Lipid Storage and Autophagy in Melanoma Cancer Cells
Abstract
:1. Introduction
2. Results
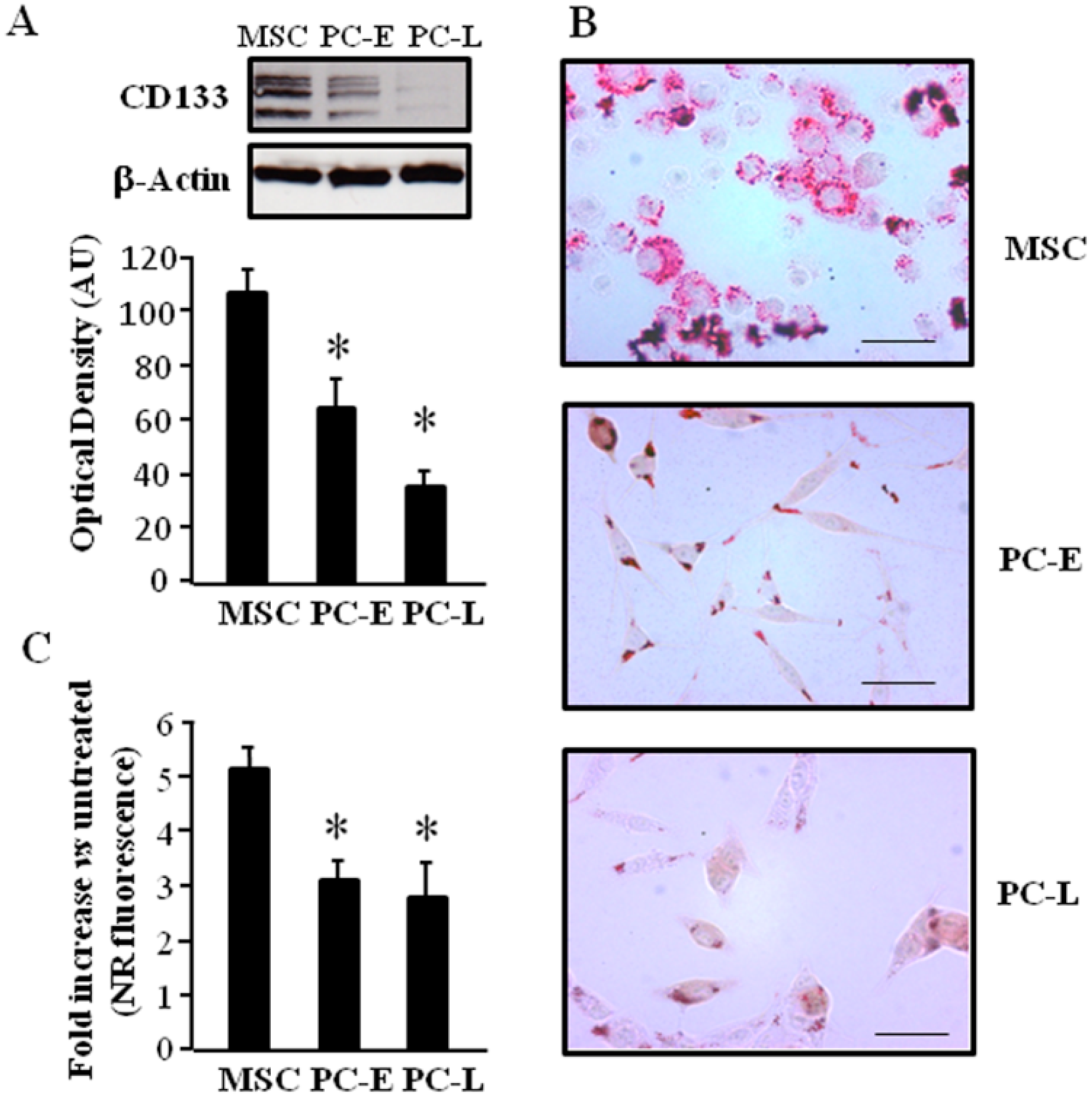
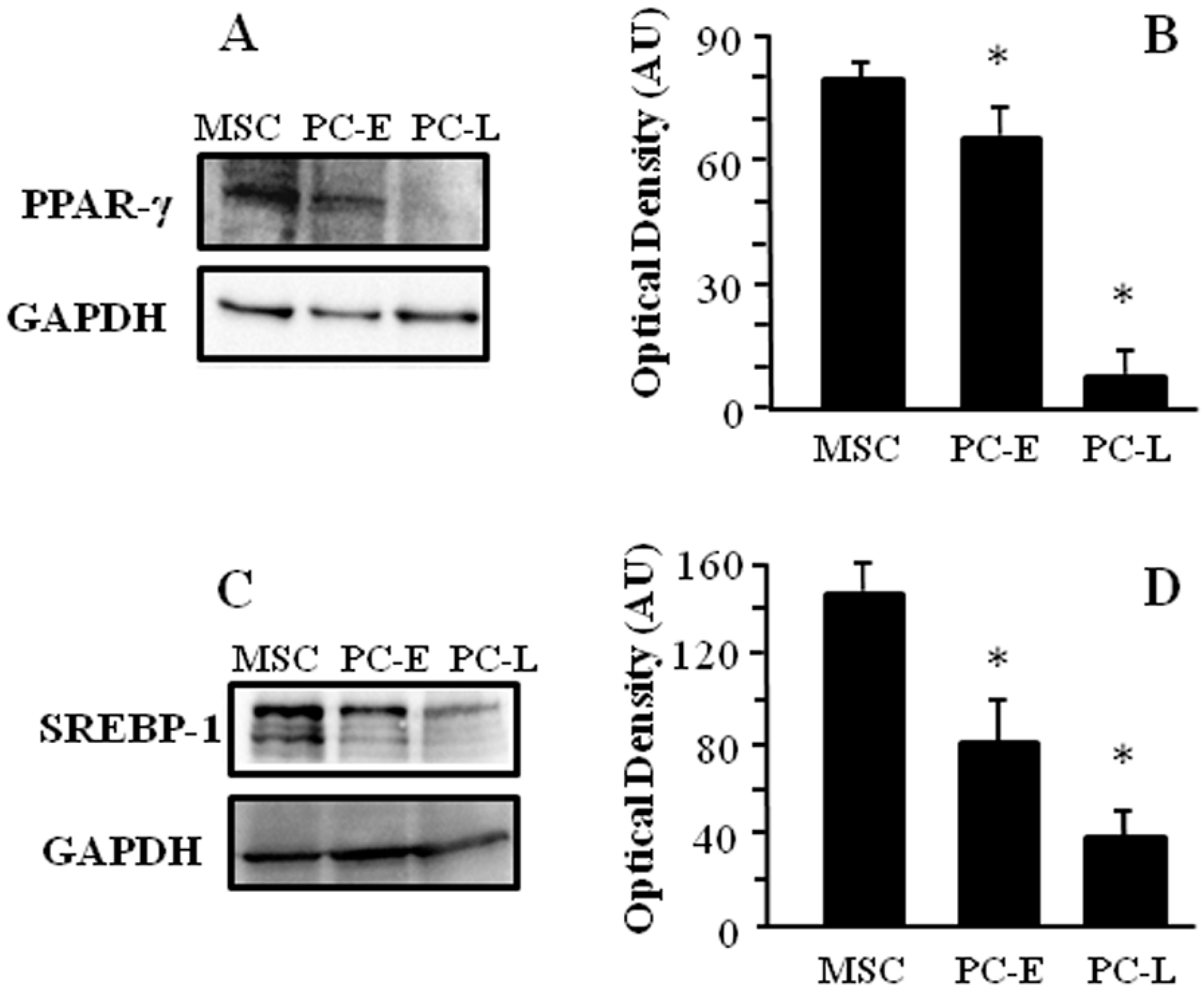
2.1. Cells of Melanospheres Isolated from Metastatic Melanoma Display Abundant LD and Lipid Uptake as well as Strong Peroxisome Proliferator-Activated Receptor-γ (PPAR-γ) and Sterol Regulatory Element-Binding Protein-1 (SREBP-1) Expression
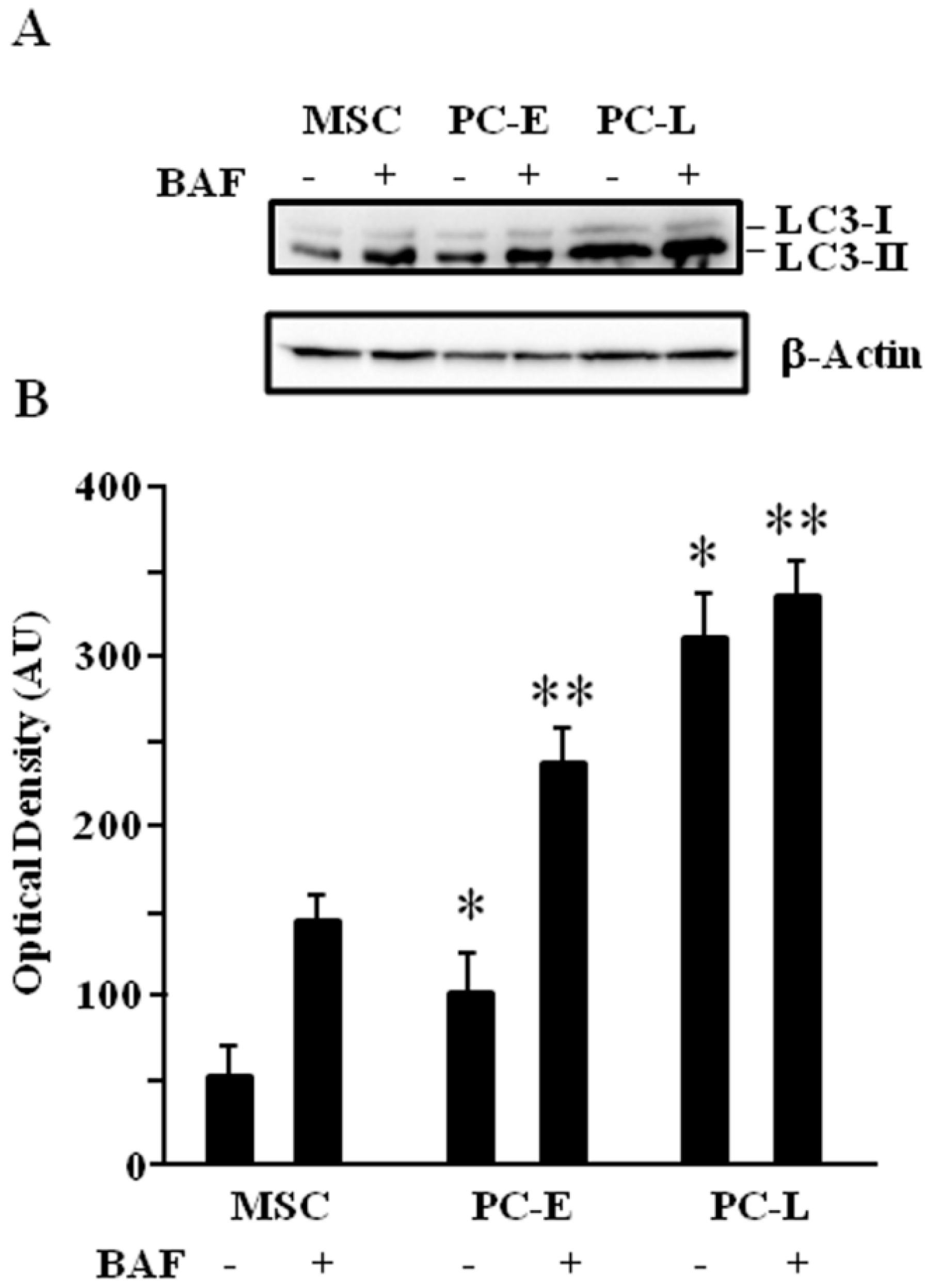
2.2. MSC Display Lower Autophagic Flux as Compared to Differentiating Melanoma Cells
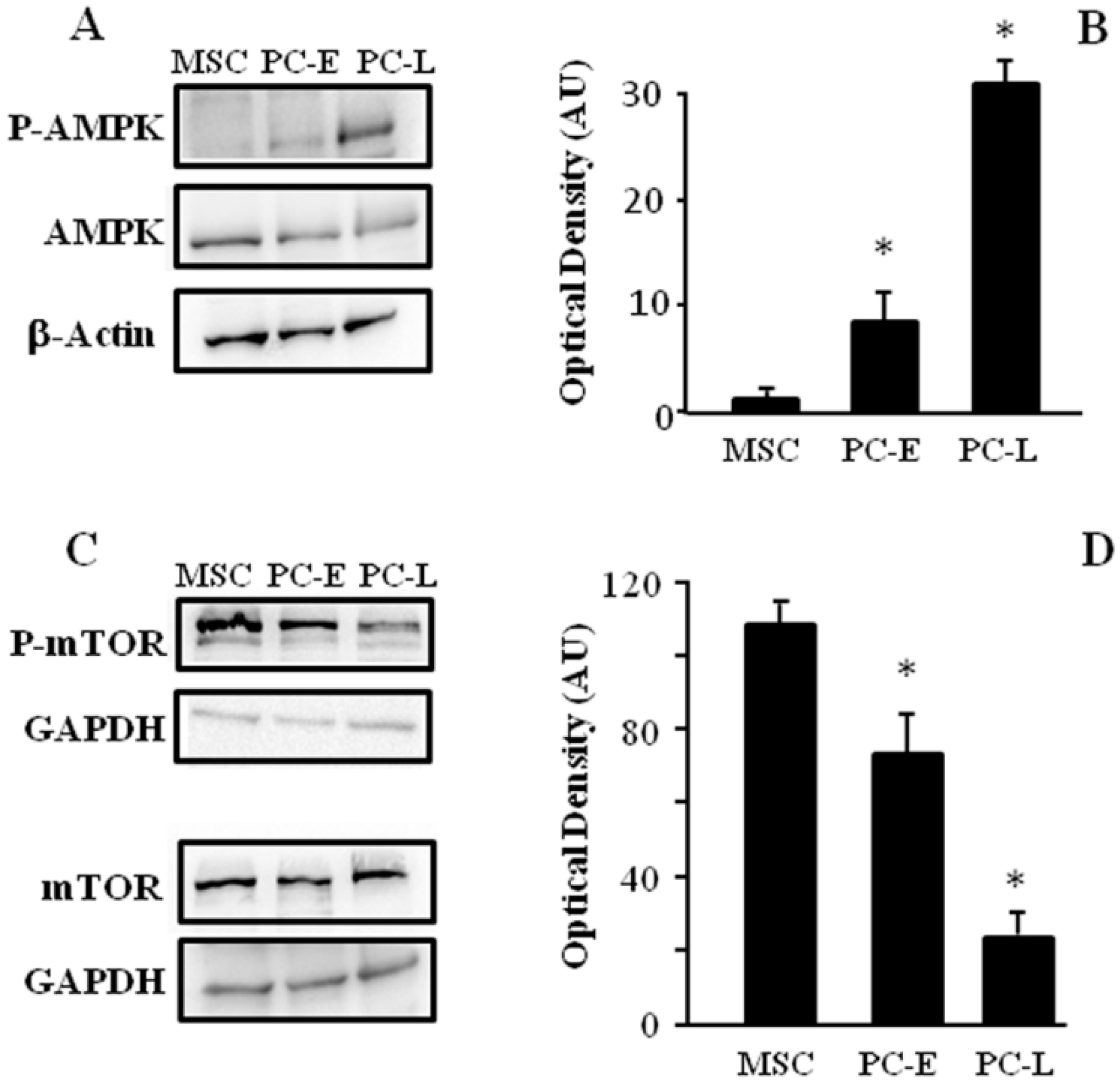
2.3. MSC Display Different AMPK and mTOR Phosphorylation as Compared to PC-E and PC-L
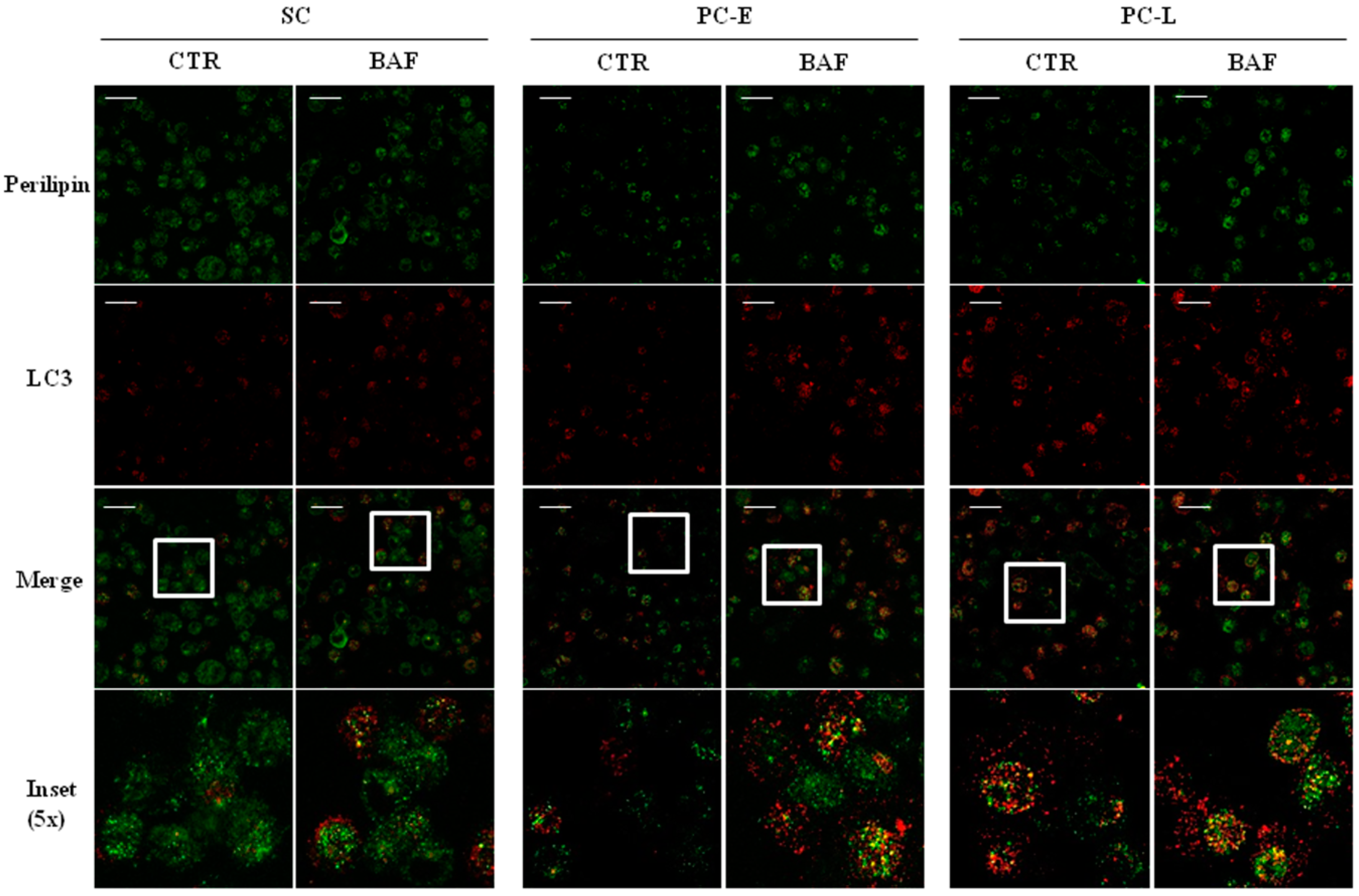
2.4. LD Co-Localization with Autophagosomes in Melanoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Oil Red O Staining
4.3. Lipid Determination by Flow Cytometry
4.4. Immunoblotting
4.5. Immunofluorescence
4.6. Data Handling and Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Singh, S.; Zafar, A.; Khan, S.; Naseem, I. Towards therapeutic advances in melanoma management: An overview. Life Sci. 2017, 174, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Sette, G.; Fecchi, K.; Salvati, V.; Lotti, F.; Pilozzi, E.; Duranti, E.; Biffoni, M.; Pagliuca, A.; Martinetti, D.; Memeo, L.; et al. Mek inhibition results in marked antitumor activity against metastatic melanoma patient-derived melanospheres and in melanosphere-generated xenografts. J. Exp. Clin. Cancer Res. 2013, 32, 91. [Google Scholar] [CrossRef] [PubMed]
- Ailles, L.E.; Weissman, I.L. Cancer stem cells in solid tumors. Curr. Opin. Biotechnol. 2007, 18, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Cojoc, M.; Mabert, K.; Muders, M.H.; Dubrovska, A. A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin. Cancer Biol. 2015, 31, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.L.; Stiles, B.L. Maf1, a new PTEN target linking RNA and lipid metabolism. Trends Endocrinol. Metab. 2016, 27, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.F.; Infante, J.R. Molecular pathways: Fatty acid synthase. Clin Cancer Res. 2015, 21, 5434–5438. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Lee, H.; Nam, M.H.; Jeong, E.; Kim, S.; Hong, Y.; Kim, N.; Yim, H.Y.; Yoo, Y.J.; Kim, J.S.; et al. Loss-of-function screens of druggable targetome against cancer stem-like cells. FASEB J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Tirinato, L.; Liberale, C.; Di Franco, S.; Candeloro, P.; Benfante, A.; La Rocca, R.; Potze, L.; Marotta, R.; Ruffilli, R.; Rajamanickam, V.P.; et al. Lipid droplets: A new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells 2015, 33, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Fargeas, C.A.; Le, T.T.; Corbeil, D.; Lorico, A. Letter to the editor: An intriguing relationship between lipid droplets, cholesterol-binding protein CD133 and Wnt/β-catenin signaling pathway in carcinogenesis. Stem Cells 2015, 33, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Menard, J.A.; Christianson, H.C.; Kucharzewska, P.; Bourseau-Guilmain, E.; Svensson, K.J.; Lindqvist, E.; Indira Chandran, V.; Kjellen, L.; Welinder, C.; Bengzon, J.; et al. Metastasis stimulation by hypoxia and acidosis-induced extracellular lipid uptake is mediated by proteoglycan-dependent endocytosis. Cancer Res. 2016, 76, 4828–4840. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Mercapide, J.; Anzanello, F.; Le, T.T.; Johlfs, M.G.; Fiscus, R.R.; Wilsch-Brauninger, M.; Corbeil, D.; Lorico, A. Wnt interaction and extracellular release of prominin-1/CD133 in human malignant melanoma cells. Exp. Cell Res. 2013, 319, 810–819. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Martinez-Lopez, N.; Otten, E.G.; Carroll, B.; Maetzel, D.; Singh, R.; Sarkar, S.; Korolchuk, V.I. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 2016, 1861, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Shatz, O.; Holland, P.; Elazar, Z.; Simonsen, A. Complex relations between phospholipids, autophagy, and neutral lipids. Trends Biochem. Sci. 2016, 41, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Giampietri, C.; Starace, D.; Petrungaro, S.; Filippini, A.; Ziparo, E. Necroptosis: Molecular signalling and translational implications. Int. J. Cell. Biol. 2014, 2014, 490275. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.P.; Koster, G.; Guillermier, C.; Hirst, E.M.; MacRae, J.I.; Lechene, C.P.; Postle, A.D.; Gould, A.P. Antioxidant role for lipid droplets in a stem cell niche of drosophila. Cell 2015, 163, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Y.; Wang, L.; Wang, N.; Li, Y.; Li, H. Transdifferentiation of fibroblasts into adipocyte-like cells by chicken adipogenic transcription factors. Comp. Biochem. Physiol. A 2010, 156, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Alers, S.; Loffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. Ampk and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell. Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Czaja, M.J. Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. 2011, 22, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Jiang, Y.; Xiao, Y.; Liu, X.D.; Yue, F.; Li, W.; Li, X.; He, Y.; Jiang, X.; Huang, H.; et al. Fast clearance of lipid droplets through MAP1S-activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival. Oncotarget 2016, 7, 6255–6265. [Google Scholar] [PubMed]
- Regad, T. Molecular and cellular pathogenesis of melanoma initiation and progression. Cell. Mol. Life Sci. 2013, 70, 4055–4065. [Google Scholar] [CrossRef] [PubMed]
- Tabolacci, C.; Cordella, M.; Turcano, L.; Rossi, S.; Lentini, A.; Mariotti, S.; Nisini, R.; Sette, G.; Eramo, A.; Piredda, L.; et al. Aloe-emodin exerts a potent anticancer and immunomodulatory activity on BRAF-mutated human melanoma cells. Eur. J. Pharmacol. 2015, 762, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Madjd, Z.; Erfani, E.; Gheytanchi, E.; Moradi-Lakeh, M.; Shariftabrizi, A.; Asadi-Lari, M. Expression of CD133 cancer stem cell marker in melanoma: A systematic review and meta-analysis. Int. J. Biol. Markers 2016, 31, e118–e125. [Google Scholar] [CrossRef] [PubMed]
- Rohwedder, A.; Zhang, Q.; Rudge, S.A.; Wakelam, M.J. Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4. J. Cell. Sci. 2014, 127, 3104–3115. [Google Scholar] [CrossRef] [PubMed]
- Shieh, Y.S.; Chang, Y.S.; Hong, J.R.; Chen, L.J.; Jou, L.K.; Hsu, C.C.; Her, G.M. Increase of hepatic fat accumulation by liver specific expression of hepatitis B virus X protein in zebrafish. Biochim. Biophys. Acta 2010, 1801, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, C.; Wang, X.; Briggs, M.R.; Admon, A.; Wu, J.; Hua, X.; Goldstein, J.L.; Brown, M.S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 1993, 75, 187–197. [Google Scholar] [CrossRef]
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 2008, 320, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.L.; et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Welte, M.A. Proteins under new management: Lipid droplets deliver. Trends Cell. Biol. 2007, 17, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Bozza, P.T.; Viola, J.P. Lipid droplets in inflammation and cancer. Prostaglandins Leukot. Essent. Fatty Acids 2010, 82, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Towers, C.G.; Thorburn, A. Therapeutic targeting of autophagy. EBioMedicine 2016, 14, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sharif, T.; Martell, E.; Dai, C.; Kennedy, B.E.; Murphy, P.; Clements, D.R.; Kim, Y.; Lee, P.W.; Gujar, S.A. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy 2017, 13, 264–284. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Bodipati, N.; Demonacos, M.K.; Peddinti, R.; Ghosh, K.; Roy, P. Long term induction by pterostilbene results in autophagy and cellular differentiation in MCF-7 cells via ROS dependent pathway. Mol. Cell. Endocrinol. 2012, 355, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Mondal, S.; Khurana, A.; Jung, D.B.; Hoffmann, R.; He, X.; Kalogera, E.; Dierks, T.; Hammond, E.; Dredge, K.; et al. Loss of HSulf-1: The missing link between autophagy and lipid droplets in ovarian cancer. Sci. Rep. 2017, 7, 41977. [Google Scholar] [CrossRef] [PubMed]
- Pavel, M.; Rubinsztein, D.C. Mammalian autophagy and the plasma membrane. FEBS J. 2017, 284, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.; Leal, R.; Roseiro, J.; Reis, A.; da Silva, T.L. Monitoring rhodosporidium toruloides ncyc 921 batch fermentations growing under carbon and nitrogen limitation by flow cytometry. World J. Microbiol. Biotechnol. 2012, 28, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Giampietri, C.; Petrungaro, S.; Conti, S.; Facchiano, A.; Filippini, A.; Ziparo, E. c-Flip KO fibroblasts display lipid accumulation associated with endoplasmic reticulum stress. Biochim. Biophys. Acta 2015, 1851, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Faroudi, M.; Utzny, C.; Salio, M.; Cerundolo, V.; Guiraud, M.; Muller, S.; Valitutti, S. Lytic versus stimulatory synapse in cytotoxic T lymphocyte/target cell interaction: Manifestation of a dual activation threshold. Proc. Natl. Acad. Sci. USA 2003, 100, 14145–14150. [Google Scholar] [CrossRef] [PubMed]
- Mancinelli, R.; Franchitto, A.; Glaser, S.; Meng, F.; Onori, P.; Demorrow, S.; Francis, H.; Venter, J.; Carpino, G.; Baker, K.; et al. GABA induces the differentiation of small into large cholangiocytes by activation of Ca(2+) /CaMK I-dependent adenylyl cyclase 8. Hepatology 2013, 58, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Francis, H.; Onori, P.; Gaudio, E.; Franchitto, A.; DeMorrow, S.; Venter, J.; Kopriva, S.; Carpino, G.; Mancinelli, R.; White, M.; et al. H3 histamine receptor-mediated activation of protein kinase cα inhibits the growth of cholangiocarcinoma in vitro and in vivo. Mol. Cancer Res. 2009, 7, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Antonangeli, F.; Giampietri, C.; Petrungaro, S.; Filippini, A.; Ziparo, E. Expression profile of a 400-bp Stra8 promoter region during spermatogenesis. Microsc. Res. Tech. 2009, 72, 816–822. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giampietri, C.; Petrungaro, S.; Cordella, M.; Tabolacci, C.; Tomaipitinca, L.; Facchiano, A.; Eramo, A.; Filippini, A.; Facchiano, F.; Ziparo, E. Lipid Storage and Autophagy in Melanoma Cancer Cells. Int. J. Mol. Sci. 2017, 18, 1271. https://doi.org/10.3390/ijms18061271
Giampietri C, Petrungaro S, Cordella M, Tabolacci C, Tomaipitinca L, Facchiano A, Eramo A, Filippini A, Facchiano F, Ziparo E. Lipid Storage and Autophagy in Melanoma Cancer Cells. International Journal of Molecular Sciences. 2017; 18(6):1271. https://doi.org/10.3390/ijms18061271
Chicago/Turabian StyleGiampietri, Claudia, Simonetta Petrungaro, Martina Cordella, Claudio Tabolacci, Luana Tomaipitinca, Antonio Facchiano, Adriana Eramo, Antonio Filippini, Francesco Facchiano, and Elio Ziparo. 2017. "Lipid Storage and Autophagy in Melanoma Cancer Cells" International Journal of Molecular Sciences 18, no. 6: 1271. https://doi.org/10.3390/ijms18061271
APA StyleGiampietri, C., Petrungaro, S., Cordella, M., Tabolacci, C., Tomaipitinca, L., Facchiano, A., Eramo, A., Filippini, A., Facchiano, F., & Ziparo, E. (2017). Lipid Storage and Autophagy in Melanoma Cancer Cells. International Journal of Molecular Sciences, 18(6), 1271. https://doi.org/10.3390/ijms18061271




