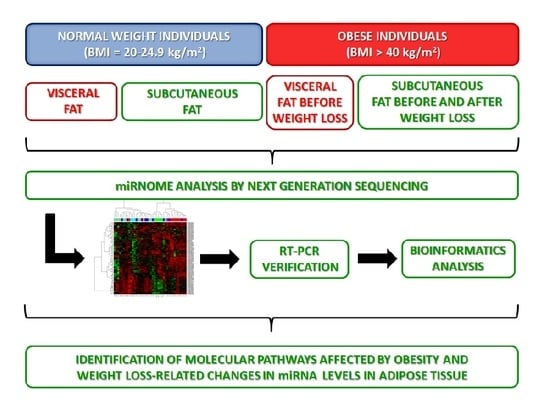
NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue
Abstract
1. Introduction
2. Results
2.1. miRNAs Expression in Adipose Tissue
2.2. Weight Loss Is Associated with Change of the SAT miRNA Profile
2.3. miRNA Profile in Adipose Tissue after Weight Loss Differs from That in Adipose Tissue of Normal-Weight Individuals
2.4. Real Time PCR Verification of the NGS Results
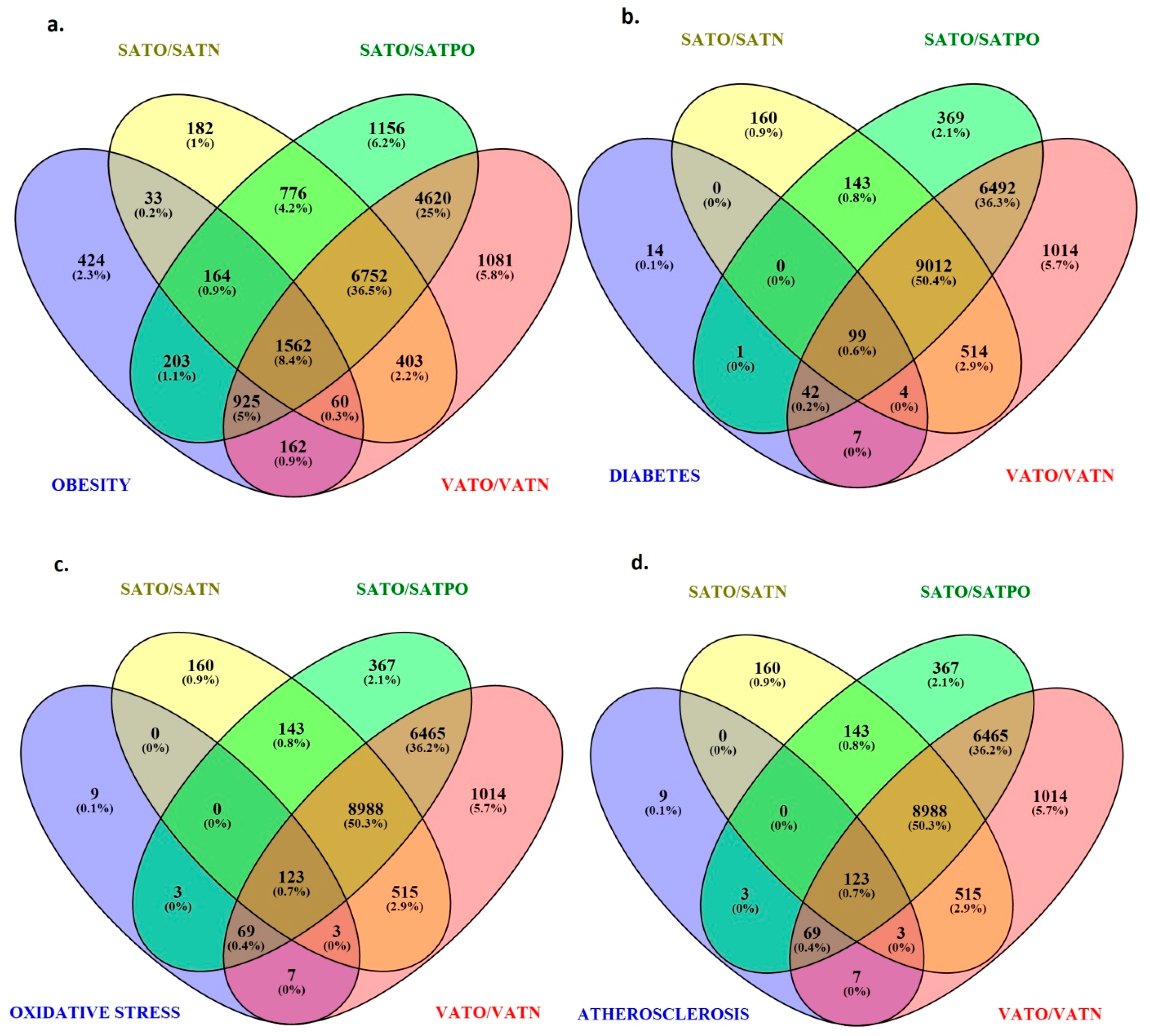
2.5. Identification of Target Genes Regulated by Differentially Expressed miRNAs in Adipose Tissues
2.6. Molecular Pathways Regulated by Differentially Expressed miRNAs
3. Discussion
4. Materials and Methods
4.1. Study Groups
4.2. miRNA Expression Analysis by Next Generation Sequencing
4.3. Validation of miRNA Expression by Real-Time PCR
4.4. Identification of miRNA Target Genes and Their Molecular Pathways
4.5. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Samaras, K.; Botelho, N.K.; Chisholm, D.J.; Lord, R.V. Subcutaneous and visceral adipose tissue gene expression of serum adipokines that predict type 2 diabetes. Obesity 2010, 18, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Neville, M.J.; Collins, J.M.; Gloyn, A.L.; McCarthy, M.I.; Karpe, F. Comprehensive human adipose tissue mRNA and microRNA endogenous control selection for quantitative real-time-PCR normalization. Obesity 2011, 19, 888–892. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.S.; Pérusse, J.R.; Lavoie, M.È.; Sladek, R.; Madiraju, S.R.; Ruderman, N.B.; Coulombe, B.; Prentki, M.; Rabasa-Lhoret, R. Increased subcutaneous adipose tissue expression of genes involved in glycerolipid-fatty acid cycling in obese insulin-resistant versus -sensitive individuals. J. Clin. Endocrinol. Metab. 2014, 99, E2518–E2528. [Google Scholar] [CrossRef] [PubMed]
- Kirby, T.J.; Walton, R.G.; Finlin, B.; Zhu, B.; Unal, R.; Rasouli, N.; Peterson, C.A.; Kern, P.A. Integrative mRNA-microRNA analyses reveal novel interactions related to insulin sensitivity in human adipose tissue. Physiol. Genom. 2016, 48, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009, 58, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Oger, F.; Gheeraert, C.; Mogilenko, D.; Benomar, Y.; Molendi-Coste, O.; Bouchaert, E.; Caron, S.; Dombrowicz, D.; Pattou, F.; Duez, H.; et al. Cell-specific dysregulation of microRNA expression in obese white adipose tissue. J. Clin. Endocrinol. Metab. 2014, 99, 2821–2833. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Nonell, L.; Puigdecanet, E.; Rodriquez-Hermosa, J.I.; Rovira, O.; Xifra, G.; Guerra, E.; Moreno, M.; et al. Surgery-Induced Weight Loss Is Associated With the Downregulation of Genes Targeted by MicroRNAs in Adipose Tissue. J. Clin. Endocrinol. Metab. 2015, 100, E1467–E1476. [Google Scholar] [CrossRef] [PubMed]
- Klöting, N.; Berthold, S.; Kovacs, P.; Schön, M.R.; Fasshauer, M.; Ruschke, K.; Stumvoll, M.; Blüher, M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE 2009, 4, e4699. [Google Scholar] [CrossRef] [PubMed]
- Swierniak, M.; Wojcicka, A.; Czetwertynska, M.; Stachlewska, E.; Maciag, M.; Wiechno, W.; Gornicka, B.; Bogdanska, M.; Koperski, L.; de la Chapelle, A.; et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, E1401–E1409. [Google Scholar] [CrossRef] [PubMed]
- Wojcicka, A.; Swierniak, M.; Kornasiewicz, O.; Gierlikowski, W.; Maciag, M.; Kolanowska, M.; Kotlarek, M.; Gornicka, B.; Koperski, L.; Niewinski, G.; et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2014, 53, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, Y.; Niu, Y.; Chen, W.; Wang, Q.; Xiao, S.; Li, A.; Xie, Y.; Li, J.; Zhao, X.; et al. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genom. 2010, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Nardelli, C.; Iaffaldano, L.; Pilone, V.; Labruna, G.; Ferrigno, M.; Carlomagno, N.; Dodaro, C.A.; Forestieri, P.; Buono, P.; Salvatore, F.; et al. Changes in the MicroRNA Profile Observed in the Subcutaneous Adipose Tissue of Obese Patients after Laparoscopic Adjustable Gastric Banding. J. Obes. 2017, 2017, 6754734. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.; Gburcik, V.; Petrovic, N.; Gallagher, I.J.; Nedergaard, J.; Cannon, B.; Timmons, J.A. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Van Dooren, E.; Mathieu, C.; Holvoet, P. Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. PLoS ONE 2012, 7, e32794. [Google Scholar] [CrossRef] [PubMed]
- Ogando, J.; Tardáguila, M.; Díaz-Alderete, A.; Usategui, A.; Miranda-Ramos, V.; Martínez-Herrera, D.J.; de la Fuente, L.; García-León, M.J.; Moreno, M.C.; Escudero, S.; et al. Notch-regulated miR-223 targets the aryl hydrocarbon receptor pathway and increases cytokine production in macrophages from rheumatoid arthritis patients. Sci. Rep. 2016, 6, 20223. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Obin, M.S. Obesity and the role of adipose tissue in inflammation and metabolism. Am. J. Clin. Nutr. 2006, 83, 461S–465S. [Google Scholar] [PubMed]
- Jonas, M.I.; Kurylowicz, A.; Bartoszewicz, Z.; Lisik, W.; Jonas, M.; Wierzbicki, Z.; Chmura, A.; Pruszczyk, P.; Puzianowska-Kuznicka, M. Interleukins 6 and 15 Levels Are Higher in Subcutaneous Adipose Tissue, but Obesity Is Associated with Their Increased Content in Visceral Fat Depots. Int. J. Mol. Sci. 2015, 16, 25817–25830. [Google Scholar] [CrossRef] [PubMed]
- Schinzari, F.; Tesauro, M.; Cardillo, C. Endothelial and perivascular adipose tissue abnormalities in obesity-related vascular dysfunction: Novel targets for treatment. J. Cardiovasc. Pharmacol. 2017, 69, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Alkhouri, N.; Gornicka, A.; Berk, M.P.; Thapaliya, S.; Dixon, L.J.; Kashyap, S.; Schauer, P.R.; Feldstein, A.E. Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis. J. Biol. Chem. 2010, 285, 3428–3438. [Google Scholar] [CrossRef] [PubMed]
- Vergoni, B.; Cornejo, P.J.; Gilleron, J.; Djedaini, M.; Ceppo, F.; Jacquel, A.; Bouget, G.; Ginet, C.; Gonzalez, T.; Maillet, J.; et al. DNA Damage and the Activation of the p53 Pathway Mediate Alterations in Metabolic and Secretory Functions of Adipocytes. Diabetes 2016, 65, 3062–3074. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A.; Jonas, M.; Lisik, W.; Jonas, M.; Wicik, Z.A.; Wierzbicki, Z.; Chmura, A.; Puzianowska-Kuznicka, M. Obesity is associated with a decrease in expression but not with the hypermethylation of thermogenesis-related genes in adipose tissues. J. Transl. Med. 2015, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Lopategi, A.; López-Vicario, C.; Alcaraz-Quiles, J.; García-Alonso, V.; Rius, B.; Titos, E.; Clària, J. Role of bioactive lipid mediators in obese adipose tissue inflammation and endocrine dysfunction. Mol. Cell. Endocrinol. 2016, 419, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Capobianco, V.; Nardelli, C.; Ferrigno, M.; Iaffaldano, L.; Pilone, V.; Forestieri, P.; Zambrano, N.; Sacchetti, L. miRNA and protein expression profiles of visceral adipose tissue reveal miR-141/YWHAG and miR-520e/RAB11A as two potential miRNA/protein target pairs associated with severe obesity. J. Proteome Res. 2012, 11, 3358–3369. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.W.; Zhao, J.Y.; Li, S.F.; Huang, J.L.; Qiu, Y.R.; Ma, X.; Wu, S.G.; Chen, Z.P.; Hu, Y.R.; Yang, J.Y.; et al. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Huang, F.; Gu, X.; Zhang, M.; Wen, J.; Wang, X.; You, L.; Cui, X.; Ji, C.; Guo, X. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 2016, 7, 40830–40845. [Google Scholar] [CrossRef] [PubMed]
- De Aguiar Vallim, T.Q.; Tarling, E.J.; Kim, T.; Civelek, M.; Baldán, Á.; Esau, C.; Edwards, P.A. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 2013, 112, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.M.; Min, K.H.; Lee, W. Induction of miR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.R.; Lee, S.; Jung, J.W.; Hong, I.S.; Kim, H.S.; Seo, Y.; Shin, T.H.; Kang, K.S. MicroRNA-141-3p plays a role in human mesenchymal stem cell aging by directly targeting ZMPSTE24. J. Cell Sci. 2013, 126, 5422–5431. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Bai, S.; Su, L.; Li, C.; Wu, J.; Xia, Z.; Xu, D. A subset of circulating microRNAs is expressed differently in patients with myocardial infarction. Mol. Med. Rep. 2015, 12, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Dezso, Z.; MacKenzie, C.; Oestreicher, J.; Agoulnik, S.; Byrne, M.; Bernier, F.; Yanagimachi, M.; Aoshima, K.; Oda, Y. Circulating miRNA biomarkers for Alzheimer’s disease. PLoS ONE 2013, 8, e69807. [Google Scholar] [CrossRef] [PubMed]
- Madden, C.J.; Tupone, D.; Cano, G.; Morrison, S.F. α2 Adrenergic receptor-mediated inhibition of thermogenesis. J. Neurosci. 2013, 33, 2017–2028. [Google Scholar] [CrossRef] [PubMed]
- Spoto, B.; Di Betta, E.; Mattace-Raso, F.; Sijbrands, E.; Vilardi, A.; Parlongo, R.M.; Pizzini, P.; Pisano, A.; Vermi, W.; Testa, A.; et al. Pro- and anti-inflammatory cytokine gene expression in subcutaneous and visceral fat in severe obesity. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Shie, F.S.; Shiao, Y.J.; Yeh, C.W.; Lin, C.H.; Tzeng, T.T.; Hsu, H.C.; Huang, F.L.; Tsay, H.J.; Liu, H.K. Obesity and Hepatic Steatosis Are Associated with Elevated Serum Amyloid Beta in Metabolically Stressed APPswe/PS1dE9 Mice. PLoS ONE 2015, 10, e0134531. [Google Scholar] [CrossRef] [PubMed]
- Young, P.; Cawthorne, M.A.; Levy, A.L.; Wilson, K. Reduced maximum capacity of glycolysis in brown adipose tissue of genetically obese, diabetic (db/db) mice and its restoration following treatment with a thermogenic beta-adrenoceptor agonist. FEBS Lett. 1984, 176, 16–20. [Google Scholar] [CrossRef]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A.; Owczarz, M.; Polosak, J.; Jonas, M.I.; Lisik, W.; Jonas, M.; Chmura, A.; Puzianowska-Kuznicka, M. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int. J. Obes. 2016, 40, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- miRWalk2.0: A Comprehensive Atlas of Predicted and Validated miRNA-Target Interactions. Available online: http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/ (accessed on 18 October 2017).
- Dweep, H.; Gretz, N.; Sticht, C. miRWalk database for miRNA-target interactions. Methods Mol. Biol. 2014, 1182, 289–305. [Google Scholar] [CrossRef] [PubMed]
- miRTarBase: The Experimentally Validated microRNA-Target Interactions Database. Available online: http://mirtarbase.mbc.nctu.edu.tw (accessed on 18 October 2017).
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
| SAT-O vs. SAT-N | SAT-PO vs. SAT-O | SAT-PO vs. SAT-N | ||||
|---|---|---|---|---|---|---|
| NGS | Real-Time PCR | NGS | Real-Time PCR | NGS | Real-Time PCR | |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Proadipogenic | ||||||
| hsa-miR-15a-5p * | −2.992 | −2.286 | ||||
| hsa-miR-21-5p * | 2.728 | |||||
| hsa-miR-107 | −3.057 | −3.734 | ||||
| hsa-miR-143-5p * | −2.126 | |||||
| hsa-miR-146b-3p | 7.253 | 2.710 | −4.722 | −2.972 | ||
| hsa-miR-150-5p * | 2.128 | |||||
| hsa-miR-194-5p | −3.029 | −3.645 | −2.109 | −7.00 | ||
| hsa-miR-199b-5p | 2.803 | |||||
| hsa-miR-210 | −2.130 | −2.617 | ||||
| hsa-miR-214-5p * | 2.266 | |||||
| hsa-miR-335-3p | 2.574 | |||||
| hsa-miR-450a-5p | 2.282 | 2.100 | ||||
| Antiadipogenic | ||||||
| hsa-miR-18a-3p | −3.703 | −3.699 | ||||
| hsa-miR-23a-5p | 2.093 | |||||
| hsa-miR-27a-5p * | 2.363 | 2.512 | ||||
| hsa-miR-29a-3p * | 2.010 | |||||
| hsa-miR-130b-3p | −4.053 | −2.787 | ||||
| hsa-miR-196a-5p | 4.031 | 3.162 | 4.047 | 2.97 | ||
| Involved in adipocyte browning | ||||||
| hsa-miR-125b-5p | 2.247 | 2.01 | ||||
| Involved in inflammation and immune response | ||||||
| hsa-miR-15b-3p * | −5.090 | −6.432 | ||||
| hsa-miR-16-2-3p * | −7.972 | −6.263 | ||||
| hsa-miR-20b-5p * | −2.936 | −3.518 | ||||
| hsa-miR-22-3p * | 3.001 | 2.011 | ||||
| hsa-miR-32-5p * | −2.452 | |||||
| hsa-miR-92b-3p * | 2.228 | |||||
| hsa-miR-93-5p | −3.602 | −3.358 | ||||
| hsa-miR-106a-5p * | −3.470 | −4.302 | ||||
| hsa-miR-122-5p * | −5.792 | |||||
| hsa-miR-125a-5p * | 2.150 | 1.96 | ||||
| hsa-miR-125b-1-3p * | 2.063 | |||||
| hsa-miR-142-3p * | −5.215 | |||||
| hsa-miR-142-5p * | −6.087 | −5.233 | ||||
| hsa-miR-146b-5p * | 5.077 | −3.986 | ||||
| hsa-miR-182-5p * | −15.039 | |||||
| hsa-miR-223-3p * | 5.804 | 3.041 | −27.825 | −8.036 | −4.794 | −2.72 |
| hsa-miR-3200-3p | −3.103 | −3.802 | ||||
| Involved in lipid metabolism | ||||||
| hsa-miR-106b-5p * | −3.778 | −3.984 | ||||
| hsa-miR-144-3p * | −11.390 | −14.187 | ||||
| hsa-miR-185-5p | −2.836 | −3.303 | ||||
| Involved in atherosclerosis | ||||||
| hsa-miR-365a-3p | 2.559 | |||||
| hsa-miR-365b-3p | 2.559 | |||||
| Involved in insulin signaling | ||||||
| hsa-miR-96-5p | −22.917 | −9.251 | −13.913 | −15.01 | ||
| hsa-miR-128 | −2.418 | |||||
| Involved in oncogenesis | ||||||
| hsa-let-7e-3p | 2.100 | |||||
| hsa-miR-16-5p | −2.755 | |||||
| hsa-miR-17-3p | −2.066 | |||||
| hsa-miR-18a-5p | −2.410 | −4.324 | ||||
| hsa-miR-18b-5p | −4.240 | −6.464 | ||||
| hsa-miR-25-3p | −2.289 | −2.448 | ||||
| hsa-miR-28-5p | 2.086 | |||||
| hsa-miR-95 | 3.118 | |||||
| hsa-miR-106b-3p | −3.636 | |||||
| hsa-miR-144-5p | −8.083 | −8.337 | ||||
| hsa-miR-183-5p | −14.823 | −3.723 | −20.421 | −7.62 | ||
| hsa-miR-192-5p | −3.234 | −3.408 | ||||
| hsa-miR-193b-5p | 2.500 | 1.97 | ||||
| hsa-miR-205-5p | −10.897 | −5.541 | −10.030 | −10.73 | ||
| hsa-miR-215 | 2.015 | |||||
| hsa-miR-223-5p | 2.894 | −5.798 | −2.003 | |||
| hsa-miR-301b | 2.972 | |||||
| hsa-miR-330-3p | 2.452 | |||||
| hsa-miR-340-3p | 2.073 | |||||
| hsa-miR-361-3p | 2.504 | |||||
| hsa-miR-363-3p | −5.212 | −4.293 | ||||
| hsa-miR-423-5p | −2.430 | |||||
| hsa-miR-424-3p | −2.020 | |||||
| hsa-miR-425-5p | −3.062 | −2.103 | ||||
| hsa-miR-451a | −9.299 | −11.488 | ||||
| hsa-miR-483-3p | 2.681 | 2.860 | ||||
| hsa-miR-489 | 2.440 | |||||
| hsa-miR-501-3p | −3.074 | |||||
| hsa-miR-518b | 2.705 | |||||
| hsa-miR-548d-5p | −3.669 | −3.766 | ||||
| hsa-miR-550a-3p | −3.100 | |||||
| hsa-miR-576-5p | −3.083 | −3.587 | ||||
| hsa-miR-584-5p | −2.741 | −2.377 | ||||
| hsa-miR-615-3p | 3.163 | |||||
| hsa-miR-628-5p | 2.569 | 2.504 | ||||
| hsa-miR-629-5p | −2.241 | −2.261 | ||||
| hsa-miR-652-3p | −2.807 | −2.653 | ||||
| hsa-miR-671-3p | 2.878 | |||||
| hsa-miR-874 | 2.070 | |||||
| hsa-miR-891a | 2.984 | |||||
| hsa-miR-941 | 2.404 | −2.239 | ||||
| hsa-miR-1260b | 2.460 | 1.524 | 4.854 | 2.30 | ||
| hsa-miR-1271-5p | 2.540 | |||||
| hsa-miR-1285-3p | −2.396 | |||||
| hsa-miR-3688-3p | −5.070 | −5.732 | ||||
| hsa-miR-4662a-5p | 3.410 | |||||
| hsa-miR-4732-3p | −3.347 | −3.124 | ||||
| hsa-miR-5683 | 5.557 | |||||
| hsa-miR-6716-3p | 2.481 | |||||
| Involved in ageing | ||||||
| hsa-miR-141-3p | −5.518 | −8.255 | ||||
| hsa-miR-369-5p | 2.554 | |||||
| Involved in cardiovascular diseases | ||||||
| hsa-miR-589-5p | 2.193 | |||||
| hsa-miR-3615 | −5.278 | −4.355 | ||||
| Involved in neurodegenerative disorders | ||||||
| hsa-miR-15b-5p | −2.811 | −2.753 | ||||
| hsa-miR-132-5p | 2.921 | |||||
| hsa-miR-151a-5p | 2.228 | |||||
| hsa-miR-548ay-5p | −3.505 | −3.731 | ||||
| hsa-miR-664a-3p | 2.102 | |||||
| hsa-miR-3607-3p | 2.706 | |||||
| hsa-miR-3909 | 2.865 | |||||
| Unknown function | ||||||
| hsa-miR-450b-5p | 3.284 | 3.723 | 2.605 | 7.13 | ||
| hsa-miR-486-3p | −12.206 | −11.022 | ||||
| hsa-miR-486-5p | −9.535 | −4.464 | −9.118 | −8.83 | ||
| VAT-O vs. SAT-O | ||
|---|---|---|
| NGS | Real-Time PCR | |
| Proadipogenic | ||
| hsa-miR-450a-5p | 2.023 | |
| Antiadipogenic | ||
| hsa-miR-23a-5p | −3.371 | |
| hsa-miR-27a-5p | −2.512 | |
| hsa-miR-29b-3p | 2.471 | |
| hsa-miR-29c-3p | 2.084 | |
| hsa-miR-196a-5p | −14.684 | −17.21 |
| Involved in adipogenesis | ||
| hsa-miR-125b-5p | 2.084 | 1.89 |
| Involved in adipocytes browning | ||
| hsa-miR-539-3p | 2.127 | |
| Involved in inflammation and immune response | ||
| hsa-miR-125a-5p | 2.615 | 1.98 |
| Insulin signaling | ||
| hsa-miR-33a-3p | 2.283 | |
| hsa-miR-33b-5p | 3.170 | |
| hsa-miR-96-5p | −2.887 | −2.23 |
| hsa-miR-133a | 2.720 | |
| hsa-miR-133b | 2.665 | |
| Involved in oncogenesis | ||
| hsa-let-7e-3p | 2.145 | |
| hsa-miR-31-5p | 10.078 | |
| hsa-miR-100-3p | 2.979 | |
| hsa-miR-100-5p | 2.647 | |
| hsa-miR-101-5p | 2.647 | |
| hsa-miR-136-5p | 2.562 | |
| hsa-miR-183-5p | −2.780 | −1.98 |
| hsa-miR-190a | 2.017 | |
| hsa-miR-193a-3p | 3.319 | 2.03 |
| hsa-miR-200a-3p | 5.333 | |
| hsa-miR-200b-3p | 5.784 | |
| hsa-miR-203a | 4.713 | |
| hsa-miR-204-5p | 6.024 | |
| hsa-miR-218-5p | 2.537 | |
| hsa-miR-337-3p | 2.051 | |
| hsa-miR-376c-3p | 2.908 | |
| hsa-miR-381-3p | 2.405 | |
| hsa-miR-424-5p | 2.573 | |
| hsa-miR-429 | 5.626 | |
| hsa-miR-486-5p | −2.872 | −2.67 |
| hsa-miR-487b | 2.002 | |
| hsa-miR-493-3p | 5.581 | |
| hsa-miR-493-5p | 5.259 | |
| hsa-miR-497-5p | 2.083 | |
| hsa-miR-532-3p | 2.006 | |
| hsa-miR-551b-3p | 4.019 | |
| hsa-miR-561-5p | 2.275 | |
| hsa-miR-615-3p | −3.246 | |
| hsa-miR-887 | 2.400 | |
| hsa-miR-1307-5p | 2.601 | |
| Involved in ageing | ||
| hsa-miR-299-3p | 3.240 | 1.98 |
| Involved in cardiovascular disorders | ||
| hsa-miR-423-3p | 2.013 | |
| hsa-miR-495-3p | 2.702 | |
| Involved in neurodegenerative disorders | ||
| hsa-miR-9-5p | 2.536 | |
| hsa-miR-101-3p | 2.979 | |
| hsa-miR-136-3p | 2.436 | |
| hsa-miR-376a-5p | 2.099 | |
| hsa-miR-382-3p | 2.184 | |
| hsa-miR-485-3p | 2.122 | |
| hsa-miR-3607-3p | 3.016 | |
| SAT-O vs. SAT-N | SAT-O vs. SAT-PO | SAT-N vs. SAT-PO | VAT-O vs. SAT-O |
|---|---|---|---|
| PPARA | PRKAR1A | VEGFA | ADRB2 |
| CASP9 | CCND1 | CREB1 | VEGFA |
| PPARGC1B | PTEN | BCL2 | BCL2 |
| IGF1R | CDKN1A | FLT1 | SCARB1 |
| FMR1 | HMGA2 | MDM2 | CDC42 |
| SLC26A2 | HMGA1 | CCR2 | PAX8 |
| RAD51B | XIAP | PCDH15 | SP1 |
| MBD4 | CCND2 | EXOC7 | SLC16A1 |
| FOXP1 | SMAD4 | ARL17A | BICD1 |
| RNASEL | CADM1 | PARD3 | CSNK1D |
| CAMTA1 | SPRED1 | BICD1 | MCL1 |
| BMPR2 | MBD1 | ADRA1A | IL1RAP |
| VIPR2 | HSPA1B | TMPO | CLTC |
| DBT | NOTCH2 | QKI | CELF2 |
| CDS1 | BRWD1 | DNAJC10 | ENAH |
| WHSC1L1 | E2F3 | ZNF496 | HSPA13 |
| CELF1 | MAPK9 | PCDH7 | |
| DAPK2 | NRG1 | HDGF | |
| PHF20 | SLC2A3 | DYNC1I1 | |
| CRYL1 | KIF6 | PPP1R12B | |
| CCDC13 | HMBOX1 | ||
| TPGS2 |
| Fold Enrichment | ||||
|---|---|---|---|---|
| SAT-O vs. SAT-N | SAT-PO vs. SAT-O | SAT-PO vs. SAT-N | VAT-O vs. SAT-O | |
| Pathways corrected * | ||||
| Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | 1.22 | −1.13 | −1.14 | 1.17 |
| Wnt signaling pathway (P00057) | −1.53 | |||
| CCKR signaling map (P06959) | 1.35 | |||
| Pathways non corrected ** | ||||
| Beta2 adrenergic receptor signaling pathway (P04378) | −2.46 | |||
| Interleukin signaling pathway (P00036) | 1.23 | |||
| Endothelin signaling pathway (P00019) | 1.46 | |||
| Apoptosis signaling pathway (P00006) | 1.25 | 1.81 | −1.23 | −1.47 |
| p53 pathway (P00059) | 1.30 | 1.78 | −1.28 | −1.68 |
| p53 pathway by glucose deprivation (P04397) | 1.63 | |||
| Alzheimer disease-presenilin pathway (P00004) | −1.28 | −1.47 | ||
| Glycolysis (P00024) | 3.55 | −2.64 | ||
| Cholesterol biosynthesis (P00014) | −2.44 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuryłowicz, A.; Wicik, Z.; Owczarz, M.; Jonas, M.I.; Kotlarek, M.; Świerniak, M.; Lisik, W.; Jonas, M.; Noszczyk, B.; Puzianowska-Kuźnicka, M. NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue. Int. J. Mol. Sci. 2018, 19, 66. https://doi.org/10.3390/ijms19010066
Kuryłowicz A, Wicik Z, Owczarz M, Jonas MI, Kotlarek M, Świerniak M, Lisik W, Jonas M, Noszczyk B, Puzianowska-Kuźnicka M. NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue. International Journal of Molecular Sciences. 2018; 19(1):66. https://doi.org/10.3390/ijms19010066
Chicago/Turabian StyleKuryłowicz, Alina, Zofia Wicik, Magdalena Owczarz, Marta Izabela Jonas, Marta Kotlarek, Michał Świerniak, Wojciech Lisik, Maurycy Jonas, Bartłomiej Noszczyk, and Monika Puzianowska-Kuźnicka. 2018. "NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue" International Journal of Molecular Sciences 19, no. 1: 66. https://doi.org/10.3390/ijms19010066
APA StyleKuryłowicz, A., Wicik, Z., Owczarz, M., Jonas, M. I., Kotlarek, M., Świerniak, M., Lisik, W., Jonas, M., Noszczyk, B., & Puzianowska-Kuźnicka, M. (2018). NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue. International Journal of Molecular Sciences, 19(1), 66. https://doi.org/10.3390/ijms19010066