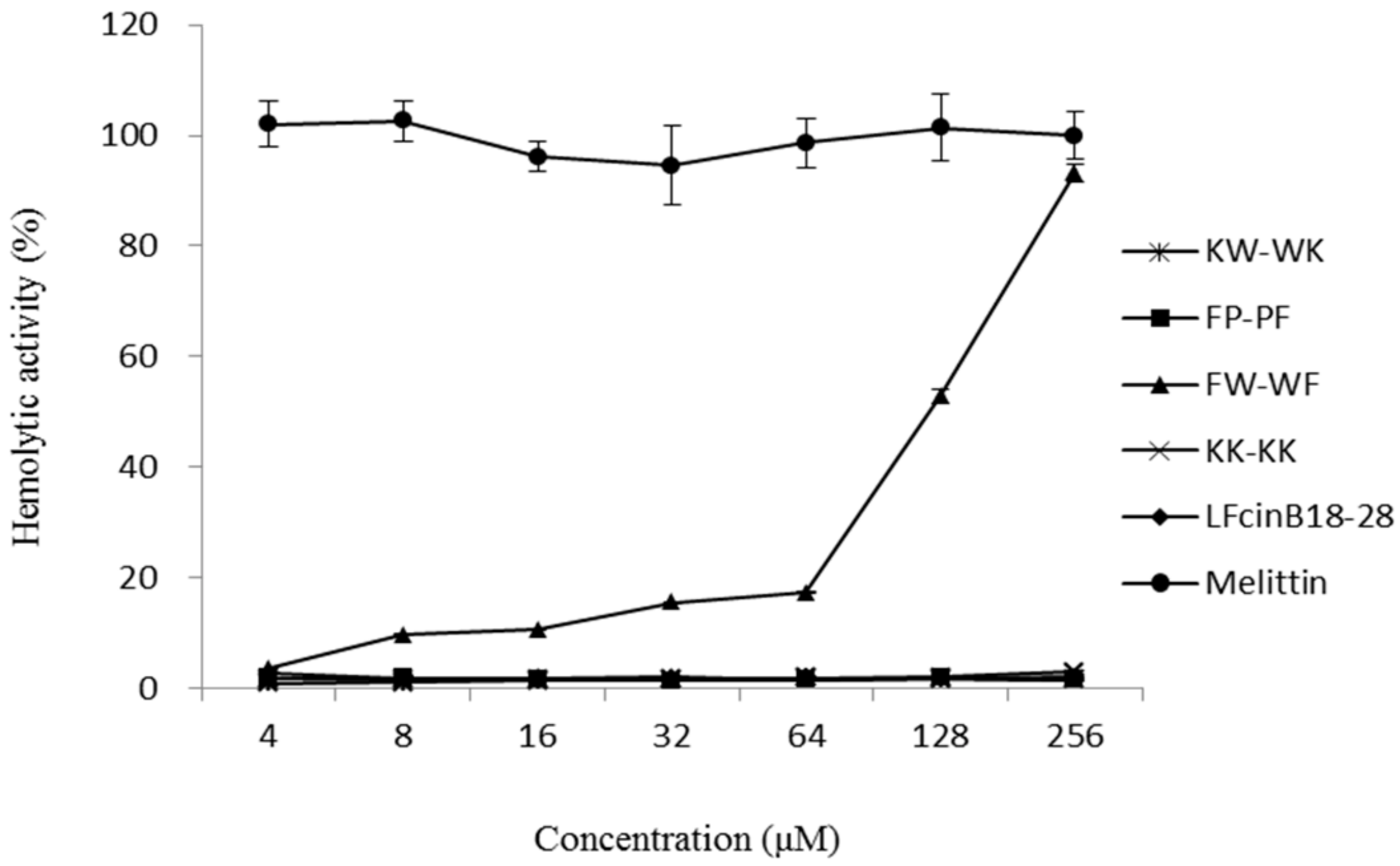
2.4. Hemolytic Activity
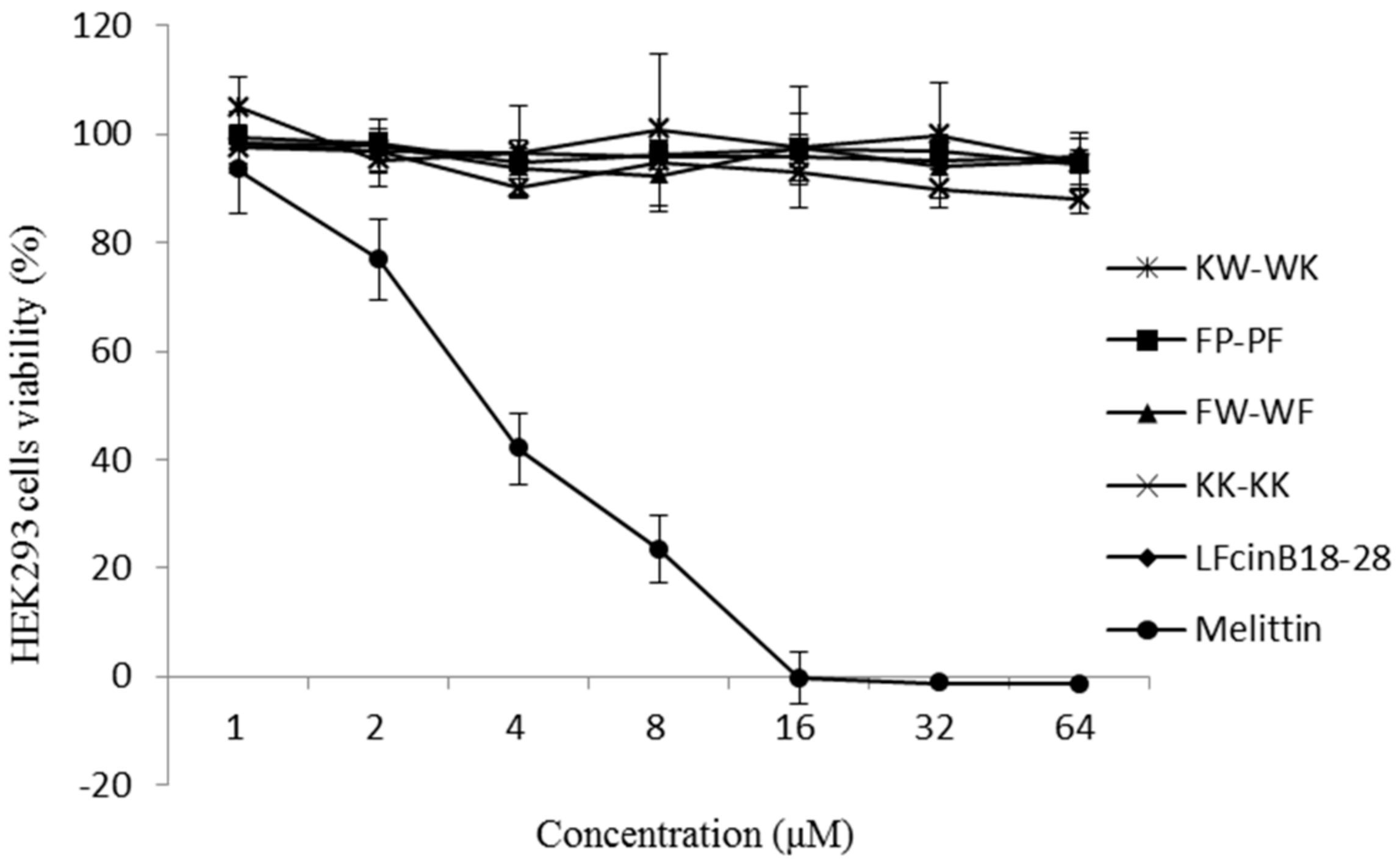
2.5. Cytotoxicity
2.6. Stability
2.7. Mechanism of Action of the Peptides
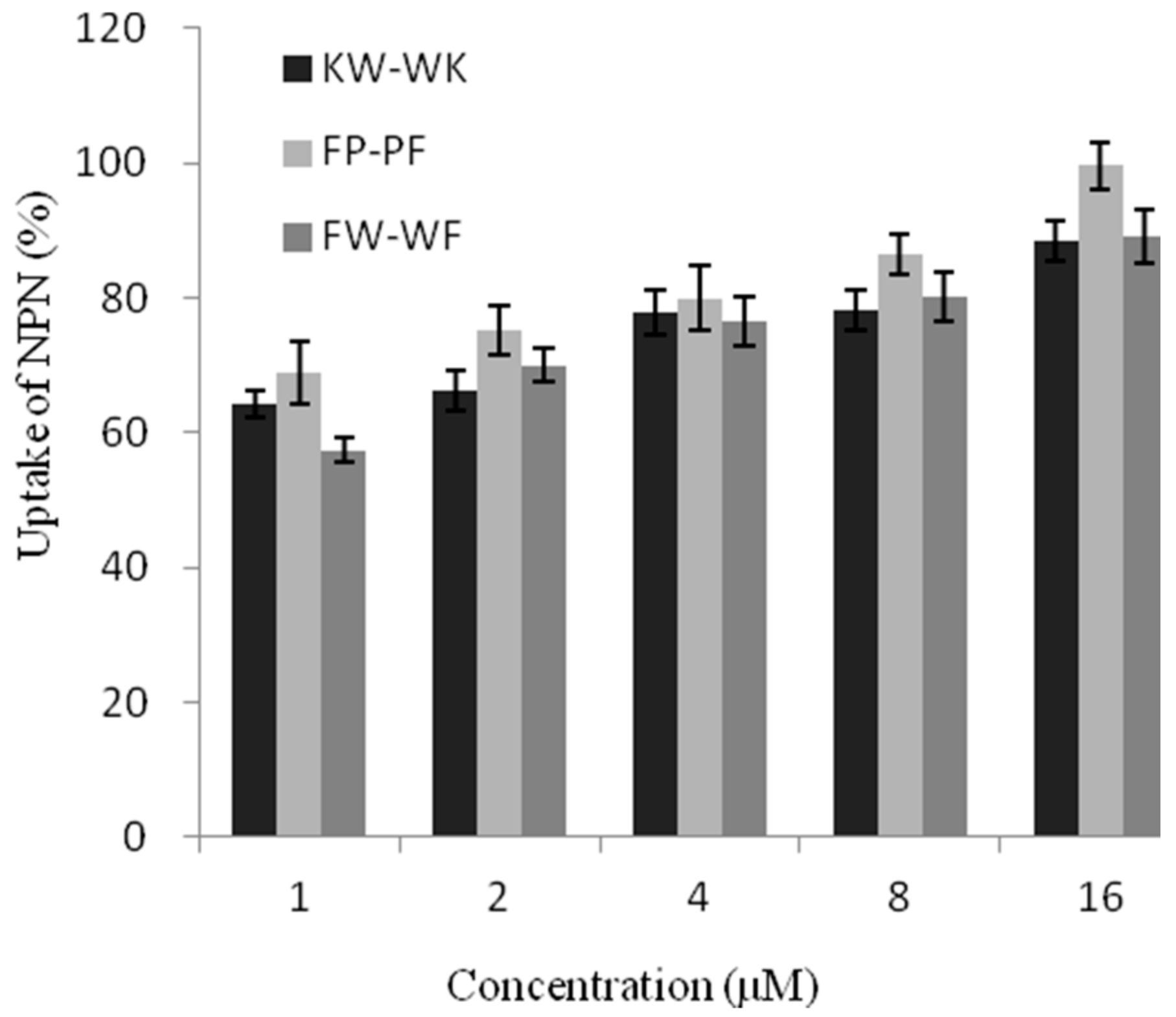
2.7.1. Outer Membrane Permeability
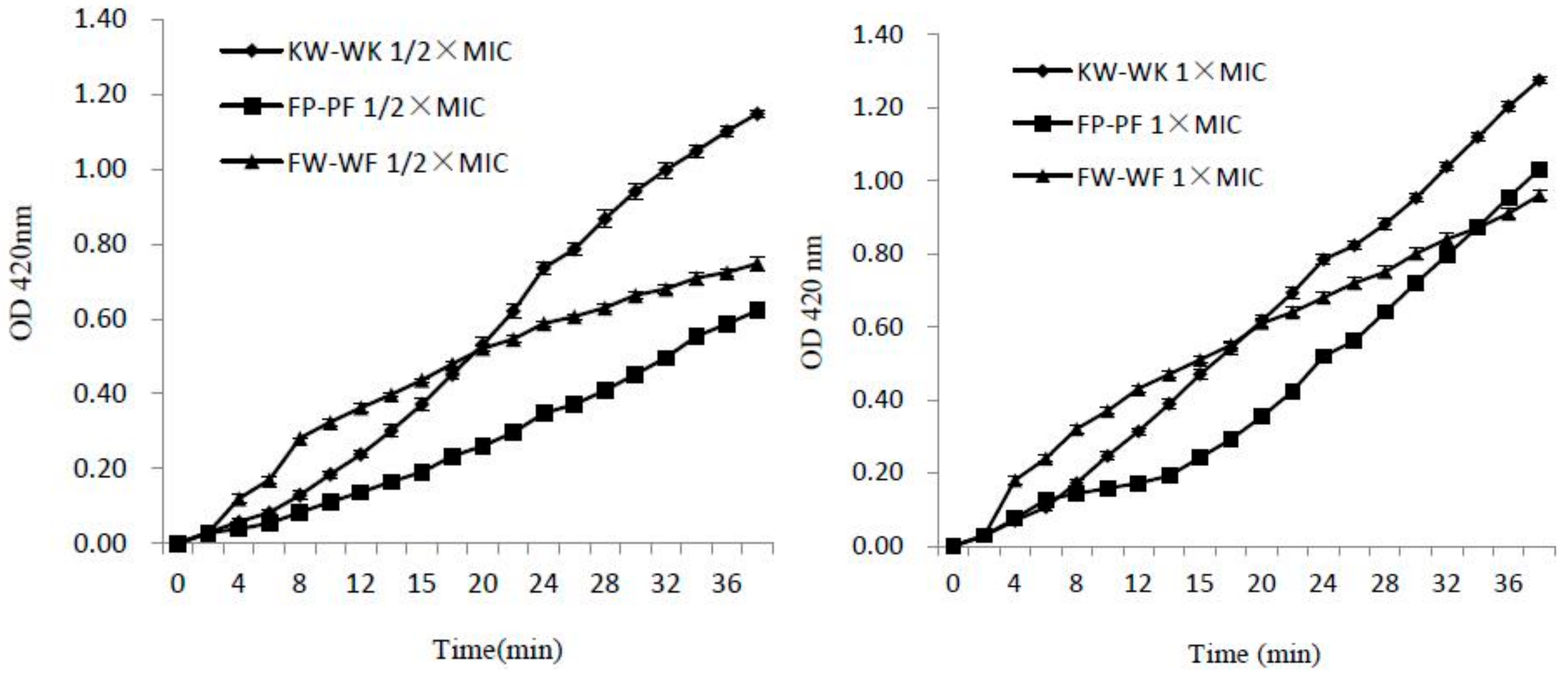
2.7.2. Inner Membrane Permeability
2.7.3. Cytoplasmic Membrane Electrical Potential
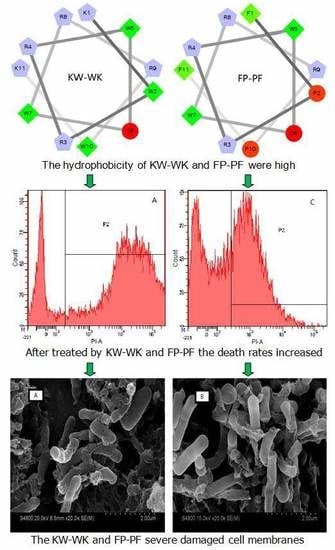
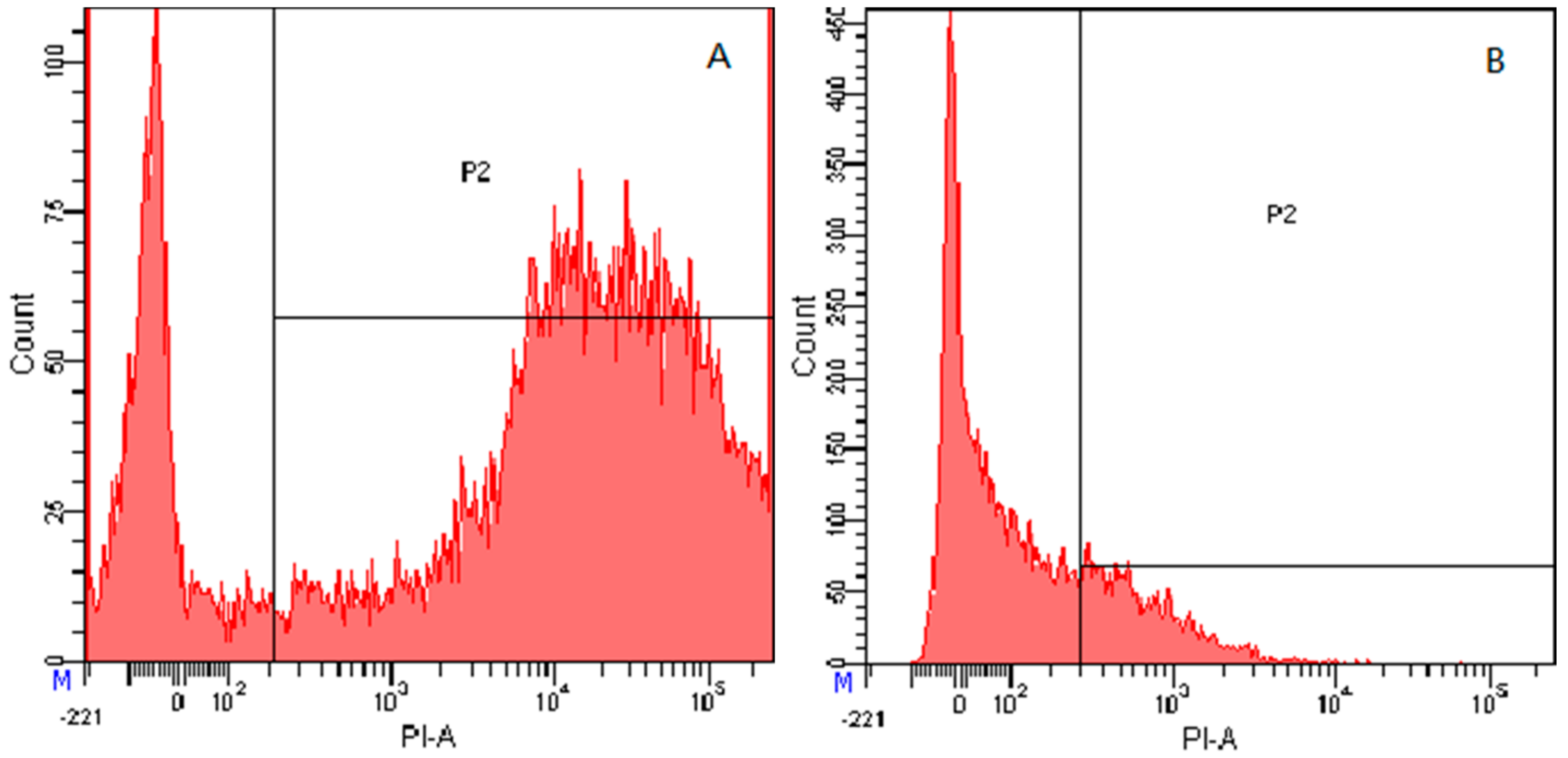
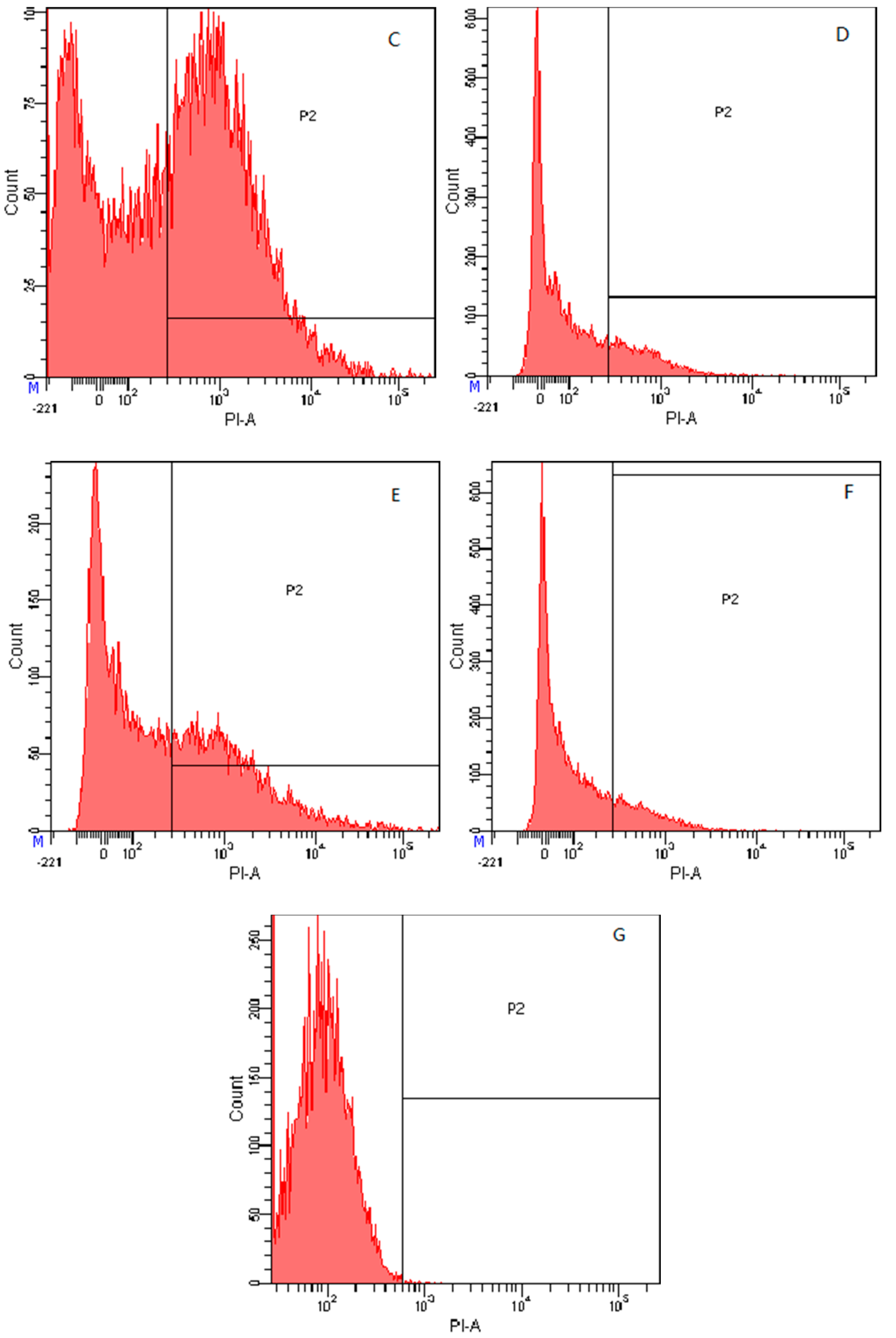
2.7.4. Flow Cytometry
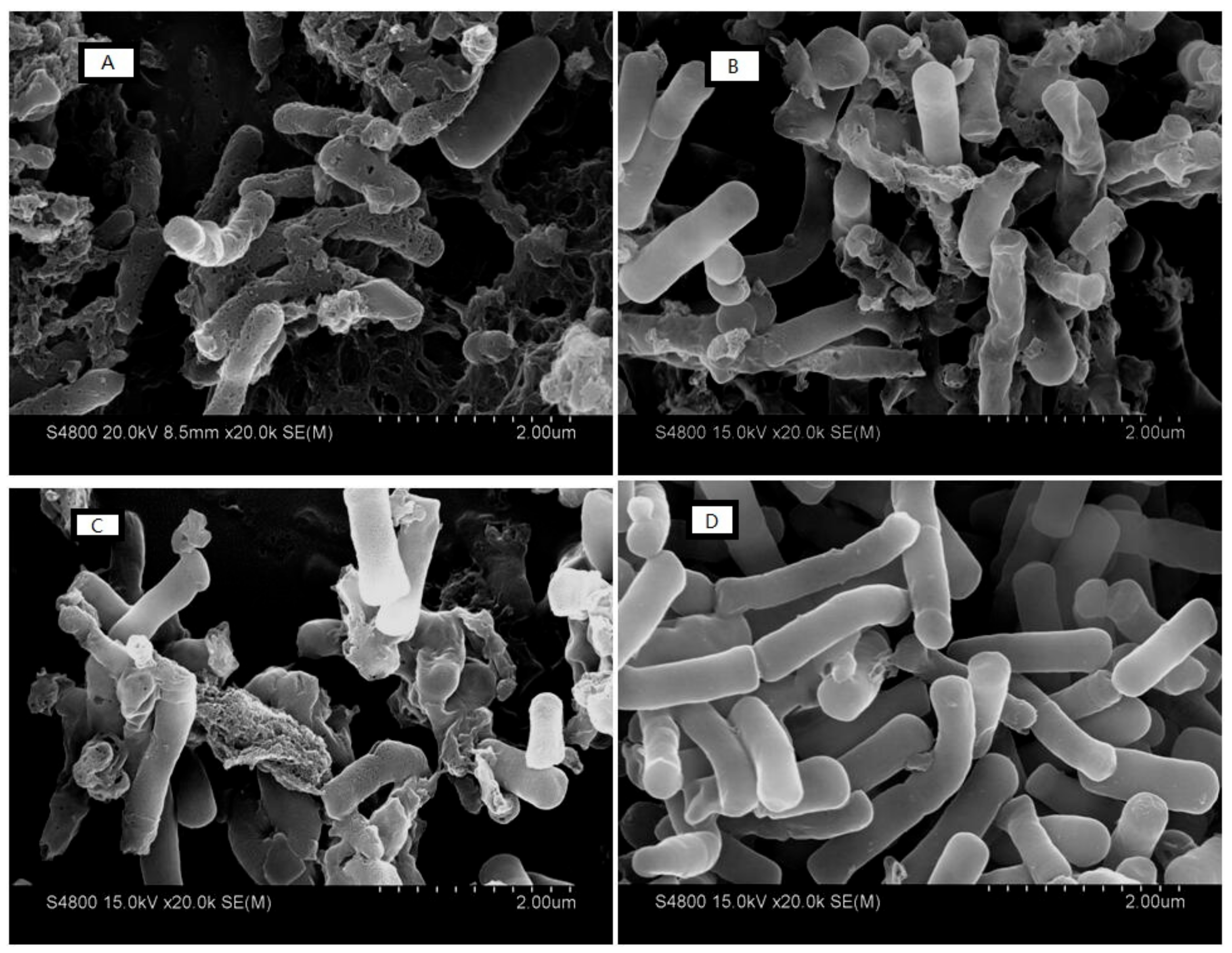
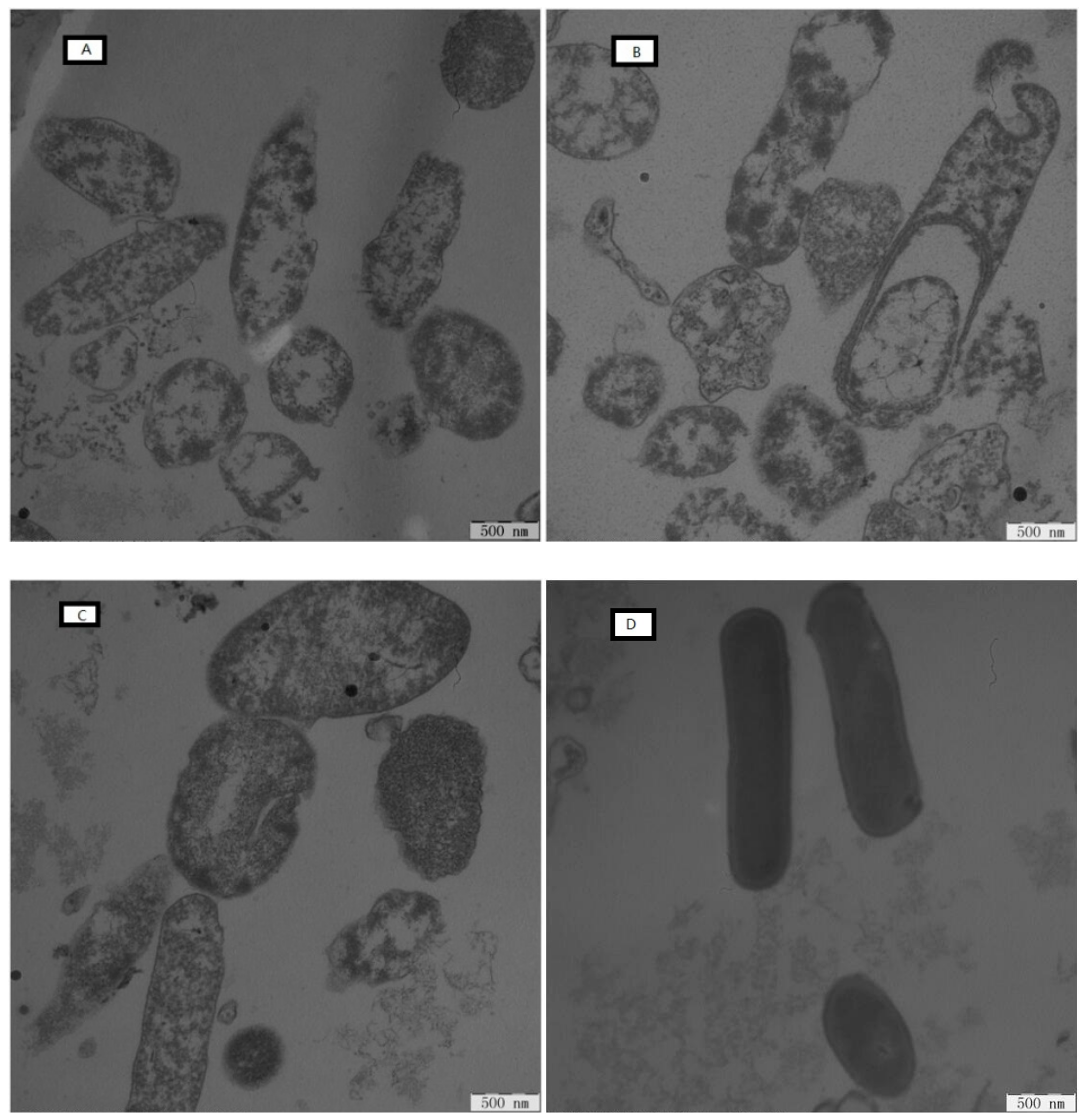
2.7.5. Electron Microscopic Studies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Design and Sequence Analysis of Peptides
4.3. Circular Dichroism (CD) Spectra
4.4. Antimicrobial Activity
4.5. Hemolytic Activity
4.6. Cytotoxicity
4.7. Stability
4.8. Mechanism of Action of the Peptides
4.8.1. Outer Membrane Permeability
4.8.2. Inner Membrane Permeability
4.8.3. Cytoplasmic Membrane Depolarization
4.8.4. Flow Cytometry
4.8.5. Electron Microscopic Studies
4.9. Statistical Methods
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPs | Antimicrobial Peptides |
| MIC | Minimum Inhibitory Concentration |
| hRBCs | Human Red Blood Cells |
| HEK293 | Human Embryonic Kidney 293 Cell |
| GM | Geometric Mean |
| TI | Treatment Index |
| MW | Molecular Weights |
| PBS | Phosphate-Buffered Saline |
| TFE | Trifluoroethanol |
| SDS | Sodium Dodecyl Sulphate |
| NPN | N-Phenyl-1-Naphthy Lamine |
| PI | Propidium Iodide |
References
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Caputo, G.A.; Vemparala, S.; Kuroda, K. Synthetic random copolymers as a molecular platform to mimic host-defense antimicrobial peptides. Bioconjugate Chem. 2017, 28, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Lohner, K. New strategies for novel antibiotics: Peptides targeting bacterial cell membranes. Gen. Physiol. Biophys. 2009, 28, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Parente, J.; Harris, S.M.; Woods, D.E.; Hancock, R.E.; Falla, T.J. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob. Agents Chemother. 2005, 49, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- Huertas, N.J.; Zjr, M.; Medina, R.F.; Jeg, C. Antimicrobial Activity of Truncated and Polyvalent Peptides Derived from the FKCRRQWQWRMKKGLA Sequence against Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923. Molecules 2017, 22, 987. [Google Scholar] [CrossRef] [PubMed]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. BBA-Gen. Subj. 2012, 1820, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Adão, R.; Nazmi, K.; Bolscher, J.G.; Bastos, M. C- and N-truncated antimicrobial peptides from LFampin 265–284: Biophysical versus microbiology results. J. Pharmacy Bioallied Sci. 2011, 3, 60. [Google Scholar]
- Liu, Y.; Han, F.; Xie, Y.; Wang, Y. Comparative antimicrobial activity and mechanism of action of bovine lactoferricin-derived synthetic peptides. Biometals 2011, 24, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.J.; Schibli, D.J.; Jing, W.; Lohmeiervogel, E.M.; Epand, R.F.; Epand, R.M. Towards a structure-function analysis of bovine lactoferricin and related tryptophan- and arginine-containing peptides. Biochem. Cell Biol. 2002, 80, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Takase, M.; Bellamy, W.; Shimamura, S. A review: The active peptide of lactoferrin. Acta Paediatr. Jpn. Overseas Ed. 1994, 36, 585–591. [Google Scholar] [CrossRef]
- Legrand, D.; Elass, E.; Carpentier, M.; Mazurier, J. Lactoferrin: A modulator of immune and inflammatory responses. Cell. Mol. Life Sci. CMLS 2005, 62, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.L.; Song, Y.M.; Park, Y.; Park, K.H.; Yang, S.T.; Kim, J.I.; Park, I.S.; Hahm, K.S.; Shin, S.Y. Substitution of the leucine zipper sequence in melittin with peptoid residues affects self-association, cell selectivity, and mode of action. Biochim. Biophys. Acta 2007, 1768, 1506. [Google Scholar] [CrossRef] [PubMed]
- Lata, S.; Sharma, B.K.; Raghava, G. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kohn, E.M.; Shirley, D.J.; Arotsky, L.; Picciano, A.M.; Ridgway, Z.; Urban, M.W.; Carone, B.R.; Caputo, G.A. Role of Cationic Side Chains in the Antimicrobial Activity of C18G. Molecules 2018, 23, 329. [Google Scholar] [CrossRef] [PubMed]
- Jean, K.D.S.; Henderson, K.D.; Chrom, C.L.; Abiuso, L.E.; Renn, L.M.; Caputo, G.A. Effects of Hydrophobic Amino Acid Substitutions on Antimicrobial Peptide Behavior. Probiotics Antimicrob. Proteins 2017, 10, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.T.; Song, Y.S.; Kim, J.I. Interaction mode of a symmetric Trp-rich undeca peptide PST11-RK with lipid bilayers. FEBS Lett. 2007, 581, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, Z.; Cao, S.; Wang, H.; Jiang, C.; Hussain, M.; Pang, S. Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein. Molecules 2018, 23, 1220. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.K.; Ahmad, A.; Asthana, N.; Azmi, S.; Srivastava, R.M.; Srivastava, S.; Verma, R.; Vishwakarma, A.L.; Ghosh, J.K. Cell-selective lysis by novel analogues of melittin against human red blood cells and Escherichia coli. Biochemistry 2010, 49, 7920–7929. [Google Scholar] [CrossRef] [PubMed]
- Unger, T.; Oren, Z.; Shai, Y. The Effect of Cyclization of Magainin 2 and Melittin Analogues on Structure, Function, and Model Membrane Interactions: Implication to Their Mode of Action. Biochemistry 2001, 40, 6388–6397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.L.; Lan, H.; Park, Y.; Yang, S.T.; Kim, J.I.; Park, I.S.; You, H.J.; Lee, J.S.; Yong, S.P.; Kim, Y. Effects of Pro → Peptoid Residue Substitution on Cell Selectivity and Mechanism of Antibacterial Action of Tritrpticin-Amide Antimicrobial Peptide. Biochemistry 2006, 45, 13007–13017. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.Y.; Andrés, M.T.; Fierro, J.F.; Yeaman, M.R. The gamma-core motif correlates with antimicrobial activity in cysteine-containing kaliocin-1 originating from transferrins. Biochim. Biophys. Acta 2007, 1768, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Ulvatne, H.; Vorland, L.H. Bactericidal kinetics of 3 lactoferricins against Staphylococcus aureus and Escherichia coli. Scand. J. Infect. Dis. 2001, 33, 507–511. [Google Scholar] [PubMed]
- Louisjeune, C.; Andradenavarro, M.A.; Pereziratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins-Struct. Funct. Bioinform. 2012, 80, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Benz, R.; Hancock, R.E. Influence of proline residues on the antibacterial and synergistic activities of alpha-helical peptides. Biochemistry 1999, 38, 8102–8111. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agent 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Shan, A.S.; Dong, N.; Gu, Y.; Sun, W.Y.; Hu, W.N.; Feng, X.J. Cell selectivity and interaction with model membranes of Val/Arg-rich peptides. J. Pept. Sci. 2011, 17, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Chau, J.K.; Perry, N.A.; De, B.L.; Zaat, S.A.; Vogel, H.J. Serum Stabilities of Short Tryptophan- and Arginine-Rich Antimicrobial Peptide Analogs. PLoS ONE 2010, 5, e12684. [Google Scholar] [CrossRef] [PubMed]
- Fernándezvidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding amphipathic helices into membranes: Amphiphilicity trumps hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Phoenix, D.A.; Harris, F.; Wallace, J.; Dennison, S.R. Amphiphilic α-Helical Antimicrobial Peptides and Their Structure/Function Relationships. Protein Pept. Lett. 2005, 12, 31–39. [Google Scholar]
- Haney, E.F.; Nazmi, K.; Bolscher, J.G.; Vogel, H.J. Structural and biophysical characterization of an antimicrobial peptide chimera comprised of lactoferricin and lactoferrampin. Biochim. Biophys. Acta (BBA)-Biomembr. 2012, 1818, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Vasil, A.I.; Hale, J.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Adv. Exp. Med. Biol. 2008, 90, 369–383. [Google Scholar]
- Soman, N.R.; Baldwin, S.L.; Hu, G.; Marsh, J.N.; Lanza, G.M.; Heuser, J.E.; Arbeit, J.M.; Wickline, S.A.; Schlesinger, P.H. Molecularly targeted nanocarriers deliver the cytolytic peptide melittin specifically to tumor cells in mice, reducing tumor growth. J. Clin. Investig. 2009, 119, 2830–2842. [Google Scholar] [CrossRef] [PubMed]
- Schnaider, L.; Brahmachari, S.; Schmidt, N.W.; Mensa, B.; Shahamniv, S.; Bychenko, D.; Adlerabramovich, L.; Shimon, L.J.W.; Kolusheva, S.; Degrado, W.F. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat. Commun. 2017, 8, 1365. [Google Scholar] [CrossRef] [PubMed]
- Frecer, V.; Ho, B.; Ding, J.L. De Novo Design of Potent Antimicrobial Peptides. Antimicrob. Agents Chemother. 2004, 48, 3349–3357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, H.; Zhang, H.; Wang, L.; Qian, H.; Qi, X. An antimicrobial peptide screened from casein hydrolyzate by Saccharomyces cerevisiae cell membrane affinity method. Food Control 2015, 50, 413–422. [Google Scholar] [CrossRef]
- Arias, M.; Mcdonald, L.J.; Haney, E.F.; Nazmi, K.; Bolscher, J.G.M.; Vogel, H.J. Bovine and human lactoferricin peptides: Chimeras and new cyclic analogs. Biometals 2014, 27, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hao, D.; Chen, Y.; Xu, Y.; Tan, J.; Huang, Y.; Li, F.; Chen, Y. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an α-helical antibacterial peptide against bacteria. Peptides 2011, 32, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Dong, N.; Shan, A.S.; Lv, Y.F.; Li, Y.Z.; Chen, Z.H.; Cheng, B.J.; Li, Z.Y. Biochemical property and membrane-peptide interactions of de novo antimicrobial peptides designed by helix-forming units. Amino Acids 2012, 43, 2527–2536. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; Lópezotín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chou, S.; Wang, J.; Shao, C.; Li, W.; Zhu, X.; Shan, A. Antimicrobial activity and membrane-active mechanism of tryptophan zipper-like β-hairpin antimicrobial peptides. Amino Acids 2015, 47, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Aquila, M.; Benedusi, M.; Koch, K.W.; Dell’Orco, D.; Rispoli, G. Divalent cations modulate membrane binding and pore formation of a potent antibiotic peptide analog of alamethicin. Cell Calcium 2013, 53, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Chaithanya, E.R.; Philip, R.; Sathyan, N.; Kumar, P.R.A. Molecular Characterization and Phylogenetic Analysis of a Histone-Derived Antimicrobial Peptide Teleostin from the Marine Teleost Fishes, Tachysurus jella and Cynoglossus semifasciatus. ISRN Mol. Biol. 2013, 2013, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Lv, Y.F.; Gu, Y.; Dong, N.; Li, D.S.; Shan, A.S. Rational design of cationic antimicrobial peptides by the tandem of leucine-rich repeat. Amino Acids 2013, 44, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Zweytick, D.; Deutsch, G.; Andrä, J.; Blondelle, S.E.; Vollmer, E.; Jerala, R.; Lohner, K. Studies on Lactoferricin-derived Escherichia coli Membrane-active Peptides Reveal Differences in the Mechanism of N-Acylated Versus Nonacylated Peptides. J. Biol. Chem. 2011, 286, 21266–21276. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Gómez, S.; Ferrer-Espada, R.; Stewart, P.S.; Pitts, B.; Lohner, K.; Tejada, G.M.D. Antimicrobial activity of synthetic cationic peptides and lipopeptides derived from human lactoferricin against Pseudomonas aeruginosa planktonic cultures and biofilms. BMC Microbiol. 2015, 15, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, L.; Wang, J.; Ma, Z.; Xu, W.; Li, J.; Shan, A. Characterization of antimicrobial activity and mechanisms of low amphipathic peptides with different α-helical propensity. Acta Biomater. 2015, 18, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Leishangthem, G.D.; Gill, K.; Singh, A.K.; Das, S.; Singh, K.; Xess, I.; Dinda, A.; Kapil, A.; Patro, I.K. A novel antimicrobial peptide derived from modified N-terminal domain of bovine lactoferrin: Design, synthesis, activity against multidrug-resistant bacteria and Candida. Biochim. Biophys. Acta (BBA)-Biomembr. 2013, 1828, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.; Wu, D.; Li, W.; Zheng, X.; Li, W.; Shan, A. High Specific Selectivity and Membrane-Active Mechanism of Synthetic Cationic Hybrid Antimicrobial Peptides Based on the Peptide FV7. Int. J. Mol. Sci. 2017, 18, 339. [Google Scholar] [CrossRef] [PubMed]
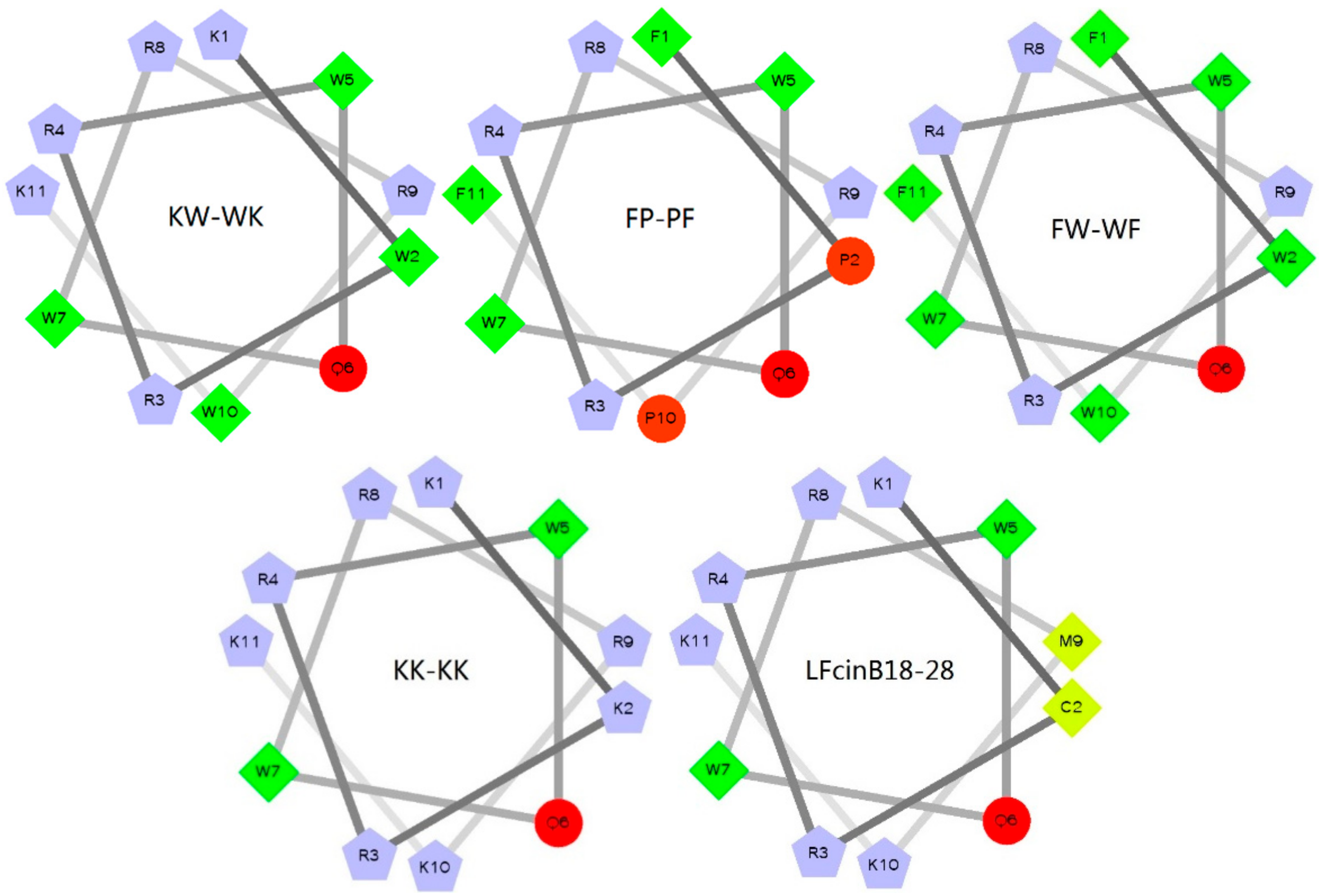

| Peptides | Sequence | Theoretical MW | Measured MW a | Net Charge | μH b | H c | Purity |
|---|---|---|---|---|---|---|---|
| KW-WK | KWRRWQWRRWK-NH2 | 1772.1 | 1771.13 | +7 | 0.441 | 0.251 | 96.94% |
| FP-PF | FPRRWQWRRPF-NH2 | 1631.9 | 1630.94 | +5 | 0.159 | 0.478 | 97.58% |
| FW-WF | FWRRWQWRRRWF-NH2 | 1810.1 | 1809.70 | +6 | 0.054 | 0.609 | 97.31% |
| KK-KK | KKRRWQWRRKK-NH2 | 1656.0 | 1655.05 | +9 | 0.010 | −0.338 | 95.21% |
| LFcinB18–28 | KCRRWQWRMKK-NH2 | 1605.9 | 1605.02 | +7 | 0.420 | 0.095 | 95.81% |
| MIC a (μM) | |||||
|---|---|---|---|---|---|
| KW-WK | FP-PF | FW-WF | KK-KK | LFcinB18–28 | |
| Gram-negative bacteria | |||||
| E. coli ATCC 25922 | 4 | 4 | 4 | 32 | 16 |
| E. coli UB 1005 | 8 | 8 | 16 | 32 | 128 |
| Salmonella typhimurium C 7731 | 16 | 64 | 16 | >128 | >128 |
| Salmonella typhimurium ATCC 14028 | 32 | 32 | 128 | >128 | >128 |
| Salmonella pullorum C 7913 | 16 | 16 | 8 | 128 | >128 |
| Salmonella enterica subsp. CMCC 50071 | 128 | 128 | >128 | 128 | >128 |
| Gram-positive bacteria | |||||
| S. aureus ATCC 29213 | 8 | 32 | >128 | >128 | >128 |
| S. aureus ATCC 25923 | 32 | 32 | 16 | 128 | 128 |
| S. epidermidis ATCC 12228 | 8 | 64 | 8 | >128 | >128 |
| MHC b (μM) | >256 | >256 | 8 | >256 | >256 |
| GM c | 28.00 | 42.22 | 78.67 | 163.55 | 200.89 |
| TI d | 9.14 | 6.06 | 0.10 | 1.56 | 1.28 |
| Peptides | Na+ | K+ | Mg2+ | Ca2+ | Zn2+ | Fe3+ | NH4+ | Control |
|---|---|---|---|---|---|---|---|---|
| KW-WK | 8 | 4 | 8 | 8 | 8 | 4 | 8 | 4 |
| FP-PF | 64 | 8 | 8 | 8 | 8 | 4 | 8 | 4 |
| FW-WF | 8 | 4 | 4 | 8 | 4 | 4 | 2 | 4 |
| KK-KK | 16 | 16 | 64 | 16 | 8 | 8 | 8 | 32 |
| LFcinB18–28 | 16 | 16 | 16 | 16 | 8 | 4 | 8 | 16 |
| Peptides | 100 °C | Control |
|---|---|---|
| KW-WK | 4 | 4 |
| FP-PF | 8 | 4 |
| FW-WF | 8 | 4 |
| KK-KK | >128 | 32 |
| LFcinB18–28 | 16 | 16 |
| Peptides | Trypsin | Pepsin | Papain | Protease K | Control |
|---|---|---|---|---|---|
| KW-WK | >128 | 16 | 8 | 8 | 4 |
| FP-PF | 128 | 16 | 8 | 4 | 4 |
| FW-WF | >128 | >128 | 4 | 16 | 4 |
| KK-KK | >128 | >128 | >128 | >128 | 32 |
| LFcinB18–28 | >128 | >128 | >128 | >128 | 16 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Li, Y.; Cao, S.; Wang, H.; Jiang, C.; Pang, S.; Hussain, M.A.; Hou, J. Antibacterial Activity and Mechanism of Action of Bovine Lactoferricin Derivatives with Symmetrical Amino Acid Sequences. Int. J. Mol. Sci. 2018, 19, 2951. https://doi.org/10.3390/ijms19102951
Sun C, Li Y, Cao S, Wang H, Jiang C, Pang S, Hussain MA, Hou J. Antibacterial Activity and Mechanism of Action of Bovine Lactoferricin Derivatives with Symmetrical Amino Acid Sequences. International Journal of Molecular Sciences. 2018; 19(10):2951. https://doi.org/10.3390/ijms19102951
Chicago/Turabian StyleSun, Changbao, Yingying Li, Songsong Cao, Haimei Wang, Chenggang Jiang, Shiyue Pang, Muhammad Altaf Hussain, and Juncai Hou. 2018. "Antibacterial Activity and Mechanism of Action of Bovine Lactoferricin Derivatives with Symmetrical Amino Acid Sequences" International Journal of Molecular Sciences 19, no. 10: 2951. https://doi.org/10.3390/ijms19102951
APA StyleSun, C., Li, Y., Cao, S., Wang, H., Jiang, C., Pang, S., Hussain, M. A., & Hou, J. (2018). Antibacterial Activity and Mechanism of Action of Bovine Lactoferricin Derivatives with Symmetrical Amino Acid Sequences. International Journal of Molecular Sciences, 19(10), 2951. https://doi.org/10.3390/ijms19102951