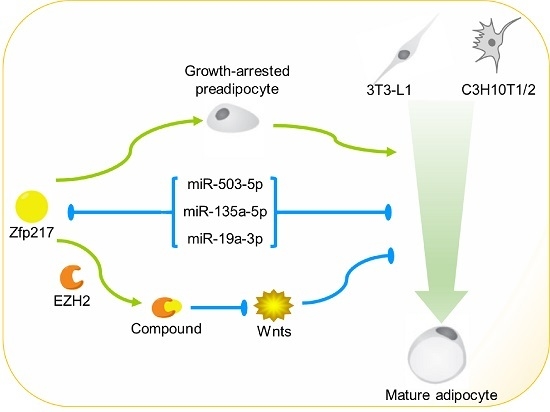
The Emerging Role of Zfp217 in Adipogenesis
Abstract
:1. Introduction
2. Results
2.1. The Potential Adipogenesis Role of Zfp217 Based on Gene Expression Omnibus (GEO) Datasets
2.2. The Association of Zfp217 Expression with Adipogenesis and Obesity
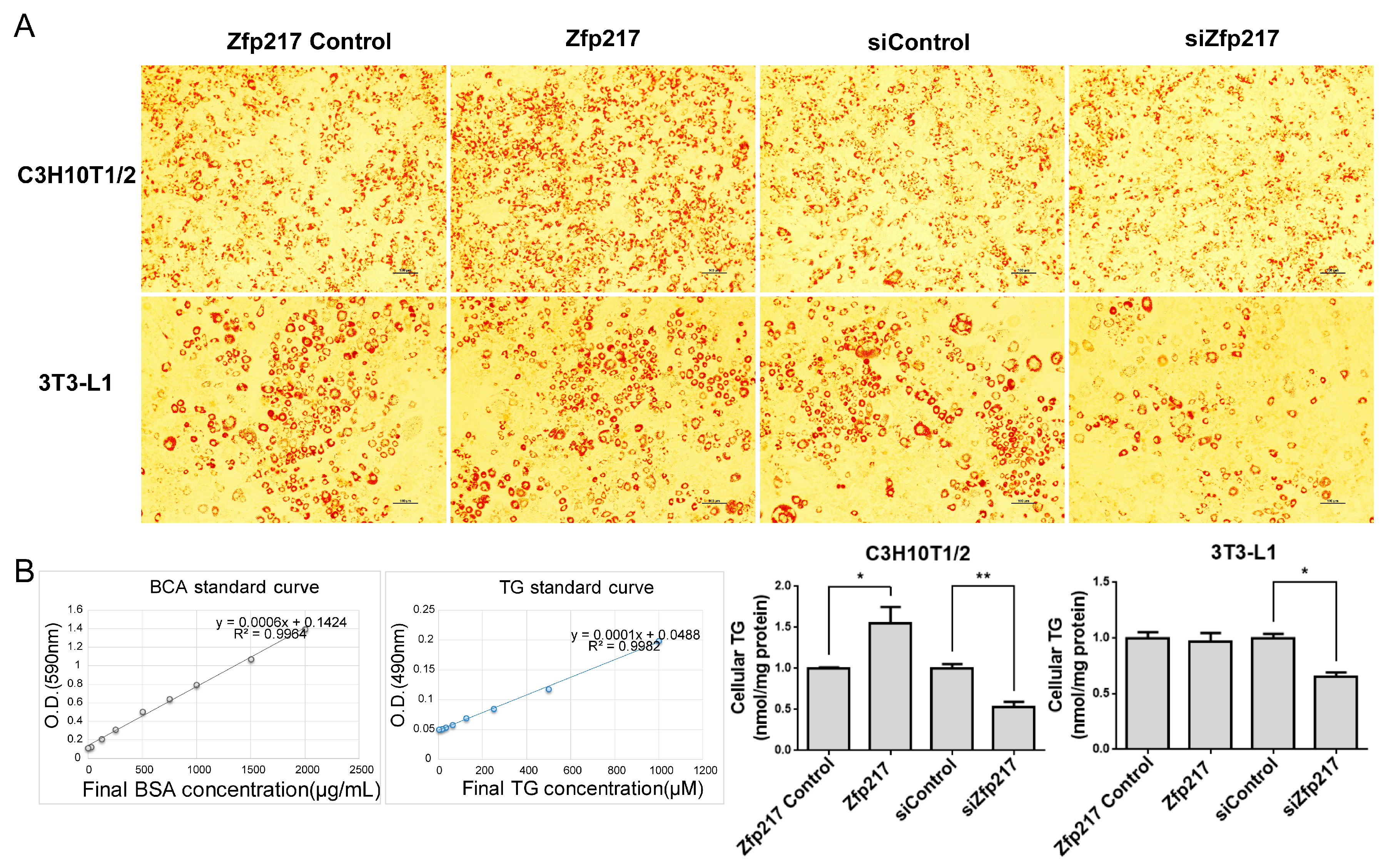
2.3. Zfp217 Regulates Adipogenesis of C3H10T1/2 and 3T3-L1
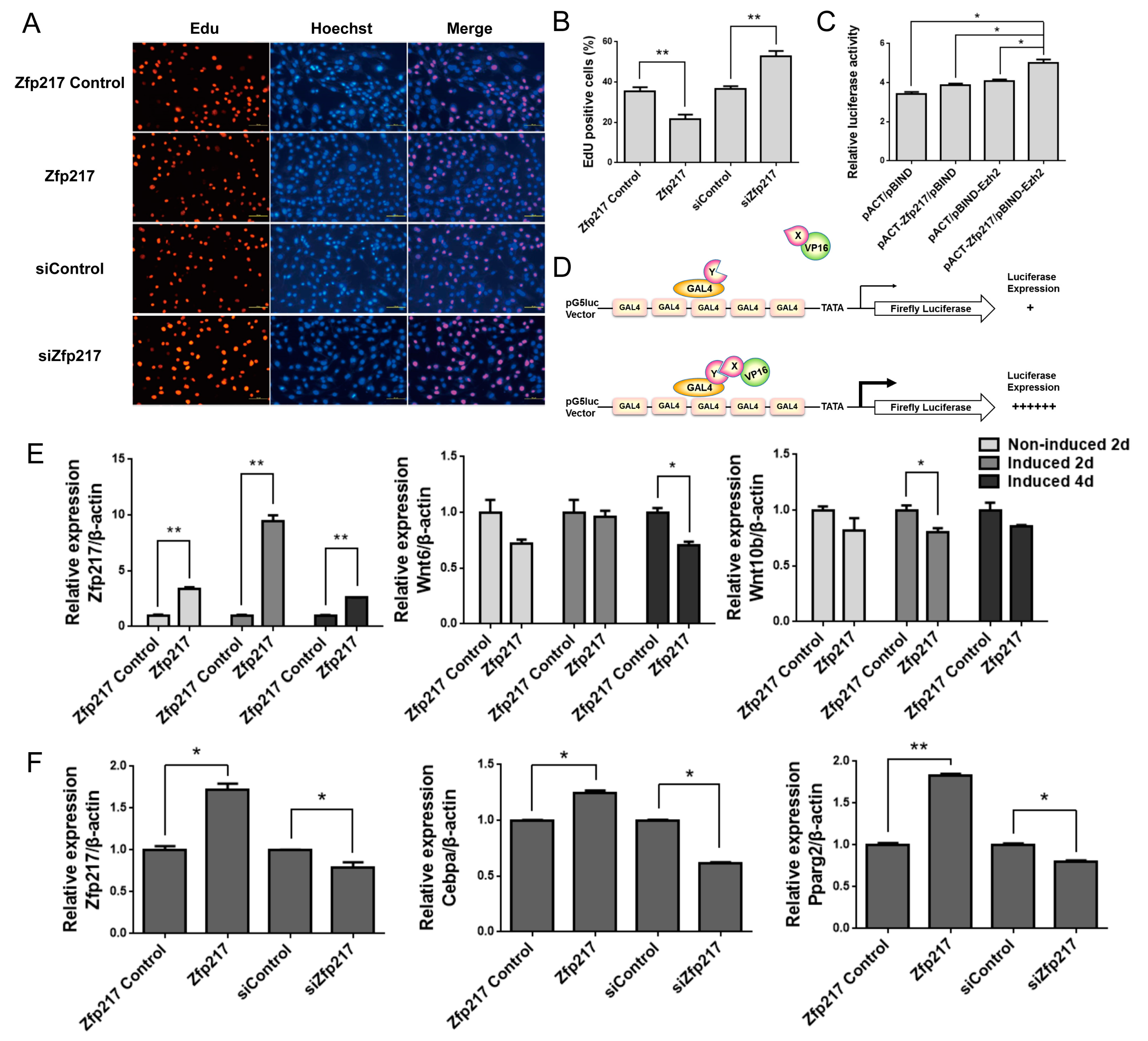
2.4. Zfp217 Positively Regulates Adipogenesis by Suppressing Cell Proliferation and Interacting with Ezh2
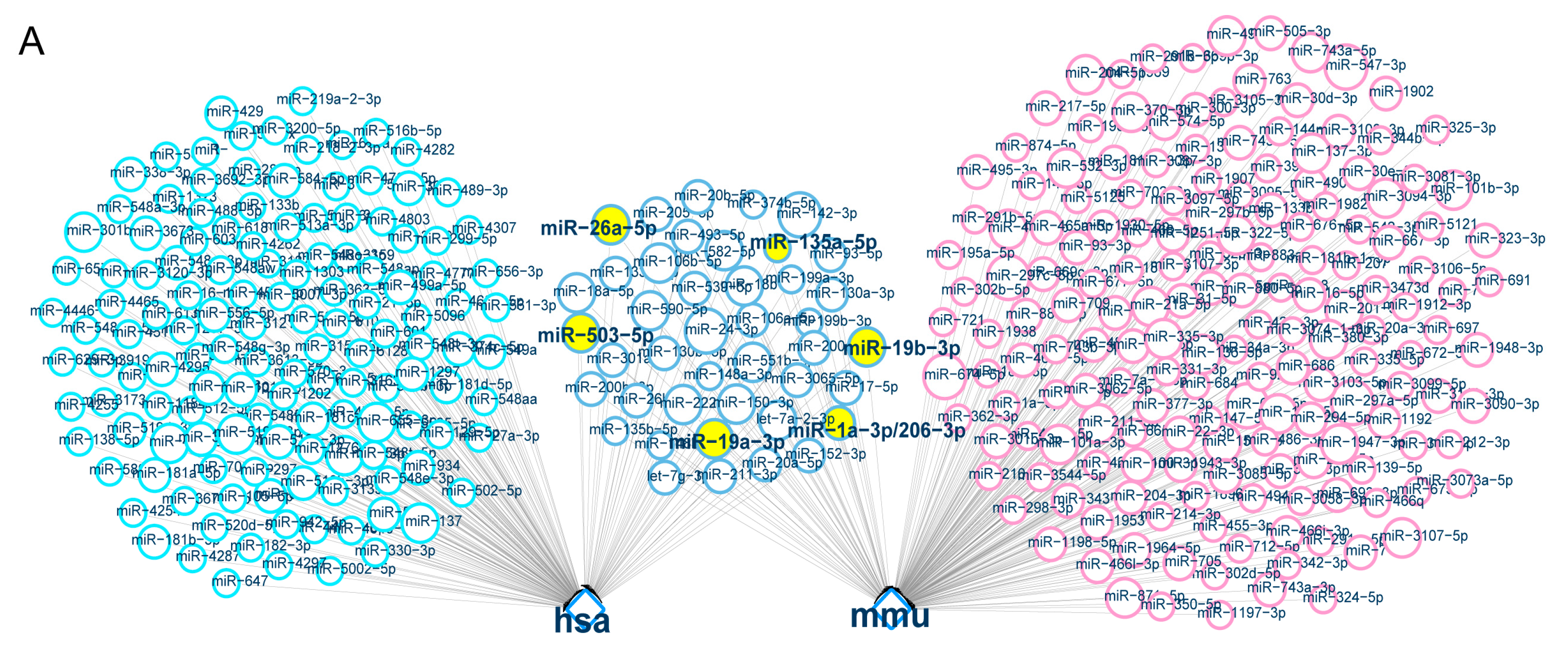
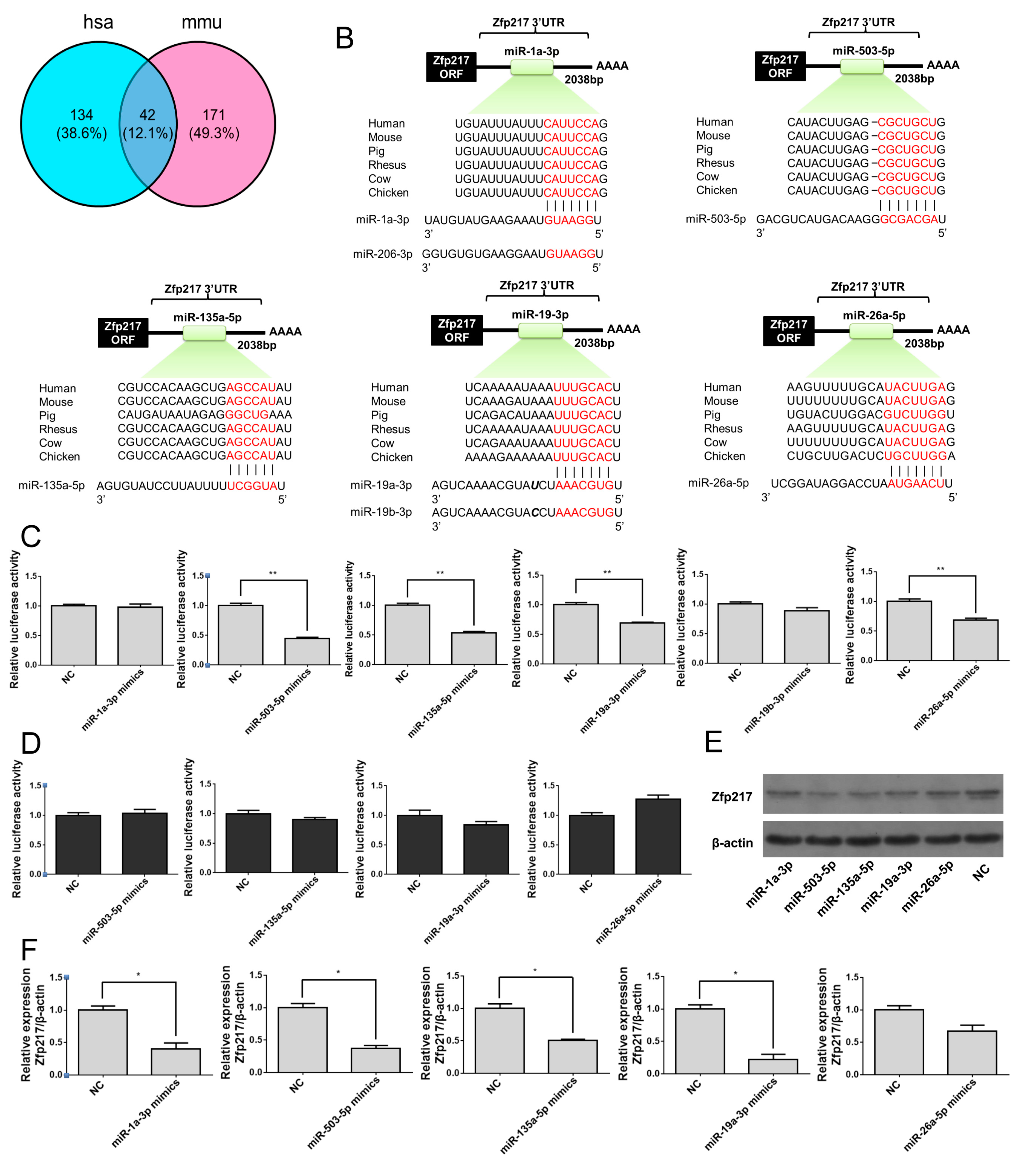
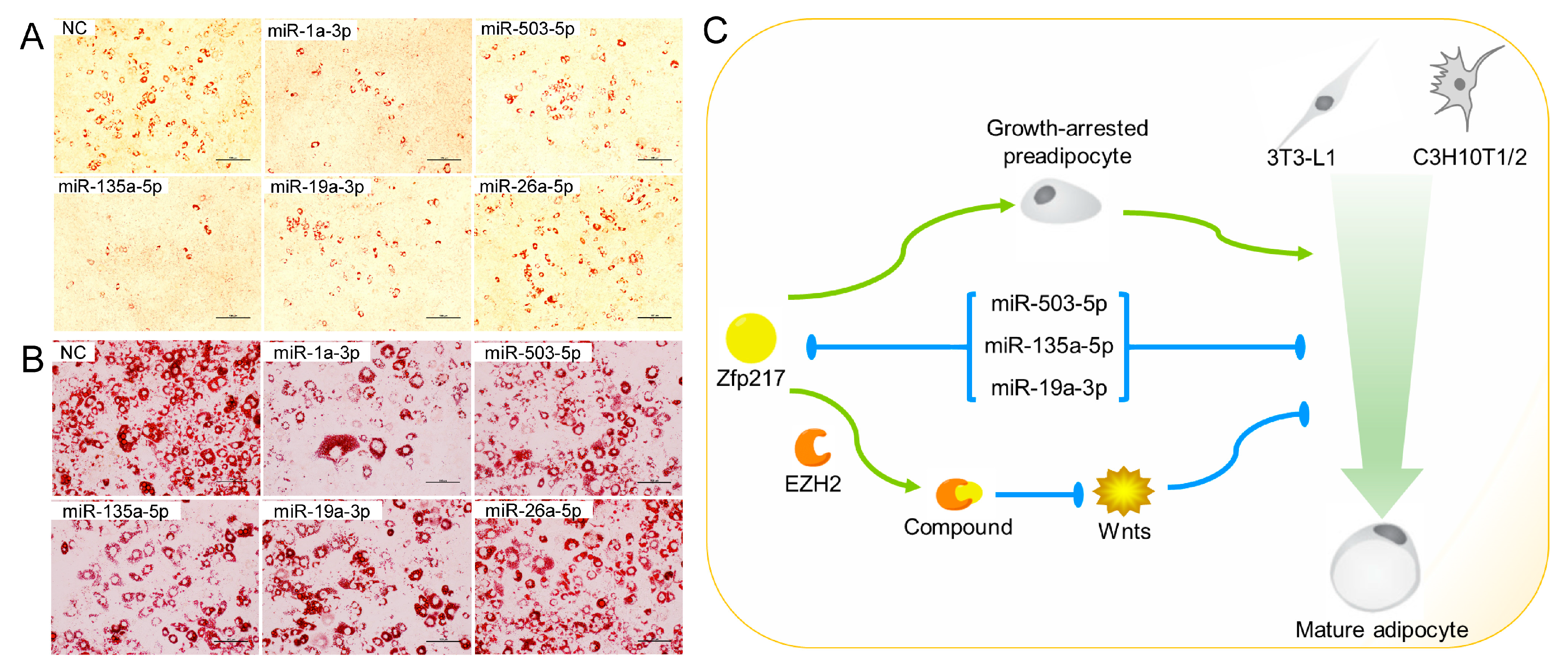
2.5. Direct Targeting of Zfp217 by miR-503-5p, miR-135a-5p and miR-19a-3p Impairs Adipocyte Differentiation
3. Discussion
4. Materials and Methods
4.1. Analysis of Gene Expression Omnibus Series (GSE) Data
4.2. Animals
4.3. Cell Culture and Adipocyte Differentiation
4.4. Oil Red O Staining
4.5. Triglyceride Assay
4.6. Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
4.7. Western Blotting
4.8. EdU Cell Proliferation Assay
4.9. Mammalian Two-Hybrid Assay
4.10. The Prediction and Screening of miRNAs
4.11. Dual-Luciferase Reporter Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Zfp217 | Zinc Finger Protein 217 |
| Pparg | Peroxisome Proliferator-Activated Receptor g |
| Cebps | CCAAT/Enhancer-Binding Proteins |
| DEGs | Differentially Expressed Genes |
| ASVC | Adipocytic Differentiation of Porcine Subcutaneous Stromal Vascular Cells |
| GEO | Gene Expression Omnibus |
| GSE | Gene Expression Omnibus Series |
| GPL | Gene Expression Omnibus Platform |
| DM | Diabetes Mellitus |
| IGT | Impaired Glucose Tolerance |
| NGT | Normal Glucose Tolerance |
| HFD | High Fat Diet Fed Mice |
| NCD | Normal Chow Diet Fed Mice |
| ORF | Open Reading Frame |
| EdU | 5-Ethynyl-2’-Deoxyuridine |
| TG | Triglycerides |
| BCA | Bicinchoninic Acid |
| wt | Wild Type |
| mut | Mutant Type |
| hsa | Homo sapiens (human) |
| mmu | Mus musculus (Mouse) |
| WAT | White Adipose Tissue |
| BAT | Brown Adipose Tissue |
| PVDF | Polyvinylidene Difluoride |
| EMT | Epithelial-to-Mesenchymal Transition |
| SBGS | Simpson-Golabi-Behmel Syndrome |
| GO | Gene Ontology |
| KM | Kun Ming |
References
- Malik, V.S.; Willett, W.C.; Hu, F.B. Global obesity: Trends, risk factors and policy implications. Nat. Rev. Endocrinol. 2013, 9, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Lazar, M.A. New developments in adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J.; Liu, X.S.; et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhang, L.; Zhou, X.; Du, M.; Jiang, Z.; Hausman, G.J.; Bergen, W.G.; Zan, L.; Dodson, M.V. Emerging roles of zinc finger proteins in regulating adipogenesis. Cell. Mol. Life Sci. 2013, 70, 4569–4584. [Google Scholar] [CrossRef] [PubMed]
- Tseng, K.-Y.; Chen, Y.-H.; Lin, S. Zinc finger protein ZFP36L1 promotes osteoblastic differentiation but represses adipogenic differentiation of mouse multipotent cells. Oncotarget 2017, 8, 20588–20601. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.M.; Zhou, Y.F.; Yang, Y.; Peng, J.; Song, T.X.; Xu, T.; Wei, H.K.; Jiang, S.W.; Peng, J. Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Laudes, M.; Christodoulides, C.; Sewter, C.; Rochford, J.J.; Considine, R.V.; Sethi, J.K.; Vidal-Puig, A.; O’Rahilly, S. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J. Biol. Chem. 2004, 279, 11711–11718. [Google Scholar] [CrossRef] [PubMed]
- Younce, C.W.; Azfer, A.; Kolattukudy, P.E. MCP-1 (monocyte chemotactic protein-1)-induced protein, a recently identified zinc finger protein, induces adipogenesis in 3T3-L1 pre-adipocytes without peroxisome proliferator-activated receptor γ. J. Biol. Chem. 2009, 284, 27620–27628. [Google Scholar] [CrossRef] [PubMed]
- Kawagishi, H.; Wakoh, T.; Uno, H.; Maruyama, M.; Moriya, A.; Morikawa, S.; Okano, H.; Sherr, C.J.; Takagi, M.; Sugimoto, M. Hzf regulates adipogenesis through translational control of C/EBP α. EMBO J. 2008, 27, 1481–1490. [Google Scholar] [PubMed]
- Sanchez-Solana, B.; Li, D.Q.; Kumar, R. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochim. Biophys. Acta 2014, 1843, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Dorronsoro, A.; Lang, V.; Jakobsson, E.; Ferrin, I.; Salcedo, J.M.; Fernandez-Rueda, J.; Fechter, K.; Rodriguez, M.S.; Trigueros, C. Identification of the NF-κB inhibitor A20 as a key regulator for human adipogenesis. Cell Death Dis. 2013, 4, e972. [Google Scholar] [CrossRef] [PubMed]
- Charrier, A.; Wang, L.; Stephenson, E.J.; Ghanta, S.V.; Ko, C.W.; Croniger, C.M.; Bridges, D.; Buchner, D.A. Zinc finger protein 407 overexpression upregulates PPAR target gene expression and improves glucose homeostasis in mice. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E869–E880. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Ihara, Y.; Tsukamoto, D.; Yasumoto, K.; Hashidume, T.; Kamimura, K.; Nakai, Y.; Hirano, S.; Shimizu, M.; Kominami, R.; et al. Identification of BCL11B as a regulator of adipogenesis. Sci. Rep. 2016, 6, 32750. [Google Scholar] [CrossRef] [PubMed]
- Heiker, J.T.; Kloting, N. Replication initiator 1 in adipose tissue function and human obesity. In Obesity; Litwack, G., Ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; Volume 91, pp. 97–105. [Google Scholar]
- Lee, Y.H.; Kim, S.H.; Lee, Y.J.; Kang, E.S.; Lee, B.W.; Cha, B.S.; Kim, J.W.; Song, D.H.; Lee, H.C. Transcription factor snail is a novel regulator of adipocyte differentiation via inhibiting the expression of peroxisome proliferator-activated receptor γ. Cell. Mol. Life Sci. 2013, 70, 3959–3971. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, S. Role of kruppel-like transcription factors in adipogenesis. Dev. Biol. 2013, 373, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Z.; Wei, H.K.; Song, T.X.; Yang, Y.; Peng, J.; Jiang, S.W. Transcriptome comparison between porcine subcutaneous and intramuscular stromal vascular cells during adipogenic differentiation. PLoS ONE 2013, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Rommens, J.M.; Kowbel, D.; Godfrey, T.; Tanner, M.; Hwang, S.I.; Polikoff, D.; Nonet, G.; Cochran, J.; Myambo, K.; et al. Positional cloning of ZNF217 and NABC1: Genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl. Acad. Sci. USA 1998, 95, 8703–8708. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.K.; Zhang, C.W.; Qi, S.Y.; Guo, S.Q.; Chen, Y.; Du, E.; Zhang, H.T.; Wang, X.M.; Liu, R.L.; Qiao, B.M.; et al. Elevated expression of ZNF217 promotes prostate cancer growth by restraining ferroportin-conducted iron egress. Oncotarget 2016, 7, 84893–84906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-C.; Zheng, L.-Q.; Pan, L.-J.; Guo, J.-X.; Yang, G.-S. Znf217 is overexpressed and enhances cell migration and invasion in colorectal carcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 2459–2463. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; Vendrell, J.A.; Poulard, C.; Győrffy, B.; Goddard-Léon, S.; Bièche, I.; Corbo, L.; le Romancer, M.; Bachelot, T.; Treilleux, I.; et al. A functional interplay between ZNF217 and estrogen receptor α exists in luminal breast cancers. Mol. Oncol. 2014, 8, 1441–1457. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Nakayama, K.; Rahman, M.; Nakayama, N.; Ishikawa, M.; Katagiri, A.; Iida, K.; Nakayama, S.; Otsuki, Y.; Shih, I.-M.; et al. Prognostic and therapeutic impact of the chromosome 20q13.2 ZNF217 locus amplification in ovarian clear cell carcinoma. Cancer 2012, 118, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Rooney, P.H.; Boonsong, A.; McFadyen, M.C.E.; McLeod, H.L.; Cassidy, J.; Curran, S.; Murray, G.I. The candidate oncogene ZNF217 is frequently amplified in colon cancer. J. Pathol. 2004, 204, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Nonet, G.H.; Stampfer, M.R.; Chin, K.; Gray, J.W.; Collins, C.C.; Yaswen, P. The ZNF217 gene amplified in breast cancers promotes immortalization of human mammary epithelial cells. Cancer Res. 2001, 61, 1250–1254. [Google Scholar] [PubMed]
- Vendrell, J.A.; Thollet, A.; Nguyen, N.T.; Ghayad, S.E.; Vinot, S.; Biéche, I.; Grisard, E.; Josserand, V.; Coll, J.-L.; Roux, P.; et al. Znf217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res. 2012, 72, 3593–3606. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Zhang, F.; Sancho, A.; Fidalgo, M.; di Cecilia, S.; Vashisht, A.; Lee, D.F.; Chen, C.H.; Rengasamy, M.; Andino, B.; et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 2015, 17, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Littlepage, L.E.; Adler, A.S.; Kouros-Mehr, H.; Huang, G.; Chou, J.; Krig, S.R.; Griffith, O.L.; Korkola, J.E.; Qu, K.; Lawson, D.A.; et al. The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov. 2012, 2, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Donini, C.F.; Nguyen, N.T.; Lincet, H.; Vendrell, J.A. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget 2015, 6, 41566–41581. [Google Scholar] [PubMed]
- Pang, F.; Zha, R.P.; Zhao, Y.J.; Wang, Q.F.; Chen, D.; Zhang, Z.F.; Chen, T.Y.; Yao, M.; Gu, J.R.; He, X.H. miR-525–3p enhances the migration and invasion of liver cancer cells by downregulating ZNF395. PLoS ONE 2014, 9, e90867. [Google Scholar] [CrossRef] [PubMed]
- Harder, L.; Puller, A.-C.; Horstmann, M.A. ZNF423: Transcriptional modulation in development and cancer. Mol. Cell. Oncol. 2014, 1, e969655. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, M.G.; Oswald, E.; Yu, W.; Xue, F.; MacKerell, A.D., Jr.; Melnick, A.M. The expanding role of the BCL6 oncoprotein as a cancer therapeutic target. Clin. Cancer Res. 2017, 23, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Spina, R.; Filocamo, G.; Iaccino, E.; Scicchitano, S.; Lupia, M.; Chiarella, E.; Mega, T.; Bernaudo, F.; Pelaggi, D.; Mesuraca, M.; et al. Critical role of zinc finger protein 521 in the control of growth, clonogenicity and tumorigenic potential of medulloblastoma cells. Oncotarget 2013, 4, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Pulkka, O.P.; Nilsson, B.; Sarlomo-Rikala, M.; Reichardt, P.; Eriksson, M.; Hall, K.S.; Wardelmann, E.; Vehtari, A.; Joensuu, H.; Sihto, H. Slug transcription factor: A pro-survival and prognostic factor in gastrointestinal stromal tumour. Br. J. Cancer 2017, 116, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Won, M.; Shin, E.; Kim, H.M.; Lee, K.; Bae, J. EGR2 is a gonadotropin-induced survival factor that controls the expression of IER3 in ovarian granulosa cells. Biochem. Biophys. Res. Commun. 2017, 482, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Mak, V.C.Y.; Wong, O.G.W.; Siu, M.K.Y.; Wong, E.S.Y.; Ng, W.-Y.; Wong, R.W.C.; Chan, K.-K.; Ngan, H.Y.S.; Cheung, A.N.Y. FBI-1 is overexpressed in gestational trophoblastic disease and promotes tumor growth and cell aggressiveness of choriocarcinoma via PI3K/Akt signaling. Am. J. Pathol. 2015, 185, 2038–2048. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Yang, J.P.; Hu, J.H.; Wang, L.N.; Zuo, J.L. MCP-1 stimulates spinal microglia via PI3K/Akt pathway in bone cancer pain. Brain Res. 2015, 1599, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.M.; Hao, A.X.; Zhang, Q.; Sui, G.C. The role of YY1 in oncogenesis and its potential as a drug target in cancer therapies. Curr. Cancer Drug Targets 2015, 15, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Motamed, N.; Rabiee, B.; Keyvani, H.; Hemasi, G.R.; Khonsari, M.; Saeedian, F.S.; Maadi, M.; Zamani, F. The best obesity indices to discriminate type 2 diabetes mellitus. Metab. Syndr. Relat. Disord. 2016, 14, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. MirWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Banck, M.S.; Li, S.; Nishio, H.; Wang, C.; Beutler, A.S.; Walsh, M.J. The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics 2009, 4, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, K.G.; Verger, A.; Yaswen, P.; Crossley, M. Amplification of zinc finger gene 217 (ZNF217) and cancer: When good fingers go bad. Biochim. Biophys. Acta 2007, 1775, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Lyssenko, V.; Almgren, P.; Anevski, D.; Orho-Melander, M.; Sjögren, M.; Saloranta, C.; Tuomi, T.; Groop, L.; Botnia Study, G. Genetic prediction of future type 2 diabetes. PLoS Med. 2005, 2, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Lehrke, M.; Lazar, M.A. The many faces of PPARγ. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Shinoda, K.; Spiegelman, B.M.; Kajimura, S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012, 15, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Lo, K.A.; Sun, L. Turning wat into bat: A review on regulators controlling the browning of white adipocytes. Biosci. Rep. 2013, 33, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Rossmeisl, M.; Barbatelli, G.; Flachs, P.; Brauner, P.; Zingaretti, M.C.; Marelli, M.; Janovska, P.; Horakova, M.; Syrovy, I.; Cinti, S.; et al. Expression of the uncoupling protein 1 from the aP2 gene promoter stimulates mitochondrial biogenesis in unilocular adipocytes in vivo. Eur. J. Biochem. 2002, 269, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Barbatelli, G.; Murano, I.; Madsen, L.; Hao, Q.; Jimenez, M.; Kristiansen, K.; Giacobino, J.P.; de Matteis, R.; Cinti, S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E1244–E1253. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Frontini, A.; Cinti, S. Convertible visceral fat as a therapeutic target to curb obesity. Nat. Rev. Drug Discov. 2016, 15, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Nov, O.; Shapiro, H.; Ovadia, H.; Tarnovscki, T.; Dvir, I.; Shemesh, E.; Kovsan, J.; Shelef, I.; Carmi, Y.; Voronov, E.; et al. Interleukin-1β regulates fat-liver crosstalk in obesity by auto-paracrine modulation of adipose tissue inflammation and expandability. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Kano, F.; Shiota, K.; Murata, M. Expression of the peroxisome proliferator activated receptor γ gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Bookout, A.L.; Lee, S.; Sun, K.; Jia, L.; Lee, C.; Udit, S.; Deng, Y.; Scherer, P.E.; Mangelsdorf, D.J.; et al. PPARγ in vagal neurons regulates high-fat diet induced thermogenesis. Cell Metab. 2014, 19, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Konopka, B.; Szafron, L.M.; Kwiatkowska, E.; Podgorska, A.; Zolocinska, A.; Pienkowska-Grela, B.; Dansonka-Mieszkowska, A.; Balcerak, A.; Lukasik, M.; Stachurska, A.; et al. The significance of c.690G>T polymorphism (rs34529039) and expression of the CEBPA gene in ovarian cancer outcome. Oncotarget 2016, 7, 67412–67424. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-F.; Wu, K.-J. Endothelial transdifferentiation of tumor cells triggered by the twist1-jagged1-KLF4 axis: Relationship between cancer stemness and angiogenesis. Stem Cells Int. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Min, J.-Y.; Min, K.-B. Replication of early B-cell factor 1 (EBF1) gene-by-psychosocial stress interaction effects on central adiposity in a Korean population. J. Prev. Med. Public Health 2016, 49, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jin, Q.; Lee, J.E.; Su, I.H.; Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7317–7322. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, W.P.; Bree, A.J.; Yao, Y.; Du, B.; Hemati, N.; Martinez-Santibanez, G.; MacDougald, O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 2012, 50, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Man, X.F.; Tan, S.W.; Tang, H.N.; Guo, Y.; Tang, C.Y.; Tang, J.; Zhou, C.L.; Zhou, H.D. miR-503 inhibits adipogenesis by targeting bone morphogenetic protein receptor 1a. Am. J. Transl. Res. 2016, 8, 2727–2737. [Google Scholar] [PubMed]
- Chen, C.; Peng, Y.D.; Peng, Y.L.; Peng, J.; Jiang, S.W. miR-135a-5p inhibits 3T3-L1 adipogenesis through activation of canonical Wnt/β-catenin signaling. J. Mol. Endocrinol. 2014, 52, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Pang, W.J.; Zhou, Y.F.; Yao, W.Y.; Xia, L.; Wang, C.; Chen, X.; Zen, K.; Zhang, C.Y.; Yuan, Y.Z. Altered profile of serum microRNAs in pancreatic cancer-associated new-onset diabetes mellitus. J. Diabetes 2016, 8, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Ma, Y.L.; Letu, G.; Wang, H.; Huang, T.H.; Ni, H.W.; Wu, M.J.; Zhou, Z.Q.; Li, M.; Zou, X.N. Expression pattern and regulation of miR-19a and CCND1 in C3H10T1/2 chondrogenic differentiation. Int. J. Clin. Exp. Med. 2016, 9, 13958–13964. [Google Scholar]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, H.; Zhong, Z.-X.; Peng, Y.-D.; Jiang, S.-W. The Emerging Role of Zfp217 in Adipogenesis. Int. J. Mol. Sci. 2017, 18, 1367. https://doi.org/10.3390/ijms18071367
Xiang H, Zhong Z-X, Peng Y-D, Jiang S-W. The Emerging Role of Zfp217 in Adipogenesis. International Journal of Molecular Sciences. 2017; 18(7):1367. https://doi.org/10.3390/ijms18071367
Chicago/Turabian StyleXiang, Hong, Zhu-Xia Zhong, Yong-Dong Peng, and Si-Wen Jiang. 2017. "The Emerging Role of Zfp217 in Adipogenesis" International Journal of Molecular Sciences 18, no. 7: 1367. https://doi.org/10.3390/ijms18071367
APA StyleXiang, H., Zhong, Z.-X., Peng, Y.-D., & Jiang, S.-W. (2017). The Emerging Role of Zfp217 in Adipogenesis. International Journal of Molecular Sciences, 18(7), 1367. https://doi.org/10.3390/ijms18071367