Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review
Abstract
1. Introduction
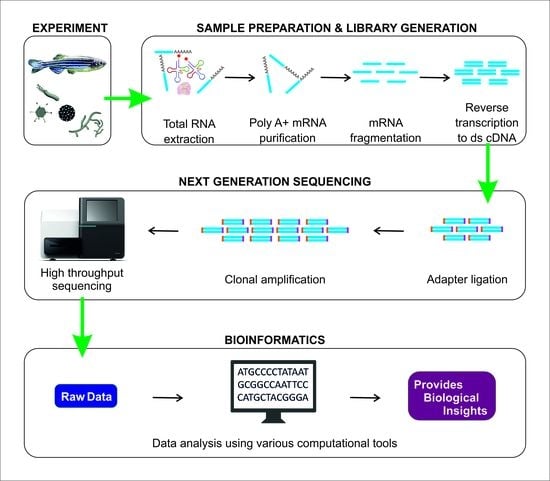
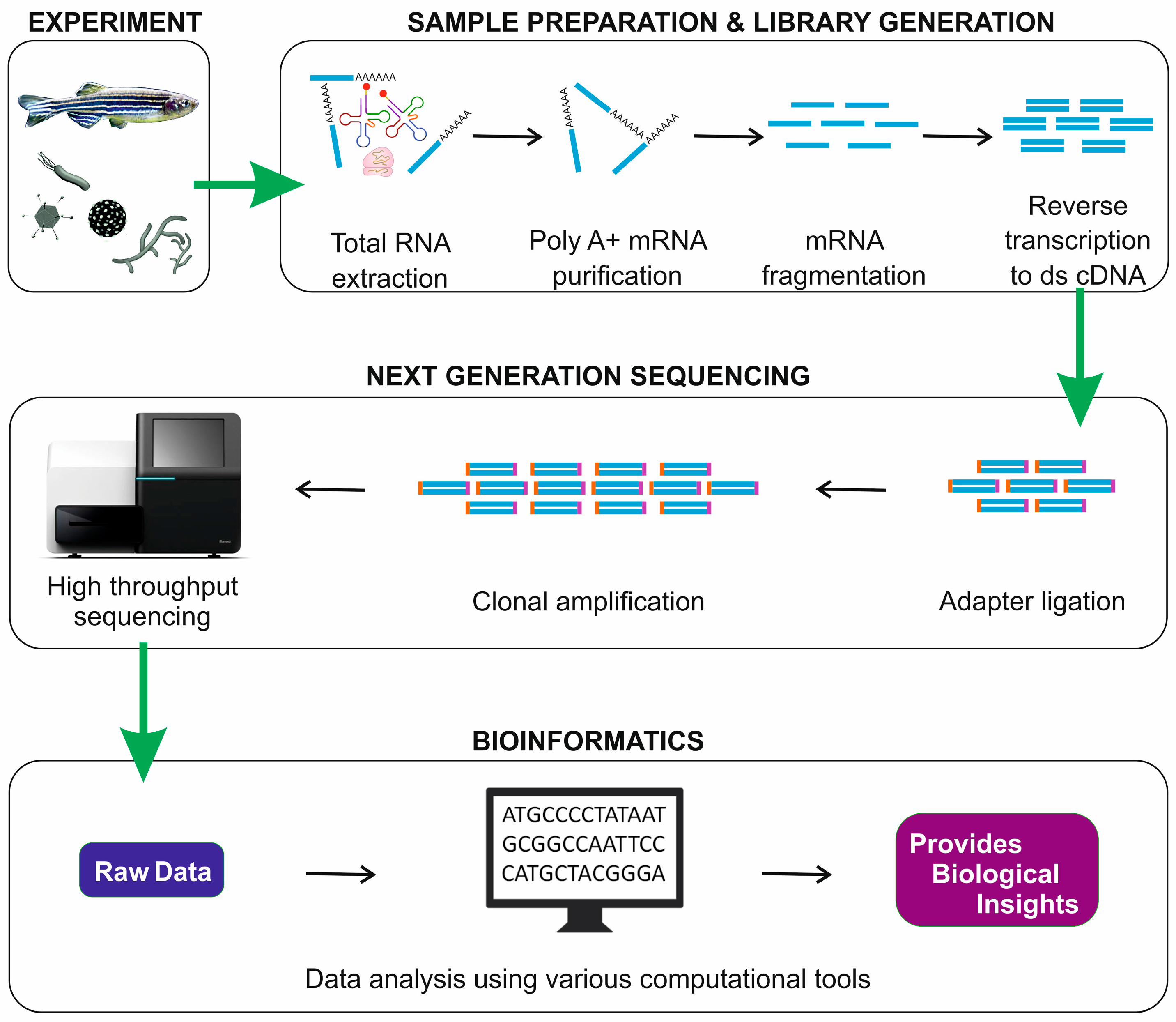
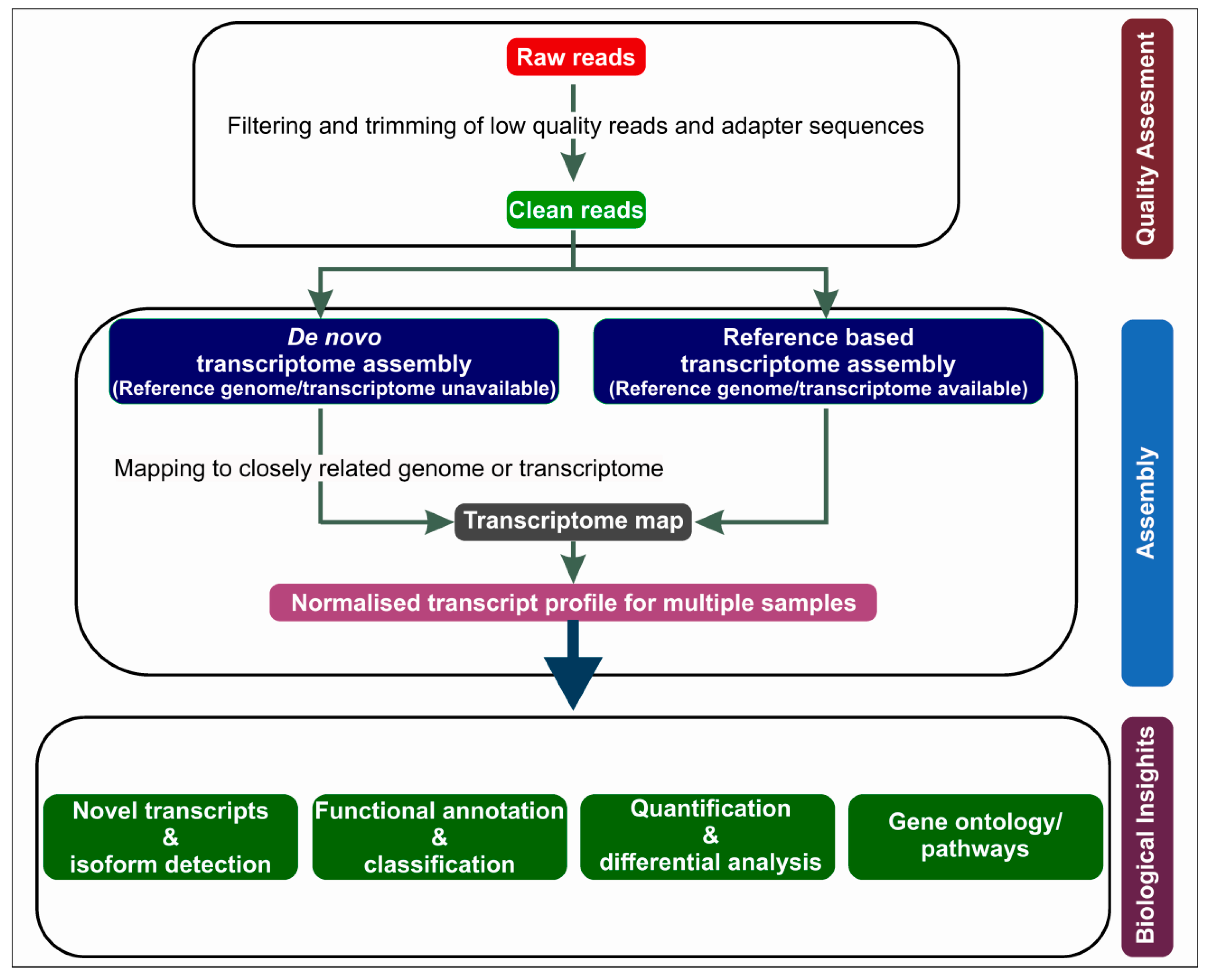
2. RNA-Seq Workflow
3. Fish Immune Response to Bacterial Pathogens
4. Fish Immune Response to Viral Pathogens
5. Fish Immune Responses to Parasites
6. RNA-Seq Analysis of Oomycetes
7. RNA-Seq Analysis of Healthy Fish
8. Conclusions and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CAGE | Cap analysis gene expression |
| CyHV-3 | Cyprinid herpesvirus-3 |
| GALT | Gut-associated lymphoid tissue |
| GCRV | Grass carp reovirus |
| HSMI | Heart and skeletal muscle inflammation |
| IHNV | Infectious hematopoietic necrosis virus |
| ISAV | Infectious salmon anaemia virus |
| JAK | Janus kinase |
| KHV | Koi herpesvirus |
| LPS | Lipopolysaccharide |
| MHC | Major histocompatibility complex |
| NCBI | National center for biotechnology information |
| NGS | Next-generation sequencing |
| NLR | Nucleotide-binding oligomerization domain-like receptor |
| NNV | Nervous necrosis virus |
| PRV | Piscine orthoreovirus |
| RBL | Rhamnose-binding lectin |
| RLR | Retinoic acid inducible gene-I like receptor |
| RIN | RNA integrity number |
| RNA-Seq | RNA sequencing |
| SAGE | Series analysis of gene expression |
| SAV | Salmonid alphavirus |
| ScRNA-Seq | Single-cell RNA-Seq |
| SGIV | Singapore grouper iridovirus |
| SNP | Single nucleotide polymorphism |
| SSR | Single sequence repeat |
| SVCV | Spring viremia of carp virus |
| TLR | Toll-like receptor |
| VER | Viral encephalopathy and retinopathy |
| VHSV | Viral hemorrhagic septicemia virus |
| VNN | Viral nervous necrosis |
References
- Food and Agriculture Organization of the United Nations (FAO). The State of World Fisheries and Aquaculture; FAO: Rome, Italy, 2016. [Google Scholar]
- Huang, L.; Li, G.; Mo, Z.; Xiao, P.; Li, J.; Huang, J. De Novo assembly of the Japanese flounder (Paralichthys olivaceus) spleen transcriptome to identify putative genes involved in immunity. PLoS ONE 2015, 10, e0117642. [Google Scholar] [CrossRef]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Casneuf, T.; Van de Peer, Y.; Huber, W. In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinform. 2007, 8, 461. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J. The beginning of the end for microarrays? Nat. Methods 2008, 5, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, 11, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mehinto, A.C.; Martyniuk, C.J.; Spade, D.J.; Denslow, N.D. Applications of next-generation sequencing in fish ecotoxicogenomics. Front. Genet. 2012, 3, 62. [Google Scholar] [CrossRef] [PubMed]
- Kroeker, A.L.; Berard, A.R.; Coombs, K.M. Fishing beyond the peer: Future omic analyses of virus-host Interactions. J. Virol. Antivir. Res. 2012, 1. [Google Scholar] [CrossRef]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. OMICS 2014, 18, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Kjällquist, U.; Moliner, A.; Zajac, P.; Fan, J.B.; Lönnerberg, P.; Linnarsson, S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-Seq. Genome Res. 2011, 21, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013, 14, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011, 12, R22. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Bähler, J. RNA-Seq: From technology to biology. Cell. Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef] [PubMed]
- Oshlack, A.; Robinson, M.D.; Young, M.D. From RNA-Seq reads to differential expression results. Genome Biol. 2010, 11, 220. [Google Scholar] [CrossRef] [PubMed]
- Kvam, V.M.; Liu, P.; Si, Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-Seq data. Am. J. Bot. 2012, 99, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Stockhammer, O.W.; Rauwerda, H.; Wittink, F.R.; Breit, T.M.; Meijer, A.H.; Spaink, H.P. Transcriptome analysis of Traf6 function in the innate immune response of zebrafish embryos. Mol. Immunol. 2010, 48, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Ordas, A.; Hegedus, Z.; Henkel, C.V.; Stockhammer, O.W.; Butler, D.; Jansen, H.J.; Racz, P.; Mink, M.; Spaink, H.P.; Meijer, A.H. Deep sequencing of the innate immune transcriptomic response of zebrafish embryos to Salmonella infection. Fish Shellfish Immunol. 2011, 31, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Z.; Su, Y.; Wang, D.; Yin, B.; Shu, B.; Zhang, J.; Zhu, X.; Jia, C. Characterization of Mycobacterium marinum infections in zebrafish wounds and sinus tracts. Wound Repair Regen. 2017, 25, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Stupka, E.; Henkel, C.V.; Jansen, H.J.; Spaink, H.P.; Verbeek, F.J. Identification of common carp innate immune genes with whole-genome sequencing and RNA-Seq data. J. Integr. Bioinform. 2011, 8, 169. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Wang, R.; Lu, J.; Nandi, S.; Mohanty, S.; Terhune, J.; Liu, Z.; Peatman, E. RNA-Seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immunol. 2012, 32, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, K.V.; Zhang, J.; Liu, S.; Kucuktas, H.; Wang, X.; Liu, H.; Sha, Z.; Terhune, J.; Peatman, E.; Liu, Z. Pathogen recognition receptors in channel catfish: I. Identification, phylogeny and expression of NOD-like receptors. Dev. Comp. Immunol. 2012, 37, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, K.V.; Zhang, J.; Liu, S.; Peatman, E.; Kucuktas, H.; Wang, X.; Liu, H.; Wood, T.; Terhune, J.; Liu, Z. Pathogen recognition receptors in channel catfish: II. Identification, phylogeny and expression of retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs). Dev. Comp. Immunol. 2012, 37, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Y.; Liu, S.; Li, C.; Sun, L.; Bao, L.; Feng, J.; Liu, Z. Analysis of 52 Rab GTPases from channel catfish and their involvement in immune responses after bacterial infections. Dev. Comp. Immunol. 2014, 45, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, S.; Rajendran, K.V.; Sun, L.; Zhang, Y.; Sun, F.; Kucuktas, H.; Liu, H.; Liu, Z. Pathogen recognition receptors in channel catfish: III Phylogeny and expression analysis of Toll-like receptors. Dev. Comp. Immunol. 2013, 40, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Sun, L.; Bao, L.; Zhang, J.; Jiang, Y.; Yao, J.; Song, L.; Feng, J.; Liu, S.; Liu, Z. Bulk segregant RNA-Seq reveals expression and positional candidate genes and allele-specific expression for disease resistance against enteric septicemia of catfish. BMC Genom. 2013, 14, 929. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Liu, X.X.; Lv, X.; Li, S.Y.; Li, Y.D.; Yu, X.J.; Wang, X.G. Deciphering transcriptome profile of the yellow catfish (Pelteobagrus fulvidraco) in response to Edwardsiella ictaluri. Fish Shellfish Immunol. 2017, 70, 593–608. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, X.; Cheng, J.; He, Y.; Wang, X.; Wang, Z.; Qi, J.; Yu, H.; Zhang, Q. Transcriptome profiling provides gene resources for understanding gill immune responses in Japanese flounder (Paralichthys olivaceus) challenged with Edwardsiella tarda. Fish Shellfish Immunol. 2018, 72, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Peatman, E.; Li, C.; Peterson, B.C.; Straus, D.L.; Farmer, B.D.; Beck, B.H. Basal polarization of the mucosal compartment in Flavobacterium columnare susceptible and resistant channel catfish (Ictalurus punctatus). Mol. Immunol. 2013, 56, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Peatman, E.; Li, C.; Liu, S.; Jiang, Y.; Zhou, Z.; Liu, Z. Transcriptomic signatures of attachment, NF-KB suppression and IFN stimulation in the catfish gill following columnaris bacterial infection. Dev. Comp. Immunol. 2012, 38, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.H.; Farmer, B.D.; Straus, D.L.; Li, C.; Peatman, E. Putative roles for a rhamnose binding lectin in Flavobacterium columnare pathogenesis in channel catfish Ictalurus punctatus. Fish Shellfish Immunol. 2012, 33, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Thongda, W.; Li, C.; Zhao, H.; Beck, B.H.; Mohammed, H.; Arias, C.R.; Peatman, E. More than just antibodies: Protective mechanisms of a mucosal vaccine against fish pathogen Flavobacterium columnare. Fish Shellfish Immunol. 2017, 71, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; He, D.; Dong, W.; Zhang, Y.; Shao, J. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genom. 2010, 11, 472. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Ding, F.; Cui, P.; Ao, J.; Hu, S.; Chen, X. Transcriptome and expression profiling analysis revealed changes of multiple signaling pathways involved in immunity in the large yellow croaker during Aeromonas hydrophila infection. BMC Genom. 2010, 11, 506. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulou, E.; Galindo-Villegas, J.; García-Alcázar, A.; Kasapidis, P.; Mulero, V. Characterization of European sea bass transcripts by RNA SEQ after oral vaccine against V. anguillarum. Mar. Biotechnol. 2012, 14, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liu, Q.; Yang, M.; Wu, H.; Wang, Q.; Xiao, J.; Zhang, Y. RNA-Seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish. BMC Genom. 2012, 13, 319. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.H.; Liu, P.; Liu, F.; Lin, G.; Sun, F.; Tu, R.; Yue, G.H. Analysis of stress-responsive transcriptome in the intestine of Asian Seabass (Lates calcarifer) using RNA-Seq. DNA Res. 2013, 20, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Huang, S.J.; Chou, H.N.; Liao, W.L.; Gong, H.Y.; Chen, J.Y. Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related complement pathway in Epinephelus coioides. BMC Genom. 2014, 15, 1102. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, C.; Ao, Q.; Tan, Y.; Luo, Y.; Guo, Y.; Lan, G.; Jiang, H.; Gan, X. Trancriptomic profiling revealed the signatures of acute immune response in tilapia (Oreochromis niloticus) following Streptococcus iniae challenge. Fish Shellfish Immunol. 2015, 46, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Chen, S.; Chen, Y.; Liu, Y.; Shao, C.; Wang, Q.; Lu, Y.; Gong, G.; Ding, S.; et al. Transcriptome analysis revealed changes of multiple genes involved in immunity in Cynoglossus semilaevis during Vibrio anguillarum infection. Fish Shellfish Immunol. 2015, 43, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.T.; Gao, Z.X.; Zhao, H.H.; Yi, S.K.; Chen, B.X.; Zhao, Y.H.; Lin, L.; Liu, X.Q.; Wang, W.M. Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish Shellfish Immunol. 2015, 45, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gan, Z.; Cai, S.; Wang, Z.; Yu, D.; Lin, Z.; Lu, Y.; Wu, Z.; Jian, J. Comprehensive identification and profiling of Nile tilapia (Oreochromis niloticus) microRNAs response to Streptococcus agalactiae infection through high-throughput sequencing. Fish Shellfish Immunol. 2016, 54, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Wang, Y.H.; Hui, C.F.; Chen, J.Y. Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related TLR5-mediated induction of cytokines in Epinephelus lanceolatus. Fish Shellfish Immunol. 2016, 52, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, P.; Wan, Z.Y.; Huang, S.Q.; Wen, Y.F.; Lin, G.; Yue, G.H. RNA-Seq revealed the impairment of immune defence of tilapia against the infection of Streptococcus agalactiae with simulated climate warming. Fish Shellfish Immunol. 2016, 55, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Fu, M.; Wang, C.; Jiao, Z.; Qiu, L. RNA-Seq analysis of immune-relevant genes in Lateolabrax japonicus during Vibrio anguillarum infection. Fish Shellfish Immunol. 2016, 52, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, H.; Li, H.; Wang, A. Transcriptome profiling of grass carp (Ctenopharyngodon idellus) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2016, 51, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Feng, S.; Zhang, S.; Liu, H.; Feng, J.; Mu, X.; Sun, X.; Xu, P. Transcriptome signatures in common carp spleen in response to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2016, 57, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, Q.; Ao, Q.; Tan, Y.; Luo, Y.; Jiang, H.; Li, C.; Gan, X. Transcriptomic profiling analysis of tilapia (Oreochromis niloticus) following Streptococcus agalactiae challenge. Fish Shellfish Immunol. 2017, 62, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sahoo, P.K.; Barat, A. Transcriptome profiling and expression analysis of immune responsive genes in the liver of Golden mahseer (Tor putitora) challenged with Aeromonas hydrophila. Fish Shellfish Immunol. 2017, 67, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Y.; Cao, Y.; Wang, D.; Liu, H.; Lu, T. Trancriptome profiles of Amur sturgeon spleen in response to Yersinia ruckeri infection. Fish Shellfish Immunol. 2017, 70, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Jozwick, A.K.S.; Graf, J.; Welch, T.J. The flagellar master operon flhDC is a pleiotropic regulator involved in motility and virulence of the fish pathogen Yersinia ruckeri. J. Appl. Microbiol. 2017, 122, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Hu, X.; Sun, B.; Bo, Y.; Wu, K.; Xiao, L.; Gong, C. A transcriptome analysis focusing on inflammation-related genes of grass carp intestines following infection with Aeromonas hydrophila. Sci. Rep. 2017, 7, 40777. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Byadgi, O.; Chen, Y.C.; Aoki, T.; Takeyama, H.; Yoshida, T.; Hikima, J.I.; Sakai, M.; Wang, P.C.; Chen, S.C. Transcriptome analysis of immune response against Vibrio harveyi infection in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2017, 70, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Ken, C.F.; Chen, C.N.; Ting, C.H.; Pan, C.Y.; Chen, J.Y. Transcriptome analysis of hybrid tilapia (Oreochromis spp.) with Streptococcus agalactiae infection identifies Toll-like receptor pathway-mediated induction of NADPH oxidase complex and piscidins as primary immune-related responses. Fish Shellfish Immunol. 2017, 70, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Gong, Q.; Wen, Z.; Yuan, D.; Shao, T.; Wang, J.; Li, H. Transcriptome analysis of the spleen of the darkbarbel catfish Pelteobagrus vachellii in response to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2017, 70, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Beck, B.; Su, B.; Terhune, J.; Peatman, E. Early mucosal responses in blue catfish (Ictalurus furcatus) skin to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2013, 34, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Menanteau-Ledouble, S.; Saleh, M.; El-Matbouli, M. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet. Res. 2015, 46, 103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Hummel, K.; Ahrens, M.; Menanteau-Ledouble, S.; Welch, T.J.; Eisenacher, M.; Razzazi-Fazeli, E.; El-Matbouli, M. Shotgun proteomic analysis of Yersinia ruckeri strains under normal and iron-limited conditions. Vet. Res. 2016, 47, 100. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Hummel, K.; Welch, T.J.; Razzazi-Fazeli, E.; El-Matbouli, M. Global proteomic profiling of Yersinia ruckeri strains. Vet. Res. 2017, 48, 55. [Google Scholar] [CrossRef] [PubMed]
- Smail, D.A.; Munro, E.S. The virology of teleosts. In Fish Pathology, 4th ed.; Roberts, R.J., Ed.; Wiley-Blackwell: Oxford, UK, 2012; pp. 186–291. ISBN 9781118222942. [Google Scholar]
- Morera, D.; Roher, N.; Ribas, L.; Balasch, J.C.; Doñate, C.; Callol, A.; Boltaña, S.; Roberts, S.; Goetz, G.; Goetz, F.W.; et al. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PLoS ONE 2011, 6, e26998. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Li, M.; Ding, F.; Ding, Y.; Ao, J.; Hu, S.; Chen, X. De novo characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PLoS ONE 2014, 9, e97471. [Google Scholar] [CrossRef] [PubMed]
- Genet, C.; Verrier, E.R.; Ciobotaru, C.; Klopp, C.; Esquerre, D.; Laloe, D.; Boudinot, P.; Quillet, E. RNA-Seq analysis of transcriptome response to VHS-V infection in two target tissues of resistant vs susceptible trout clonal lines. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Shi, M.; Huang, R.; Du, F.; Pei, Y.; Liao, L.; Zhu, Z.; Wang, Y. RNA-Seq profiles from grass carp tissues after reovirus (GCRV) infection based on singular and modular enrichment analyses. Mol. Immunol. 2014, 61, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Miranda, D.; Boltaña, S.; Cabrejos, M.E.; Yáñez, J.M.; Gallardo-Escárate, C. High-throughput transcriptome analysis of ISAV-infected Atlantic salmon Salmo salar unravels divergent immune responses associated to head-kidney, liver and gills tissues. Fish Shellfish Immunol. 2015, 45, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Su, J. Transcriptome analysis provides insights into the regulatory function of alternative splicing in antiviral immunity in grass carp (Ctenopharyngodon idella). Sci. Rep. 2015, 5, 12946. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Gao, Y.; Xu, G.; Wu, C.; Xu, T. Transcriptome comparative analysis revealed poly(I:C) activated RIG-I/MDA5-mediated signaling pathway in miiuy croaker. Fish Shellfish Immunol. 2015, 47, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Zhong, L.; Liu, Q.L.; Xiao, T.Y.; Su, J.M.; Chen, K.J.; Wang, H.Q.; Dai, Y.J.; Chen, J. Characterization of grass carp spleen transcriptome during GCRV infection. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Kwon, M.G.; Jung, S.H.; Park, M.A.; Kim, D.W.; Cho, W.S.; Park, J.W.; Son, M.H. RNA-Seq transcriptome analysis of the olive flounder (Paralichthys olivaceus) kidney response to vaccination with heat-inactivated viral hemorrhagic septicemia virus. Fish Shellfish Immunol. 2017, 62, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Dettleff, P.; Moen, T.; Santi, N.; Martinez, V. Transcriptomic analysis of spleen infected with infectious salmon anemia virus reveals distinct pattern of viral replication on resistant and susceptible Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 2017, 61, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Li, D.; Lian, F.; Yang, S.; Wu, J.; Liu, H.; Bu, G.; Meng, F.; Cao, X.; et al. Transcriptome profiling of spleen provides insights into the antiviral mechanism in Schizothorax prenanti after poly (I:C) challenge. Fish Shellfish Immunol. 2017, 62, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Ho, L.P.; Yang, C.H.; Kao, T.Y.; Chou, H.Y.; Pai, T.W. Comparison of grouper infection with two different iridoviruses using transcriptome sequencing and multiple reference species selection. Fish Shellfish Immunol. 2017, 71, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.W.; Ngou, F.H.; Chao, Y.M.; Lai, Y.S.; Chen, N.Y.; Lee, F.Y.; Chiou, P.P. Transcriptome characterization and gene expression of Epinephelus spp in endoplasmic reticulum stress-related pathway during betanodavirus infection in vitro. BMC Genom. 2012, 13, 651. [Google Scholar] [CrossRef] [PubMed]
- Polinski, M.P.; Bradshaw, J.C.; Inkpen, S.M.; Richard, J.; Fritsvold, C.; Poppe, T.T.; Rise, M.L.; Garver, K.A.; Johnson, S.C. De novo assembly of sockeye salmon kidney transcriptomes reveal a limited early response to piscine reovirus with or without infectious hematopoietic necrosis virus superinfection. BMC Genom. 2016, 17, 848. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Evensen, Ø.; Munang’andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and salmonid alphavirus subtype-3 infection. BMC Genom. 2015, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Pozo, E.; Valero, Y.; Esteve-Codina, A.; Gómez-Garrido, J.; Dabad, M.; Alioto, T.; Meseguer, J.; Esteban, M.Á.; Cuesta, A. Innate cell-mediated cytotoxic activity of European sea bass leucocytes against nodavirus-infected cells: A functional and RNA-Seq study. Sci. Rep. 2017, 7, 15396. [Google Scholar] [CrossRef] [PubMed]
- Lee, X.; Yi, Y.; Weng, S.; Zeng, J.; Zhang, H.; He, J.; Dong, C. Transcriptomic analysis of koi (Cyprinus carpio) spleen tissue upon cyprinid herpesvirus 3 (CyHV3) infection using next generation sequencing. Fish Shellfish Immunol. 2016, 49, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Tang, B.P.; Jiang, S.H.; Zhang, H.B.; Zhou, C.L.; Chai, X.Y.; et al. Transcriptome analysis of yellow catfish (Pelteobagrus fulvidraco) liver challenged with polyriboinosinic polyribocytidylic acid (poly I:C). Fish Shellfish Immunol. 2017, 68, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Lu, Y.; Wang, F.; Liu, L.; Liu, J.; Liu, X. Comparative transcriptome analysis of zebrafish (Danio rerio) brain and spleen infected with spring viremia of carp virus (SVCV). Fish Shellfish Immunol. 2017, 69, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Kwon, Y.K.; Nam, M.; Vaidya, B.; Kim, S.R.; Lee, S.; Kwon, J.; Kim, D.; Hwang, G.S. Integrated profiling of global metabolomic and transcriptomic responses to viral hemorrhagic septicemia virus infection in olive flounder. Fish Shellfish Immunol. 2017, 71, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, X.; Yan, Y.; Cai, J.; Ouyang, Z.; Cui, H.; Wang, P.; Qin, Q. Transcriptome analysis of orange-spotted grouper (Epinephelus coioides) spleen in response to Singapore grouper iridovirus. BMC Genom. 2011, 12, 556. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, L. Transcriptome analysis reveals temperature-regulated antiviral response in turbot Scophthalmus maximus. Fish Shellfish Immunol. 2017, 68, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K. Fish Viruses and Fish Viral Diseases; Cornell University Press: New York, NY, USA, 1988; ISBN 0801412595. [Google Scholar]
- Lenoir, G.; de Kinkelin, P. Fish rhabdoviruses: Comparative study of protein structure. J. Virol. 1975, 16, 259–262. [Google Scholar] [PubMed]
- Bernard, J.; Lecocq-Xhonneux, F.; Rossius, M.; Thiry, M.E.; Dekinkelin, P. Cloning and sequencing the messenger RNA of the N gene of viral hemorrhagic septicemia virus. J. Gen. Virol. 1990, 71, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Mjaaland, S.; Rimstad, E.; Falk, K.; Dannevig, B.H. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): An orthomyxo-like virus in a teleost. J. Virol. 1997, 71, 7681–7686. [Google Scholar] [PubMed]
- Aamelfot, M.; Dale, O.B.; Weli, S.C.; Koppang, E.O.; Falk, K. Expression of the infectious salmon anemia virus receptor on Atlantic salmon endothelial cells correlates with the cell tropism of the virus. J. Virol. 2012, 86, 10571–10578. [Google Scholar] [CrossRef] [PubMed]
- Palacios, G.; Lovoll, M.; Tengs, T.; Hornig, M.; Hutchison, S.; Hui, J.; Kongtorp, R.T.; Savji, N.; Bussetti, A.V.; Solovyov, A.; et al. Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PLoS ONE 2010, 5, e11487. [Google Scholar] [CrossRef] [PubMed]
- Garver, K.A.; Marty, G.D.; Cockburn, S.N.; Richard, J.; Hawley, L.M.; Müller, A.; Thompson, R.L.; Purcell, M.K.; Saksida, S. Piscine reovirus, but not jaundice syndrome, was transmissible to chinook salmon, Oncorhynchus tshawytscha (Walbaum), sockeye salmon, Oncorhynchus nerka (Walbaum), and Atlantic salmon, Salmo salar L. J. Fish Dis. 2016, 39, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Rudakova, S.L.; Kurath, G.; Bochkova, E.V. Occurrence and genetic typing of infectious hematopoietic necrosis virus in Kamchatka, Russia. Dis. Aquat. Org. 2007, 75, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.M.; Rodger, H.D.; Drinan, E.M.; Gannon, F.; Kruse, P.; Korting, W. The sequential pathology of pancreas disease in Atlantic salmon farms in Ireland. J. Fish Dis. 1992, 15, 401–408. [Google Scholar] [CrossRef]
- Waltzek, T.B.; Kelley, G.O.; Stone, D.M.; Way, K.; Hanson, L.; Fukuda, H.; Hirono, I.; Aoki, T.; Davison, A.J.; Hedrick, R.P. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 2005, 86, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Rathore, G.; Kumar, G.; Raja Swaminathan, T.; Swain, P. Koi herpes virus: A review and risk assessment of Indian aquaculture. Indian J. Virol. 2012, 23, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Y.; Yeap, S.K.; Omar, A.R.; Tan, W.S. Advances in the study of nodavirus. PeerJ 2017, 5, e3841. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Fenner, B.J.; Goh, W.; Kwang, J. Dissection of double-stranded RNA binding protein B2 from betanodavirus. J. Virol. 2007, 81, 5449–5459. [Google Scholar] [CrossRef] [PubMed]
- Fenner, B.J.; Thiagarajan, R.; Chua, H.K.; Kwang, J. Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J. Virol. 2006, 80, 85–94. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fenner, B.J.; Goh, W.; Kwang, J. Sequestration and protection of double-stranded RNA by the betanodavirus B2 protein. J. Virol. 2006, 80, 6822–6833. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Kileng, Ø.; Skugor, S.; Jørgensen, S.M.; Afanasyev, S.; Timmerhaus, G.; Sommer, A.I.; Jensen, I. Genomic analysis of the host response to nervous necrosis virus in Atlantic cod (Gadus morhua) brain. Mol. Immunol. 2013, 54, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Li, J.; Hu, C.; Wang, F.; Wang, B.; Shi, X.; Hou, Q.; Huang, W.; Lin, G. Transcriptome data analysis of grass carp (Ctenopharyngodon idella) infected by reovirus provides insights into two immune-related genes. Fish Shellfish Immunol. 2017, 64, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Flegr, J. Evolutionary parasitology: The development of invasion, evasion, and survival mechanisms used by bacterial, viral, protozoan, and metazoan parasites. In Food Consumption and Disease Risk: Consumer-Pathogen Interactions, 1st ed.; Potter, M., Ed.; Woodhead Publishing Ltd. Abington: Cambridge, UK, 2006; pp. 251–270. ISBN 9781845692025. [Google Scholar]
- Robledo, D.; Ronza, P.; Harrison, P.W.; Losada, A.; Bermúdez, R.; Pardo, B.G.; José Redondo, M.; Sitjà-Bobadilla, A.; Quiroga, M.; Martínez, P. RNA-Seq analysis reveals significant transcriptome changes in turbot (Scophthalmus maximus) suffering severe enteromyxosis. BMC Genom. 2014, 15, 1149. [Google Scholar] [CrossRef] [PubMed]
- Haase, D.; Rieger, J.K.; Witten, A.; Stoll, M.; Bornberg-Bauer, E.; Kalbe, M.; Reusch, T.B.H. Specific gene expression responses to parasite genotypes reveal redundancy of innate immunity in vertebrates. PLoS ONE 2014, 9, e108001. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Nuñez-Acuña, G. RNA-Seq analysis using de novo transcriptome assembly as a reference for the salmon louse Caligus rogercresseyi. PLoS ONE 2014, 9, e92239. [Google Scholar] [CrossRef] [PubMed]
- Haase, D.; Rieger, J.K.; Witten, A.; Stoll, M.; Bornberg-Bauer, E.; Kalbe, M.; Reusch, T.B.H. Immunity comes first: The effect of parasite genotypes on adaptive immunity and immunization in three-spined sticklebacks. Dev. Comp. Immunol. 2016, 54, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, J.; Su, Y.Q.; Mao, Y.; Zhang, J.S.; Wu, C.W.; Ke, Q.Z.; Han, K.H.; Zheng, W.Q.; Xu, N.D. Transcriptome analysis of the Larimichthys crocea liver in response to Cryptocaryon irritans. Fish Shellfish Immunol. 2016, 48, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ronza, P.; Robledo, D.; Bermúdez, R.; Losada, A.P.; Pardo, B.G.; Sitjà-Bobadilla, A.; Quiroga, M.I.; Martínez, P. RNA-Seq analysis of early enteromyxosis in turbot (Scophthalmus maximus): New insights into parasite invasion and immune evasion strategies. Int. J. Parasitol. 2016, 46, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Gao, Q.; Tang, B.; Sun, P.; Han, K.; Huang, W. Transcriptome and analysis on the complement and coagulation cascades pathway of large yellow croaker (Larimichthys crocea) to ciliate ectoparasite Cryptocaryon irritans infection. Fish Shellfish Immunol. 2016, 50, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Sun, P.; Wang, J.; Gao, Q. Transcriptome analysis of dormant tomonts of the marine fish ectoparasitic ciliate Cryptocaryon irritans under low temperature. Parasites Vectors 2016, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.Q.; Li, Y.W.; Wang, H.Q.; Wang, J.L.; Ni, L.Y.; Yang, M.; Lao, G.F.; Luo, X.C.; Li, A.X.; Dan, X.M. Comparative transcriptional profile of the fish parasite Cryptocaryon irritans. Parasites Vectors 2016, 9, 630. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, A.; Xu, Y.; Jiang, B.; Lu, G.; Luo, X. Transcriptomic variation of locally-infected skin of Epinephelus coioides reveals the mucosal immune mechanism against Cryptocaryon irritans. Fish Shellfish Immunol. 2017, 66, 398–410. [Google Scholar] [CrossRef] [PubMed]
- El-Matbouli, M.; Hoffmann, R.W. Influence of water quality on the outbreak of proliferative kidney disease—Field studies and exposure experiments. J. Fish Dis. 2002, 25, 459–467. [Google Scholar] [CrossRef]
- Grabner, D.S.; El-Matbouli, M. Comparison of the susceptibility of brown trout (Salmo trutta) and four rainbow trout (Oncorhynchus mykiss) strains to the myxozoan Tetracapsuloides bryosalmonae, the causative agent of proliferative kidney disease (PKD). Vet. Parasitol. 2009, 165, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Abd-Elfattah, A.; Saleh, M.; El-Matbouli, M. Fate of Tetracapsuloides bryosalmonae (Myxozoa) after infection of brown trout Salmo trutta and rainbow trout Oncorhynchus mykiss. Dis. Aquat. Org. 2013, 107, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Abd-Elfattah, A.; El-Matbouli, M. Identification of differentially expressed genes of brown trout (Salmo trutta) and rainbow trout (Oncorhynchus mykiss) in response to Tetracapsuloides bryosalmonae (Myxozoa). Parasitol. Res. 2015, 114, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Minor, K.L.; Anderson, V.L.; Davis, K.S.; Van Den Berg, A.H.; Christie, J.S.; Löbach, L.; Faruk, A.R.; Wawra, S.; Secombes, C.J.; van West, P. A putative serine protease, SpSsp1, from Saprolegnia parasitica is recognised by sera of rainbow trout, Oncorhynchus mykiss. Fungal Biol. 2014, 118, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Majeed, M.; Kumar, G.; Schlosser, S.; El-Matbouli, M.; Saleh, M. In vitro investigations on extracellular proteins secreted by Aphanomyces invadans, the causative agent of epizootic ulcerative syndrome. Acta Vet. Scand. 2017, 59, 78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.H.Y.; de Bruijn, I.; Haas, B.J.; Belmonte, R.; Löbach, L.; Christie, J.; van den Ackerveken, G.; Bottin, A.; Bulone, V.; Díaz-Moreno, S.M.; et al. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet. 2013, 9, e1003272. [Google Scholar] [CrossRef] [PubMed]
- Press, C.M.L.; Evensen, O. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999, 9, 309–318. [Google Scholar] [CrossRef]
- Rauta, P.R.; Nayak, B.; Das, S. Immune system and immune responses in fish and their role in comparative immunity study: A model for higher organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Rexroad, C.; Thorgaard, G.; Yao, J.; Salem, M. Characterization of the rainbow trout spleen transcriptome and identification of immune-related genes. Front. Genet. 2014, 5, 348. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Zhang, C.; Zhang, R.; Zhao, K. Transcriptome profiling analysis of naked carp (Gymnocypris przewalskii) provides insights into the immune-related genes in highland fish. Fish Shellfish Immunol. 2015, 46, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Tian, F.; Zhao, K. Genomic signature of highland adaptation in fish: A case study in Tibetan Schizothoracinae species. BMC Genom. 2017, 18, 948. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, X.; Wang, J.; Zhou, L.; Zhang, Y.; Zhang, G.; Lu, G.; Wang, C. An integrated and comprehensive transcriptome reveals immune-related genes and signal pathways in topmouth culter (Culter alburnus). Aquac. Res. 2017, 48, 2231–2242. [Google Scholar] [CrossRef]
- Li, G.; Zhao, Y.; Wang, J.; Liu, B.; Sun, X.; Guo, S.; Feng, J. Transcriptome profiling of developing spleen tissue and discovery of immune-related genes in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2017, 60, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Liu, S.; Zhang, B.; Wang, H.; Sun, H.; Song, S.; Qiu, X.; Liu, Y.; Wang, X.; Jiang, Z.; et al. Transciptome analysis of the gill and swimbladder of Takifugu rubripes by RNA-Seq. PLoS ONE 2014, 9, e85505. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhong, H.; Liu, Z.; Yu, F.; Luo, Y.; Gan, X.; Zhou, Y. Transcriptome analysis revealed positive selection of immune-related genes in tilapia. Fish Shellfish Immunol. 2015, 44, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Nuñez Ortiz, N.; Gerdol, M.; Stocchi, V.; Marozzi, C.; Randelli, E.; Bernini, C.; Buonocore, F.; Picchietti, S.; Papeschi, C.; Sood, N.; et al. T cell transcripts and T cell activities in the gills of the teleost fish sea bass (Dicentrarchus labrax). Dev. Comp. Immunol. 2014, 47, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Hang, X.Y.; Yin, L.; He, Y.Q.; Chen, J.; Shi, Y.H.; Li, C.H. Sequencing of the first ayu (Plecoglossus altivelis) macrophage transcriptome and microarray development for investigation the effect of LECT2 on macrophages. Fish Shellfish Immunol. 2013, 34, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, E.I.; Andres, H.; Botthof, J.G.; Ferreira, L.; Lio, P.; Cvejic, A. Single-cell RNA-Sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 2017, 8, 2045. [Google Scholar] [CrossRef] [PubMed]
- Carmona, S.J.; Teichmann, S.A.; Ferreira, L.; Macaulay, C.; Stubbington, M.J.T.; Cvejic, A.; Gfeller, D. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017, 27, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Li, Q.; Zhou, B.; Song, G.; Li, T.; Cui, Z. De novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PLoS ONE 2013, 8, e56998. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, E.; Li, Y.; Li, X.; Ding, C. Transcriptome analysis reveals increases in visceral lipogenesis and storage and activation of the antigen processing and presentation pathway during the mouth-opening stage in zebrafish larvae. Int. J. Mol. Sci. 2017, 18, 1634. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Magadán, S. Omics in fish mucosal immunity. Dev. Comp. Immunol. 2017, 75, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, C.; Su, B.; Beck, B.H.; Peatman, E. Short-term feed deprivation alters immune status of surface mucosa in channel catfish (Ictalurus punctatus). PLoS ONE 2013, 8, e74581. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Bernatchez, L.; Beheregaray, L.B. RNA-Seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genom. 2013, 14, 375. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; You, F.; Wang, Q.; Weng, S.; Liu, H.; Wang, L.; Zhang, P.J.; Tan, X. Transcriptional responses of olive flounder (Paralichthys olivaceus) to low temperature. PLoS ONE 2014, 9, e108582. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, G.; Palti, Y.; Cleveland, B.M.; Weber, G.M.; Rexroad, C.E. RNA-Seq analysis of early hepatic response to handling and confinement stress in rainbow trout. PLoS ONE 2014, 9, e88492. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Bao, P.; Tang, B. Transcriptome analysis and discovery of genes involved in immune pathways in large yellow croaker (Larimichthys crocea) under high stocking density stress. Fish Shellfish Immunol. 2017, 68, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Mu, Y.; Xiang, L.X.; Fan, D.D.; Feng, M.J.; Zhang, S.; Shi, Q.; Zhu, L.Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Dai, S.; Liu, H.; Cao, Q.; Yin, S.; Lai, K.P.; Tse, W.K.F.; Wong, C.K.C.; Shi, H. Identification of immune-related genes in gill cells of Japanese eels (Anguilla japonica) in adaptation to water salinity changes. Fish Shellfish Immunol. 2018, 73, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.N.; Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Jiang, S.H.; Chai, X.Y.; Wang, Z.F.; Zhang, H.B.; Bian, X.G.; Zhou, C.L.; et al. De novo transcriptome assembly and analysis of differential gene expression following lipopolysaccharide challenge in Pelteobagrus fulvidraco. Fish Shellfish Immunol. 2017, 73, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Galina, J.; Yin, G.; Ardó, L.; Jeney, Z. The use of immunostimulating herbs in fish. An overview of research. Fish Physiol. Biochem. 2009, 35, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Wang, S.; Guo, W.; Xie, Z.; Zheng, Y.; Cao, Z.; Zhou, Y. Transcriptome analysis provides insights into the immune responsive pathways and genes in the head kidney of tiger grouper (Epinephelus fuscoguttatus) fed with Spatholobus suberectus, Phellodendron amurense, or Eclipta prostrata. Fish Shellfish Immunol. 2018, 73, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-Sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
| Sl No | Fish | Bacterial Species | Sample | Sequencing Platform | Sequencing Depth/Library | Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Japanese sea bass | Vibrio harveyi | Head kidney & spleen | Illumina Genome Analyzer II | Control: 33.03 M | GSE21721 | [38] |
| Challenged: 34.59 M | |||||||
| 2 | Zebrafish | Salmonella typhimurium containing DsRed vector | Whole embryo | Illumina Genome Analyzer II | 15 M a | GSE21024 | [22] |
| 3 | large yellow croaker | Aeromonas hydrophila | Spleen | Solexa/Illumina Genome Analyzer | 13.6 M | SRA010789 | [39] |
| 4 | Zebrafish | Salmonella typhimurium | Whole embryo | Illumina Genome Analyzer II | 15 M | GSE21024 | [23] |
| 5 | Common carp | Mycobacterium marinum | Whole embryo & adult animals | Illumina Genome Analyzer II | NA | NA | [25] |
| 6 | European sea bass | Oral vaccination against Vibrio anguillarum | Head kidney & hind gut | Genome Sequencer FLX Titanium | 0.269 M | SRA050000 | [40] |
| 7 | Zebrafish | Edwardsiella tarda live attenuated vaccine | Liver | Illumina HiSeq 2000 | Control: 18.13 M a Vaccinated: 14.27 M a | SRA048658 | [41] |
| 8 | Channel catfish | Edwardsiella ictaluri | Intestine | Illumina HiSeq 2000 | 197.6 M | SRP009069 | [26] |
| 9 | Channel catfish | Flavobacterium columnare | Gill | Illumina HiSeq 2000 | 203.2 M | NA | [35] |
| 10 | Channel catfish | Flavobacterium columnare | Gill | Illumina HiSeq 2000 | 350 M | SRP017689 | [34] |
| 11 | Hybrid of channel catfish & blue catfish | Edwardsiella ictaluri | Liver | Illumina HiSeq 2500 | 400 M | SRP028159 | [31] |
| 12 | Asian seabass | Vibrio harveyi | Intestine | Genome Sequencer FLX Titanium | 1.0 M | DRR002185–DRR002190 | [42] |
| 13 | Orange spotted grouper | Vibrio alginolyticus | Whole animal | Illumina HiSeq 2000 | 114.8 M * | GSE63148 | [43] |
| 14 | Tilapia | Streptococcus iniae | Spleen | Illumina HiSeq 2000 | 223 M | NA | [44] |
| 15 | Half-smooth tongue sole | Vibrio anguillarum | Liver, head kidney, spleen & intestine | Illumina HiSeq 2000 | 80.8 M | NA | [45] |
| 16 | Blunt snout bream | Aeromonas hydrophila | Blood, liver, gill, intestine, spleen & kidney | Illumina HiSeq 2000 | 114.5 M | SRX731259 | [46] |
| 17 | Nile tilapia | Streptococcus agalactiae | Spleen | Illumina Genome Analyzer | 91.4 M | NA | [47] |
| 18 | Giant grouper | Vibrio alginolyticus | Whole animal | Illumina HiSeq 2000 | 28.7 M * | NA | [48] |
| 19 | Mozambique tilapia | Streptococcus agalactiae | Spleen | Illumina HiSeq 2000 | 200 M * | SAMD00020886–SAMD00020893 | [49] |
| (with stimulated climate warming) | |||||||
| 20 | Japanese sea bass | Vibrio anguillarum | Head kidney, liver & spleen | Illumina HiSeq 2000 | 334.3 M * | SRA296398 | [50] |
| 21 | Grass carp | Aeromonas hydrophila | Spleen | Illumina HiSeq 2500 | 63 M *,a | SRR3045340; SRR3045341 | [51] |
| 22 | Common carp | Aeromonas hydrophila | Spleen | Illumina HiSeq 2000 | 545 M | SRP072018 | [52] |
| 23 | Nile tilapia | Streptococcus agalactiae | Spleen | Illumina HiSeq 2000 | 208 M | NA | [53] |
| 24 | Golden mahseer | Aeromonas hydrophila | Liver | Illumina NextSeq 500 | Control: 25.7 M | SRS1117943; SRS1143332; SRS1152837 | [54] |
| Treatment: 27.5 M & 43.1 M | |||||||
| 25 | Amur sturgeon | Yersinia ruckeri | Spleen | Illumina HiSeq 4000 | Infected: 137.76 M | SRP110853 | [55] |
| Non infected: 139.49 M | |||||||
| 26 | - | Yersinia ruckeri | - | Illumina NextSeq | 250 M | NA | [56] |
| 27 | Grass carp | Aeromonas hydrophila | Intestine | Illumina HiSeq 2000 | 45.8 M * | SAMN04569528 | [57] |
| 28 | Zebra fish | Mycobacterium marinum | Muscle | Illumina HiSeq 2500 | NA | NA | [24] |
| 29 | Orange spotted grouper | Vibrio harveyi | Head kidney & spleen | Illumina HiSeq 4000 | 235.3 M * | NA | [58] |
| 30 | Yellow catfish | Edwardsiella ictaluri | Spleen | Illumina HiSeq 2000 | Control: 50.9 M | SRX2746577; SRX2746578. | [32] |
| Treatment: 51.4 M | |||||||
| 31 | Japanese flounder | Edwardsiella tarda | Gill | Illumina HiSeq 4000 | 417.1 M | PRJNA359626 | [33] |
| 32 | Hybrid tilapia | Streptococcus agalactiae | Liver, intestine & brain | Illumina HiSeq 2000 | NA | NA | [59] |
| 33 | Channel catfish | Flavobacterium columnare | Gill | Illumina HiSeq 2000 | 23 M a | SRP070957 | [37] |
| 34 | Darkbarbel catfish | Aeromonas hydrophila | Spleen | Illumina HiSeq 3000 | 439.9 M * | PRJNA383309 | [60] |
| Sl No | Fish | Virus | Sample | Sequencing Platform | Sequencing Depth/Library | Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Rainbow trout | Poly I:C (viral mimic) | Erythrocytes | ABI SOLiD3 | 80 M *,a | NA | [66] |
| 2 | Large yellow croaker | Poly I:C (viral mimic) | Spleen | Illumina Genome Analyzer II | 56.3 M | SRP035897 | [67] |
| 3 | Rainbow trout | Viral Hemorrhagic Septicemia Virus | Spleen & rayed fin | Illumina HiSeq 2000 | 3.119 B | NA | [68] |
| 4 | Grass carp | Grass carp reovirus | Gill, intestine, liver & spleen | Illumina HiSeq 2000 | 203 M | SRA099702 | [69] |
| 5 | Atlantic salmon | Infectious salmon anaemia virus | Head kidney, liver & gills | Illumina MiSeq | 196 M | SRX658605 | [70] |
| 6 | Grass carp | Grass carp reovirus | Head kidney & spleen | Illumina MiSeq | 107.9 M * | SRP049081 | [71] |
| 7 | Miiuy croaker | Poly I:C (viral mimic) | Spleen | Illumina HiSeq 2000 | Control: 50.3 M * | NA | [72] |
| Treated: 48.2 M * | |||||||
| 8 | Grass carp | Grass carp reovirus | Spleen | Illumina HiSeq 2000 | Control: 2.72 M * | NA | [73] |
| Infected: 2.75 M * | |||||||
| 9 | Olive flounder | Heat inactivated viral hemorrhagic septicemia virus | Kidney | Illumina HiSeq 2500 | Control: 23.7 M * | NA | [74] |
| Day 1: 24.4 M * | |||||||
| Day 3: 23.8 M * | |||||||
| Day 7: 25.9 M * | |||||||
| 10 | Atlantic salmon | Infectious salmon anaemia virus | Spleen | Illumina MiSeq | 92.8 M * | NA | [75] |
| 11 | Schizothorax | Poly I:C (viral mimic) | Spleen | Illumina Hiseq 2500 | Control: 53.8 M * | NA | [76] |
| Treated: 55.1 M * | |||||||
| 12 | Orange spotted grouper | Megalocytivirus and Ranavirus | Spleen | Illumina HiSeq | NA | NA | [77] |
| 13 | Grouper | Betanodavirus | Kidney cell line | Illumina HiSeq 2000 | 51 M | JR139431–JR139455 | [78] |
| JX644070 | |||||||
| 14 | Sockeye salmon | Piscine reovirus along with superinfection of infectious hematopoietic necrosis virus | Kidney | Illumina HiSeq 2000 | 2.3 B | SRP0740078 | [79] |
| 15 | Atlantic salmon | Salmonid | TO-cells, a macrophage/dendritic like cell line | Illumina HiSeq 2000 | Interferon treated: 23.1 M * | GSE64095 | [80] |
| SAV3 infection: 24.6 M * | |||||||
| alphavirus subtype-3 | Control: 23.4 M * | ||||||
| 16 | Gilthead seabream & European seabass | Nervous necrosis virus (Betanodavirus) | Leucocytes | Illumina HiSeq 2000 | 53 M a | GSE101662 | [81] |
| 17 | Koi carp | Cyprinid Herpesvirus 3 | Spleen | Illumina HiSeq 2500 | 111.2 M | NA | [82] |
| 18 | Yellow catfish | Poly I:C (viral mimic) | Liver | Illumina HiSeq 2000 | Control: 42.2 M * | NA | [83] |
| Treated: 41.5 M * | |||||||
| 19 | Zebrafish | Spring viremia of carp virus | Brain & spleen | Illumina HiSeq 4000 | 360.9 M * | NA | [84] |
| 20 | Olive flounder | Viral hemorrhagic septicemia virus | Kidney | Illumina HiSeq 2000 | 215.8 M | NA | [85] |
| 21 | Orange spotted grouper | Singapore grouper iridovirus | Spleen | Roche 454 Genome | Control: 0.428 M | SRA040065.1 | [86] |
| Sequencer FLX | Infected: 0.446 M | ||||||
| 22 | Turbot | Megalocytivirus | Spleen | Illumina HiSeq 2000 | S16-4d: 82.9 M | SAMN06186733 to SAMN06186736 | [87] |
| S16-8d: 86.5 M | |||||||
| S16-4d: 81.4 M | |||||||
| S16-4d: 79.1 M |
| Sl No | Fish | Parasite Species | Sample | Sequencing Platform | Sequencing Depth/Library | Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Large yellow croaker | Enteromyxum scophthalmi | Head kidney, spleen & pyloric caeca | Illumina HiSeq 2000 | 170 M | GSE63911 | [107] |
| 2 | Three spined sticklebacks | Diplostomum pseudospathaceum | Head kidney & gills | Illumina HiScan SQ | 486 M | PRJNA253091 | [108] |
| 3 | _ | Caligus rogercresseyi | Nauplius I, nauplius II, copepodid, chalimus, adult male & female | Illumina MiSeq | 154.84 M | SRR1106551 | [109] |
| 4 | Three-spined sticklebacks | Diplostomum pseudospathaceum | Head kidney & gills | Illumina HiScan SQ | 990 M | PRJNA276419 | [110] |
| 5 | Large yellow croaker | Cryptocaryon irritans | Liver | Illumina HiSeq 2000 | Control: 51.9 M | NA | [111] |
| Infected: 54.8 M | |||||||
| 6 | Turbot | Enteromyxum scophthalmi | Spleen, head kidney & pyloric caeca | Illumina HiSeq 2000 | 170 M | PRJNA300347 | [112] |
| 7 | Large yellow croaker | Cryptocaryon irritans | Gill, skin, spleen, head kidney & liver | Illumina MiSeq | 49.5 M | NA | [113] |
| 8 | _ | Cryptocaryon irritans | Tomont stage | Illumina MiSeq | 80.8 M | NA | [114] |
| 9 | _ | Cryptocaryon irritans | Trophonts, tomonts, and theronts stages | Illumina HiSeq 2500 | Trophonts: 79.35 M | SUB1416064 | [115] |
| Tomont: 66.42 M | SUB1416075 | ||||||
| Theronts:123.62 M | SUB1416142 | ||||||
| 10 | Orange spotted grouper | Cryptocaryon irritans | Skin | Illumina HiSeq 2500 | 506.6 M | GSE97397 | [116] |
| Sl No | Fish | Sample | Sequencing Platform | Sequencing Depth/Library | Accession Number | Reference |
|---|---|---|---|---|---|---|
| 1 | Ayu | Macrophage | Illumina HiSeq 2000 | 27.96 M | SRA047923.1 | [135] |
| JP722270–JP772077 | ||||||
| 2 | Mud loach | Skin | Illumina Genome Analyzer II | 111 M | SRA057415 | [138] |
| 3 | European sea bass | Gills | Illumina HiSeq2000 | 68.6 M | NA | [133] |
| 4 | Fugu | Gills & swimbladder | Illumina HiSeq 2000 | Gill: 55 M | SRA109280 | [131] |
| Swimbladder: 44.7 M | SRA109284 | |||||
| 5 | Rainbow trout | Spleen | Illumina Genome Analyzer | 93.5 M | NA | [126] |
| 6 | Japanese flounder | Spleen | Illumina MiSeq | 14.6 M | SRR1515192 | [2] |
| 7 | Naked carp | Gills & kidney | Illumina HiSeq 2000 | Gill: 90.5 M | SRX673786 | [127] |
| Kidney: 90.4 M | SRX673788 | |||||
| 8 | Tilapia | Brain, pituitary, gill, heart, liver, spleen, kidney, intestine, muscle, testis & ovary | Illumina Genome Analyzer | 52.4 M | SRS676061 | [132] |
| 9 | Topmouth culter | Brain, heart, gill, liver, intestines, muscle, gonads, kidney & pancreas | Illumina HiSeq 4000 | 44.9 M | SRX1502706 | [129] |
| 10 | Grass carp | Spleen | Illumina HiSeq 4000 | 10.1 M | SRP078553 | [130] |
| 11 | Tibetan Schizothoracinae | Gills & kidney | Illumina HiSeq 2000 | Gill: 85.3 M | SRX673793 | [128] |
| Kidney: 88.7 M | SRX673788 | |||||
| 12 | Zebrafish | Whole animal | Illumina HiSeq 2000 | 142.9 M | SRR4045953 | [139] |
| 13 | Zebrafish | Kidney-derived blood cells | Illumina HiSeq 2500 | 1 M a | E-MTAB-5530 | [136] |
| E-MTAB-4617 | ||||||
| E-MTAB-3947 | ||||||
| 14 | Zebrafish | Spleen-derived lck:GFP cells | Illumina HiSeq 2000 | 2.1 M *,a | E-MTAB-4617 | [137] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. https://doi.org/10.3390/ijms19010245
Sudhagar A, Kumar G, El-Matbouli M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. International Journal of Molecular Sciences. 2018; 19(1):245. https://doi.org/10.3390/ijms19010245
Chicago/Turabian StyleSudhagar, Arun, Gokhlesh Kumar, and Mansour El-Matbouli. 2018. "Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review" International Journal of Molecular Sciences 19, no. 1: 245. https://doi.org/10.3390/ijms19010245
APA StyleSudhagar, A., Kumar, G., & El-Matbouli, M. (2018). Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. International Journal of Molecular Sciences, 19(1), 245. https://doi.org/10.3390/ijms19010245