Epigenetic Bases of Aberrant Glycosylation in Cancer
Abstract
:1. Introduction
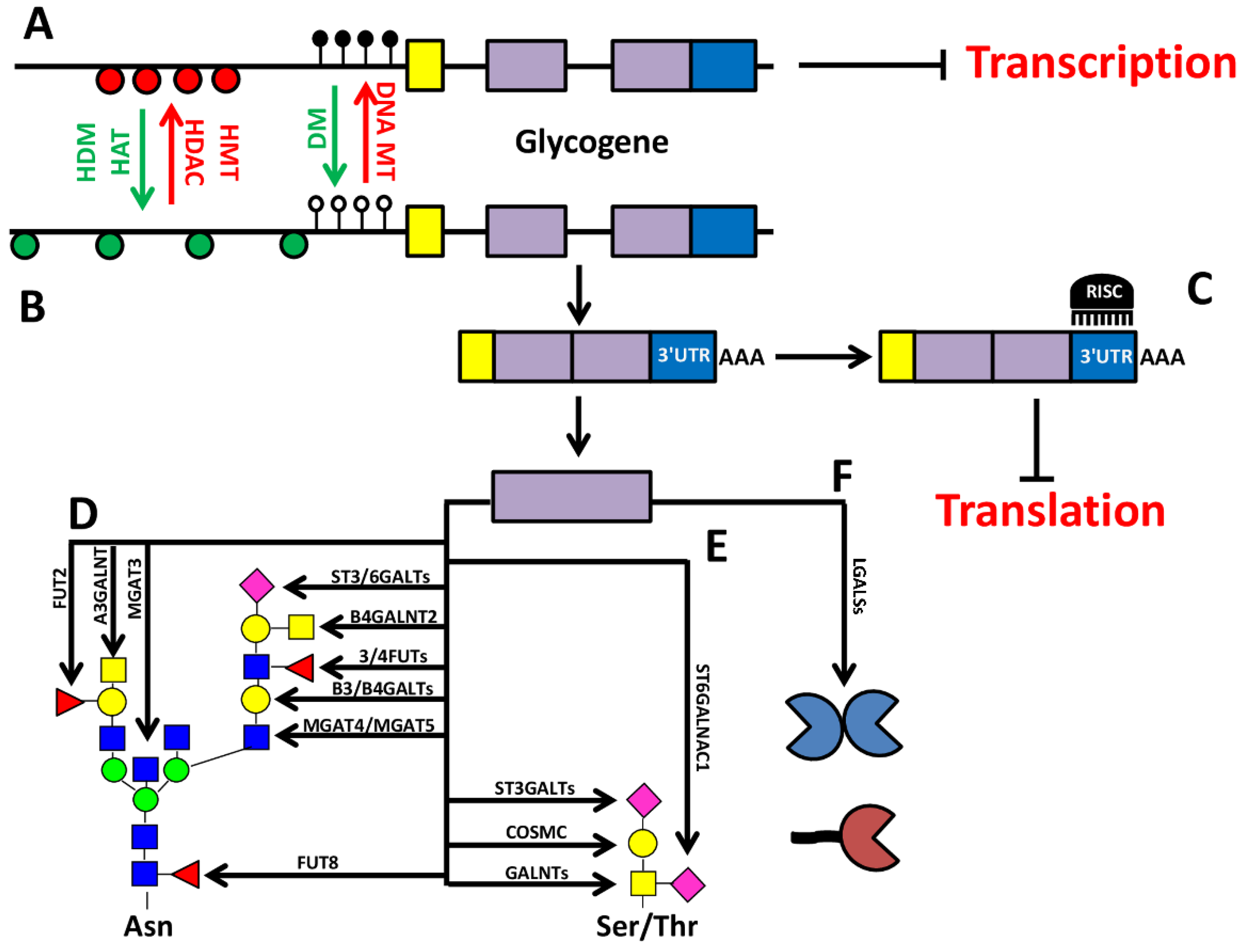
2. Epigenetic-Regulation of Cancer Associated Structures
2.1. Core Glycosylation of N-Linked Chains
2.1.1. Core Fucosylation
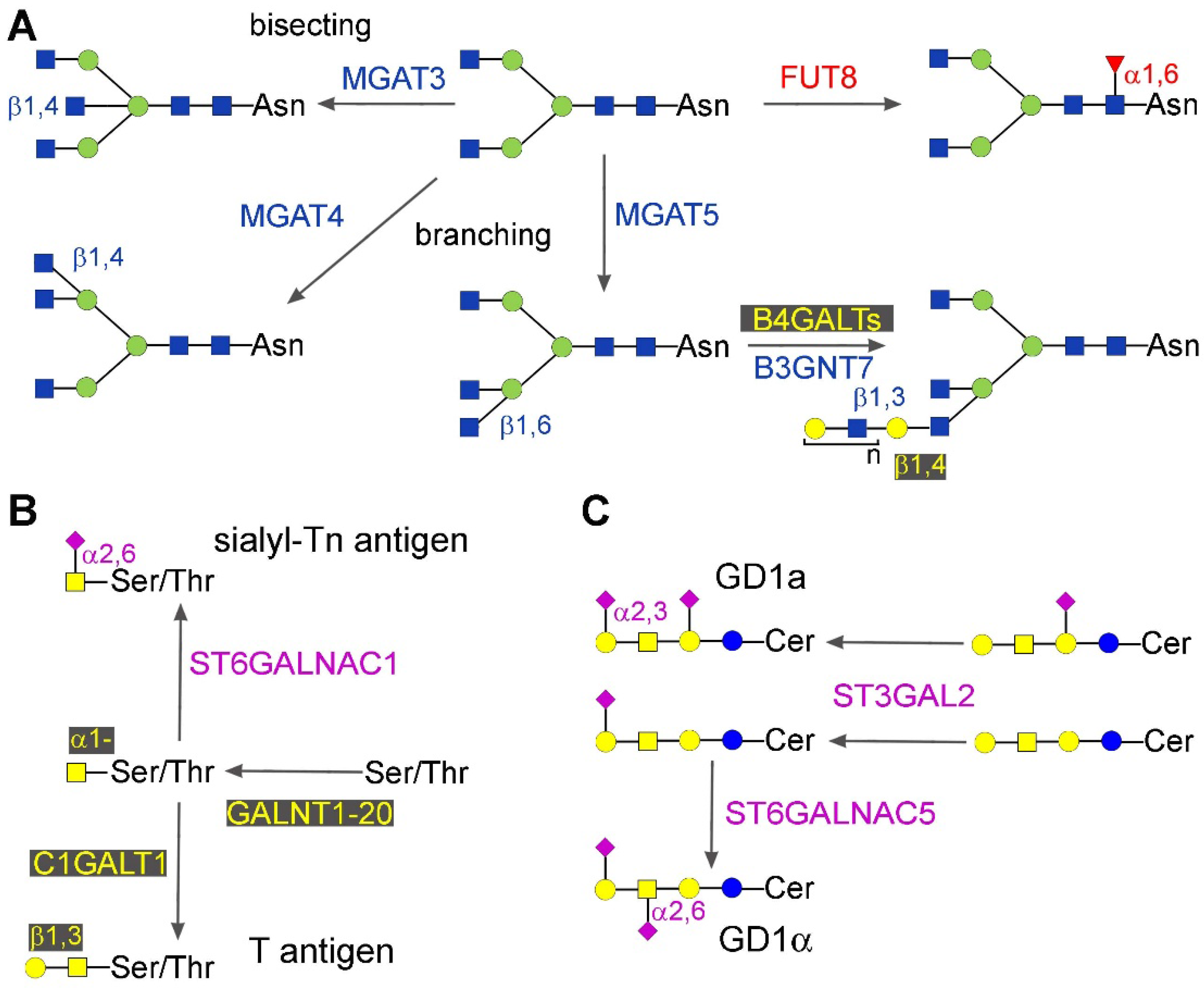
2.1.2. Bisecting GlcNAc, β1,6 and β1,4 Branching
2.2. Mucin-Type O-Glycosylation
2.3. Gangliosides
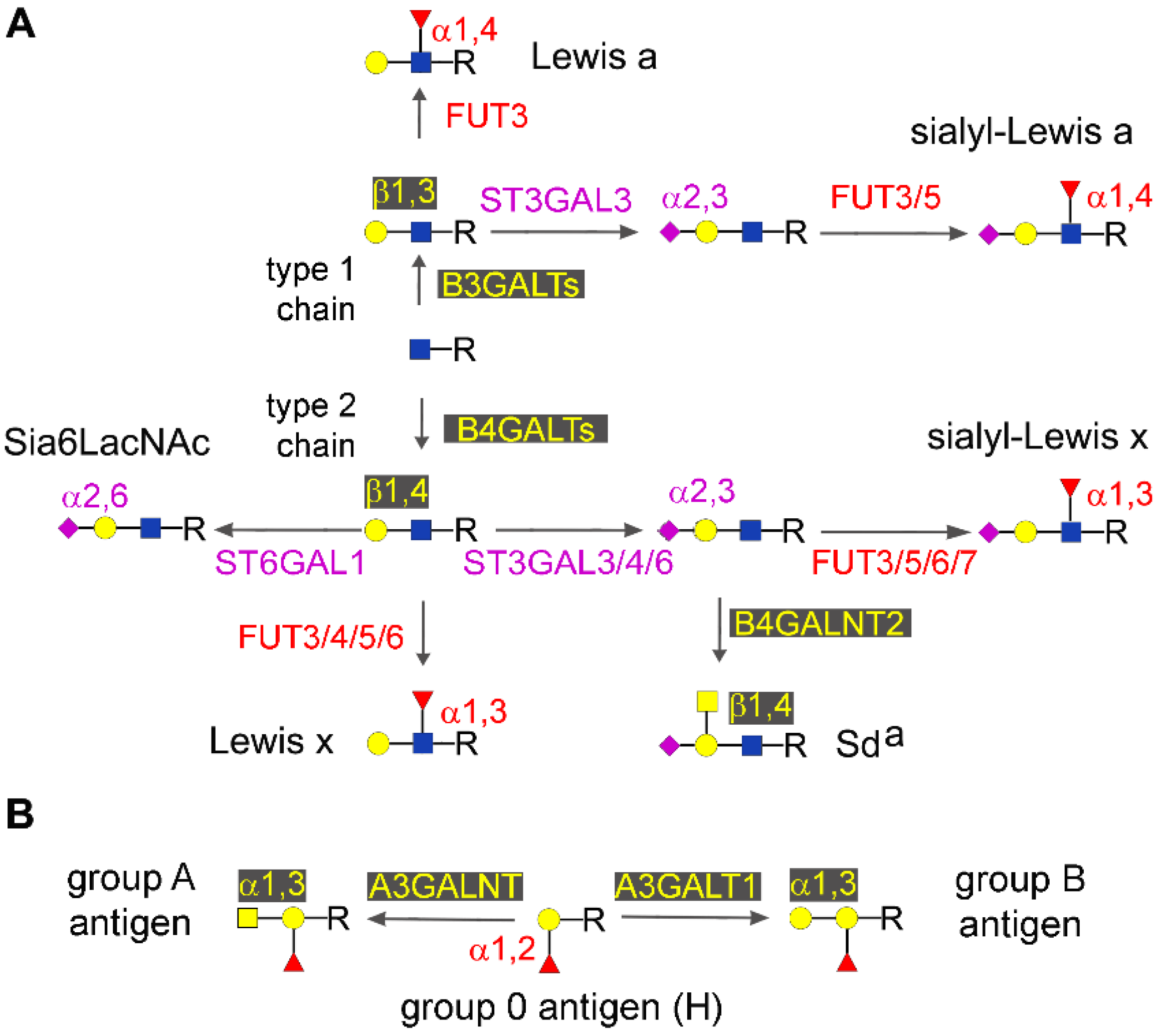
2.4. Type 1 and Type 2 Chain Elongation
2.5. Type 1 and type 2 Chain Termination
2.5.1. Major Glycosyltransferases Involved in Chain Termination
2.5.2. Lewis and Sda Antigens
2.5.3. Sia6LAcNAc and ST6GAL1
2.5.4. Other Sialyltransferases
2.5.5. AB0
2.6. Glycosaminoglycans
3. Epigenetic Regulation of Sugar binding Molecules: The Galectins
3.1. Galectin 1
3.2. Galectin 3
3.3. Galectin 7
3.4. Galectin 9
4. Glycosylation as a Part of the Epigenetic Code: O-GlcNAc
5. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 5-AZA | 5-Aza-2′-deoxycytidine |
| EMT | Epithelial to mesenchymal transition |
References
- Lauc, G.; Zoldos, V. Epigenetic regulation of glycosylation could be a mechanism used by complex organisms to compete with microbes on an evolutionary scale. Med. Hypotheses 2009, 73, 510–512. [Google Scholar] [CrossRef] [PubMed]
- Lauc, G.; Vojta, A.; Zoldos, V. Epigenetic regulation of glycosylation is the quantum mechanics of biology. Biochim. Biophys. Acta 2014, 1840, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Zoldos, V.; Novokmet, M.; Beceheli, I.; Lauc, G. Genomics and epigenomics of the human glycome. Glycoconj. J. 2013, 30, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Zoldos, V.; Horvat, T.; Novokmet, M.; Cuenin, C.; Muzinic, A.; Pucic, M.; Huffman, J.E.; Gornik, O.; Polasek, O.; Campbell, H.; et al. Epigenetic silencing of HNF1A associates with changes in the composition of the human plasma N-glycome. Epigenetics 2012, 7, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Zoldos, V.; Horvat, T.; Lauc, G. Glycomics meets genomics, epigenomics and other high throughput omics for system biology studies. Curr. Opin. Chem. Biol. 2013, 17, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Vojta, A.; Samarzija, I.; Bockor, L.; Zoldos, V. Glyco-genes change expression in cancer through aberrant methylation. Biochim. Biophys. Acta 2016, 1860, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Narisada, M.; Imai, T.; Shinzaki, S.; Miyoshi, E. The effect of epigenetic regulation of fucosylation on TRAIL-induced apoptosis. Glycoconj. J. 2010, 27, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Nakano, M.; Miura, Y.; Taniguchi, N. Epigenetic regulation of neural N-glycomics. Proteomics 2016, 16, 2854–2863. [Google Scholar] [CrossRef] [PubMed]
- Horvat, T.; Muzinic, A.; Barisic, D.; Bosnar, M.H.; Zoldos, V. Epigenetic modulation of the HeLa cell membrane N-glycome. Biochim. Biophys. Acta 2012, 1820, 1412–1419. [Google Scholar] [CrossRef] [PubMed]
- Horvat, T.; Dezeljin, M.; Redzic, I.; Barisic, D.; Herak, B.M.; Lauc, G.; Zoldos, V. Reversibility of Membrane N-Glycome of HeLa Cells upon Treatment with Epigenetic Inhibitors. PLoS ONE 2013, 8, e54672. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Yanagisawa, M.; Ariga, T.; Yu, R.K. Histone acetylation-mediated glycosyltransferase gene regulation in mouse brain during development. J. Neurochem. 2011, 116, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Kurcon, T.; Pilobello, K.T.; Rakus, J.F.; Koppolu, S.; Liu, Z.; Batista, B.S.; Eng, W.S.; Hsu, K.L.; Liang, Y.; et al. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc. Natl. Acad. Sci. USA 2014, 111, 4338–4343. [Google Scholar] [CrossRef] [PubMed]
- Kasper, B.T.; Koppolu, S.; Mahal, L.K. Insights into miRNA regulation of the human glycome. Biochem. Biophys. Res. Commun. 2014, 445, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Yanagidani, S.; Miyoshi, E.; Ihara, Y.; Sakuma, T.; Gao, C.X.; Teshima, T.; Fujii, S.; Shiba, T.; Taniguchi, N. Purification and cDNA cloning of porcine brain GDP-L-Fuc: N-acetyl-β-D-glucosaminide a1-->6fucosyltransferase. J. Biol. Chem. 1996, 271, 27810–27817. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Inoue, S.; Gu, J.; Miyoshi, E.; Noda, K.; Li, W.; Mizuno-Horikawa, Y.; Nakano, M.; Asahi, M.; Takahashi, M.; et al. Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. USA 2005, 102, 15791–15796. [Google Scholar] [CrossRef] [PubMed]
- Comunale, M.A.; Lowman, M.; Long, R.E.; Krakover, J.; Philip, R.; Seeholzer, S.; Evans, A.A.; Hann, H.W.; Block, T.M.; Mehta, A.S. Proteomic Analysis of Serum Associated Fucosylated Glycoproteins in the Development of Primary Hepatocellular Carcinoma. J. Proteome Res. 2006, 5, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Li, Q.K.; Peskoe, S.B.; Zhang, B.; Choi, C.; Platz, E.A.; Zhang, H. Overexpression of α (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 2014, 24, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.P.; Xu, X.Y.; Fang, M.; Wang, H.; You, Q.; Yi, C.H.; Ji, J.; Gu, X.; Zhou, P.T.; Cheng, C.; et al. Decreased core-fucosylation contributes to malignancy in gastric cancer. PLoS ONE 2014, 9, e94536. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, J.; Kong, X.; Chen, H.; Wang, Y.; Qin, M.; Lin, Y.; Chen, H.; Xu, J.; Hong, J.; et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci. Rep. 2014, 4, 6145. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, C.; Soffientini, U.; Piacente, F.; Tonetti, M.G. Effects of microRNAs on fucosyltransferase 8 (FUT8) expression in hepatocarcinoma cells. PLoS ONE 2013, 8, e76540. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Gao, S.; Song, X.; Dong, W.; Zhou, H.; Zhao, L.; Jia, L. Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget 2016, 7, 61199–61214. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Sato, Y.; Kariya, Y.; Isaji, T.; Taniguchi, N.; Fukuda, T. A mutual regulation between cell-cell adhesion and N-glycosylation: Implication of the bisecting GlcNAc for biological functions. J. Proteome Res. 2009, 8, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A.; Paredes, J.; Magalhaes, A.M.; Ferreira, A.C.; Figueiredo, J.; Xiaogang, W.; Carneiro, F.; Gartner, F.; Seruca, R. The role of N-acetylglucosaminyltransferase III and V in the post-transcriptional modifications of E-cadherin. Hum. Mol. Genet. 2009, 18, 2599–2608. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Seruca, R.; Gartner, F.; Yamaguchi, Y.; Gu, J.; Taniguchi, N.; Reis, C.A. Modulation of E-cadherin function and dysfunction by N-glycosylation. Cell Mol. Life Sci. 2011, 68, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Oliveira, P.; Cabral, J.; Carvalho, S.; Huntsman, D.; Gartner, F.; Seruca, R.; Reis, C.A.; Oliveira, C. Loss and recovery of MGAT3 and GnT-III Mediated E-cadherin N-glycosylation is a mechanism involved in epithelial-mesenchymal-epithelial transitions. PLoS ONE 2012, 7, e33191. [Google Scholar] [CrossRef] [PubMed]
- Kohler, R.S.; Anugraham, M.; Lopez, M.N.; Xiao, C.; Schoetzau, A.; Hettich, T.; Schlotterbeck, G.; Fedier, A.; Jacob, F.; Heinzelmann-Schwarz, V. Epigenetic activation of MGAT3 and corresponding bisecting GlcNAc shortens the survival of cancer patients. Oncotarget 2016, 7, 51674–51686. [Google Scholar] [CrossRef] [PubMed]
- Anugraham, M.; Jacob, F.; Nixdorf, S.; Everest-Dass, A.V.; Heinzelmann-Schwarz, V.; Packer, N.H. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: Glycan structures reflect gene expression and DNA methylation status. Mol. Cell Proteom. 2014, 13, 2213–2232. [Google Scholar] [CrossRef] [PubMed]
- Saldova, R.; Dempsey, E.; Perez-Garay, M.; Marino, K.; Watson, J.A.; Blanco-Fernandez, A.; Struwe, W.B.; Harvey, D.J.; Madden, S.F.; Peracaula, R.; et al. 5-AZA-2′-deoxycytidine induced demethylation influences N-glycosylation of secreted glycoproteins in ovarian cancer. Epigenetics 2011, 6, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Klasic, M.; Kristic, J.; Korac, P.; Horvat, T.; Markulin, D.; Vojta, A.; Reiding, K.R.; Wuhrer, M.; Lauc, G.; Zoldos, V. DNA hypomethylation upregulates expression of the MGAT3 gene in HepG2 cells and leads to changes in N-glycosylation of secreted glycoproteins. Sci. Rep. 2016, 6, 24363. [Google Scholar] [CrossRef] [PubMed]
- Granovsky, M.; Fata, J.; Pawling, J.; Muller, W.J.; Khokha, R.; Dennis, J.W. Suppression of tumor growth and metastasis in MGAT5-deficient mice. Nat. Med. 2000, 6, 306–312. [Google Scholar] [PubMed]
- Lau, K.S.; Partridge, E.A.; Grigorian, A.; Silvescu, C.I.; Reinhold, V.N.; Demetriou, M.; Dennis, J.W. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 2007, 129, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.K.; Sousa, J.F.; Chakraborty, D.; Funasaka, Y.; Bhattacharya, M.; Chatterjee, A.; Pawelek, J. GnT-V expression and metastatic phenotypes in macrophage-melanoma fusion hybrids is down-regulated by 5-Aza-dC: Evidence for methylation sensitive, extragenic regulation of GnT-V transcription. Gene 2006, 374, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Vaiana, C.A.; Kurcon, T.; Mahal, L.K. MicroRNA-424 Predicts a Role for b1,4 Branched Glycosylation in Cell Cycle Progression. J. Biol. Chem. 2016, 291, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Dyrskjot, L.; Ostenfeld, M.S.; Bramsen, J.B.; Silahtaroglu, A.N.; Lamy, P.; Ramanathan, R.; Fristrup, N.; Jensen, J.L.; Andersen, C.L.; Zieger, K.; et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009, 69, 4851–4860. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Yang, L.; Wu, Q.; Liu, W.; Fu, Q.; Zhang, W.; Zhang, H.; Xu, J.; Gu, J. Loss of N-acetylgalactosaminyltransferase-4 orchestrate oncogenic microRNA-9 in hepatocellular carcinoma. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.Q.; Wan, H.Y.; Li, H.F.; Liu, M.; Li, X.; Tang, H. MicroRNA-214 Suppresses Growth and Invasiveness of Cervical Cancer Cells by Targeting UDP-N-acetyl-α-D-galactosamine:Polypeptide N-Acetylgalactosaminyltransferase 7. J. Biol. Chem. 2012, 287, 14301–14309. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, H.; Sun, J. MicroRNA34a/c function as tumor suppressors in Hep2 laryngeal carcinoma cells and may reduce GALNT7 expression. Mol. Med. Rep. 2014, 9, 1293–1298. [Google Scholar] [PubMed]
- Lu, Q.; Xu, L.; Li, C.; Yuan, Y.; Huang, S.; Chen, H. miR-214 inhibits invasion and migration via downregulating GALNT7 in esophageal squamous cell cancer. Tumour Biol. 2016, 37, 14605–14614. [Google Scholar] [CrossRef] [PubMed]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.W.; Fang, L.; Shatseva, T.; Rutnam, Z.J.; Yang, X.; Du, W.; Lu, W.Y.; Xuan, J.W.; Deng, Z.; Yang, B.B. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J. Cell Sci. 2013, 126, 1517–1530. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, H.O.; Liu, Y.D.; Liu, W.S.; Pan, D.; Zhang, W.J.; Yang, L.; Fu, Q.; Xu, J.J.; Gu, J.X. Decreased Expression of Hepatocyte Nuclear Factor 4α (Hnf4α)/MicroRNA-122 (miR-122) Axis in Hepatitis B Virus-associated Hepatocellular Carcinoma Enhances Potential Oncogenic GALNT10 Protein Activity. J. Biol. Chem. 2015, 290, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, G.; Zhang, K. MiR-125a regulates ovarian cancer proliferation and invasion by repressing GALNT14 expression. Biomed. Pharmacother. 2016, 80, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Mi, R.; Song, L.; Wang, Y.; Ding, X.; Zeng, J.; Lehoux, S.; Aryal, R.P.; Wang, J.; Crew, V.K.; van Die, I.; et al. Epigenetic silencing of the chaperone cosmc in human leukocytes expressing tn antigen. J. Biol. Chem. 2012, 287, 41523–41533. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Mi, R.; Wang, Y.; Li, Y.; Lin, L.; Yao, B.; Song, L.; van Die, I.; Chapman, A.B.; Cummings, R.D.; et al. Promoters of human Cosmc and T-synthase are similar in structure, yet different in epigenetic regulation. J. Biol. Chem. 2015, 290, 19018–19033. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lee, K.M.; Han, W.; Choi, J.Y.; Lee, J.Y.; Kang, G.H.; Park, S.K.; Noh, D.Y.; Yoo, K.Y.; Kang, D. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum. Mol. Genet. 2010, 19, 4273–4277. [Google Scholar] [CrossRef] [PubMed]
- Iwaya, T.; Sawada, G.; Amano, S.; Kume, K.; Ito, C.; Endo, F.; Konosu, M.; Shioi, Y.; Akiyama, Y.; Takahara, T.; et al. Downregulation of ST6GALNAC1 is associated with esophageal squamous cell carcinoma development. Int. J. Oncol. 2017, 50, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Daniotti, J.L.; Lardone, R.D.; Vilcaes, A.A. Dysregulated Expression of Glycolipids in Tumor Cells: From Negative Modulator of Anti-tumor Immunity to Promising Targets for Developing Therapeutic Agents. Front Oncol. 2015, 5, 300. [Google Scholar] [CrossRef] [PubMed]
- Hatano, K.; Miyamoto, Y.; Mori, M.; Nimura, K.; Nakai, Y.; Nonomura, N.; Kaneda, Y. Androgen-regulated transcriptional control of sialyltransferases in prostate cancer cells. PLoS ONE 2012, 7, e31234. [Google Scholar] [CrossRef] [PubMed]
- Oster, B.; Thorsen, K.; Lamy, P.; Wojdacz, T.K.; Hansen, L.L.; Birkenkamp-Demtroder, K.; Sorensen, K.D.; Laurberg, S.; Orntoft, T.F.; Andersen, C.L. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int. J. Cancer 2011, 129, 2855–2866. [Google Scholar] [CrossRef] [PubMed]
- Kurcon, T.; Liu, Z.; Paradkar, A.V.; Vaiana, C.A.; Koppolu, S.; Agrawal, P.; Mahal, L.K. miRNA proxy approach reveals hidden functions of glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 7327–7332. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, H.; Miao, Y.; Li, N.; Zhao, L.; Jia, L. MiRNA expression profiles reveal the involvement of miR-26a, miR-548l and miR-34a in hepatocellular carcinoma progression through regulation of ST3GAL5. Lab. Investig. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mare, L.; Trinchera, M. Comparative Analysis of Retroviral and Native Promoters Driving Expression of b1,3-Galactosyltransferase b3Gal-T5 in Human and Mouse Tissues. J. Biol. Chem. 2007, 282, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Caretti, A.; Sirchia, S.M.; Tabano, S.; Zulueta, A.; Dall’Olio, F.; Trinchera, M. DNA methylation and histone modifications modulate the β1,3 galactosyltransferase β3Gal-T5 native promoter in cancer cells. Int. J. Biochem. Cell Biol. 2012, 44, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Aronica, A.; Avagliano, L.; Caretti, A.; Tosi, D.; Bulfamante, G.P.; Trinchera, M. Unexpected distribution of CA19.9 and other type 1 chain Lewis antigens in normal and cancer tissues of colon and pancreas: Importance of the detection method and role of glycosyltransferase regulation. Biochim. Biophys. Acta 2016, 1861, 3210–3220. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.A.; van de Lagemaat, L.N.; Baillie, G.J.; Mager, D.L. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: The case of primate β3GAL-T5. Gene 2005, 364, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Zulueta, A.; Caretti, A.; Signorelli, P.; Dall’Olio, F.; Trinchera, M. Transcriptional control of the B3GALT5 gene by a retroviral promoter and methylation of distant regulatory elements. FASEB J. 2014, 28, 946–955. [Google Scholar] [CrossRef] [PubMed]
- Poeta, M.L.; Massi, E.; Parrella, P.; Pellegrini, P.; de Robertis, M.; Copetti, M.; Rabitti, C.; Perrone, G.; Muda, A.O.; Molinari, F.; et al. Aberrant promoter methylation of β-1,4 galactosyltransferase 1 as potential cancer-specific biomarker of colorectal tumors. Genes Chromosomes Cancer 2012, 51, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, X.; Liu, M.; Tang, H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer Lett. 2016, 375, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Wu, W.Y.; Lai, Y.J.; Yang, C.M.; Yu, L.C. Suppression of B3GNT7 gene expression in colon adenocarcinoma and its potential effect in the metastasis of colon cancer cells. Glycobiology 2014, 24, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Chiricolo, M. Sialyltransferases in cancer. Glycoconj. J. 2001, 18, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 2014, 1840, 2752–2764. [Google Scholar] [CrossRef] [PubMed]
- Harduin-Lepers, A.; Vallejo-Ruiz, V.; Krzewinski-Recchi, M.; Samyn-Petit, B.; Julien, S.; Delannoy, P. The human sialyltransferase family. Biochimie 2001, 83, 727–737. [Google Scholar] [CrossRef]
- De Vries, T.; Knegtel, R.M.; Holmes, E.H.; Macher, B.A. Fucosyltransferases: Structure/function studies. Glycobiology 2001, 11, 119R–128R. [Google Scholar] [CrossRef] [PubMed]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology (Basel) 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Mollicone, R.; Candelier, J.J.; Reguigne, I.; Couillin, P.; Fletcher, A.; Oriol, R. Molecular genetics of a-L-fucosyltransferase genes (H, Se, Le, FUT4, FUT5 and FUT6). Transfus. Clin. Biol. 1994, 1, 91–97. [Google Scholar] [CrossRef]
- Terraneo, L.; Avagliano, L.; Caretti, A.; Bianciardi, P.; Tosi, D.; Bulfamante, G.P.; Samaja, M.; Trinchera, M. Expression of carbohydrate-antigen sialyl-Lewis a on colon cancer cells promotes xenograft growth and angiogenesis in nude mice. Int. J. Biochem. Cell Biol. 2013, 45, 2796–2800. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Ohmori, K.; Izawa, M.; Koike, T.; Kumamoto, K.; Furukawa, K.; Ando, T.; Kiso, M.; Yamaji, T.; Hashimoto, Y.; et al. Loss of disialyl Lewisa the ligand for lymphocyte inhibitory receptor sialic acid-binding immunoglobulin-like lectin-7 (Siglec-7) associated with increased sialyl Lewis a expression on human colon cancers. Cancer Res. 2004, 64, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Serpa, J.; Mesquita, P.; Mendes, N.; Oliveira, C.; Almeida, R.; Santos-Silva, F.; Reis, C.A.; Lependu, J.; David, L. Expression of Lea in gastric cancer cell lines depends on FUT3 expression regulated by promoter methylation. Cancer Lett. 2006, 242, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.M.; Dhas, K.; Nair, J.; Palve, V.; Bagwan, J.; Siddappa, G.; Suresh, A.; Kekatpure, V.D.; Kuriakose, M.A.; Panda, B. A Minimal DNA Methylation Signature in Oral Tongue Squamous Cell Carcinoma Links Altered Methylation with Tumor Attributes. Mol. Cancer Res. 2016, 14, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Walton, E.; Pingault, J.B.; Cecil, C.A.; Gaunt, T.R.; Relton, C.L.; Mill, J.; Barker, E.D. Epigenetic profiling of ADHD symptoms trajectories: A prospective, methylome-wide study. Mol. Psychiatry 2017, 22, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Chachadi, V.B.; Cheng, H.; Klinkebiel, D.; Christman, J.K.; Cheng, P.W. 5-Aza-2′-deoxycytidine increases sialyl Lewis X on MUC1 by stimulating b-galactoside:a2,3-sialyltransferase 6 gene. Int. J. Biochem. Cell Biol. 2011, 43, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Yusa, A.; Miyazaki, K.; Kimura, N.; Izawa, M.; Kannagi, R. Epigenetic silencing of the sulfate transporter gene DTDST induces sialyl Lewisx expression and accelerates proliferation of colon cancer cells. Cancer Res. 2010, 70, 4064–4073. [Google Scholar] [CrossRef] [PubMed]
- Donald, A.S.; Yates, A.D.; Soh, C.P.; Morgan, W.T.; Watkins, W.M. A blood group Sda-active pentasaccharide isolated from Tamm-Horsfall urinary glycoprotein. Biochem. Biophys. Res. Commun. 1983, 115, 625–631. [Google Scholar] [CrossRef]
- Presti, L.; Cabuy, E.; Chiricolo, M.; Dall’Olio, F. Molecular Cloning of the Human b1,4 N-Acetylgalactosaminyltransferase Responsible for the Biosynthesis of the Sda Histo-Blood Group Antigen: The Sequence Predicts a Very Long Cytoplasmic Domain. J. Biochem. (Tokyo) 2003, 134, 675–682. [Google Scholar] [CrossRef]
- Montiel, M.D.; Krzewinski-Recchi, M.A.; Delannoy, P.; Harduin-Lepers, A. Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Aca2-3Galb-R b1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: Evidence for an unusual extended cytoplasmic domain. Biochem. J. 2003, 373, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N.; Chiricolo, M.; Trinchera, M.; Harduin-Lepers, A. The expanding roles of the Sda/Cad carbohydrate antigen and its cognate glycosyltransferase B4GALNT2. Biochim. Biophys. Acta 2014, 1840, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Groux-Degroote, S.; Wavelet, C.; Krzewinski-Recchi, M.A.; Portier, L.; Mortuaire, M.; Mihalache, A.; Trinchera, M.; Delannoy, P.; Malagolini, N.; Chiricolo, M.; et al. B4GALNT2 gene expression controls the biosynthesis of Sda and sialyl Lewis X antigens in healthy and cancer human gastrointestinal tract. Int. J. Biochem. Cell Biol. 2014, 53, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Malagolini, N.; Dall’Olio, F.; Di Stefano, G.; Minni, F.; Marrano, D.; Serafini-Cessi, F. Expression of UDP-GalNAc:NeuAc α2,3Gal β-R β 1,4(GalNAc to Gal) N-acetylgalactosaminyltransferase involved in the synthesis of Sda antigen in human large intestine and colorectal carcinomas. Cancer Res. 1989, 49, 6466–6470. [Google Scholar] [PubMed]
- Malagolini, N.; Santini, D.; Chiricolo, M.; Dall’Olio, F. Biosynthesis and expression of the Sda and sialyl Lewis x antigens in normal and cancer colon. Glycobiology 2007, 17, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.R.; Hsieh, C.Y.; Twu, Y.C.; Yu, L.C. Expression of the human Sda β-1,4-N-acetylgalactosaminyltransferase II gene is dependent on the promoter methylation status. Glycobiology 2008, 18, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.I.; Toyota, M.; Kawashima, R.; Hagiwara, T.; Suzuki, H.; Imai, K.; Shinomura, Y.; Tokino, T.; Kannagi, R.; Dohi, T. DNA hypermethylation contributes to incomplete synthesis of carbohydrate determinants in gastrointestinal cancer. Gastroenterology 2008, 135, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Mare, L.; Caretti, A.; Albertini, R.; Trinchera, M. CA19.9 antigen circulating in the serum of colon cancer patients: Where is it from? Int. J. Biochem. Cell Biol. 2013, 45, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Trinchera, M.; Malagolini, N.; Chiricolo, M.; Santini, D.; Minni, F.; Caretti, A.; Dall’Olio, F. The biosynthesis of the selectin-ligand sialyl Lewis x in colorectal cancer tissues is regulated by fucosyltransferase VI and can be inhibited by an RNA interference-based approach. Int. J. Biochem. Cell Biol. 2011, 43, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tong, S.; Liu, J.; Han, L.; Yang, X.; Hou, H.; Yan, Q.; Wang, X.Q. Differential fucosyltransferase IV expression in squamous carcinoma cells is regulated by promoter methylation. Cell Mol. Biol. Lett. 2012, 17, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhao, L.; Gao, S.; Song, X.; Dong, W.; Zhao, Y.; Zhou, H.; Cheng, L.; Miao, X.; Jia, L. Increased fucosylation has a pivotal role in multidrug resistance of breast cancer cells through miR-224-3p targeting FUT4. Gene 2016, 578, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Feng, X.; Song, X.; Zhou, H.; Zhao, Y.; Cheng, L.; Jia, L. miR-493-5p attenuates the invasiveness and tumorigenicity in human breast cancer by targeting FUT4. Oncol. Rep. 2016, 36, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, Y.; Miao, Y.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life 2016, 68, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.; Lee, E.U.; McEntee, K.; Lai, P.H.; Paulson, J.C. Primary structure of β-galactoside α 2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J. Biol. Chem. 1987, 262, 17735–17743. [Google Scholar] [PubMed]
- Seales, E.C.; Jurado, G.A.; Brunson, B.A.; Wakefield, J.K.; Frost, A.R.; Bellis, S.L. Hypersialylation of b1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 2005, 65, 4645–4652. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.B.; Nasirikenari, M.; Feng, L.; Migliore, M.T.; Choi, K.S.; Kazim, L.; Lau, J.T. Role for hepatic and circulatory ST6Gal-1 sialyltransferase in regulating myelopoiesis. J. Biol. Chem. 2010, 285, 25009–25017. [Google Scholar] [CrossRef] [PubMed]
- Nasirikenari, M.; Chandrasekaran, E.V.; Matta, K.L.; Segal, B.H.; Bogner, P.N.; Lugade, A.A.; Thanavala, Y.; Lee, J.J.; Lau, J.T. Altered eosinophil profile in mice with ST6Gal-1 deficiency: An additional role for ST6Gal-1 generated by the P1 promoter in regulating allergic inflammation. J. Leukoc. Biol. 2010, 87, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Nasirikenari, M.; Veillon, L.; Collins, C.C.; Azadi, P.; Lau, J.T. Remodeling of Marrow Hematopoietic Stem and Progenitor Cells by Non-self ST6Gal-1 Sialyltransferase. J. Biol. Chem. 2014, 289, 7178–7189. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F. The sialyl-a2,6-lactosaminyl-structure: Biosynthesis and functional role. Glycoconj. J 2000, 17, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N.; Di Stefano, G.; Minni, F.; Marrano, D.; Serafini-Cessi, F. Increased CMP-NeuAc:Galb1,4GlcNAc-R a 2,6 sialyltransferase activity in human colorectal cancer tissues. Int. J. Cancer 1989, 44, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.J.; Holdbrooks, A.T.; Chakraborty, A.; Grizzle, W.E.; Landen, C.N.; Buchsbaum, D.J.; Conner, M.G.; Arend, R.C.; Yoon, K.J.; Klug, C.A.; et al. The Tumor-Associated Glycosyltransferase ST6Gal-I Regulates Stem Cell Transcription Factors and Confers a Cancer Stem Cell Phenotype. Cancer Res. 2016, 76, 3978–3988. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Y.; Chammas, R.; Bellis, S.L. Sialylation of b1 integrins blocks cell adhesion to galectin-3 and protects cells against galectin-3-induced apoptosis. J. Biol. Chem. 2008, 283, 22177–22185. [Google Scholar] [CrossRef] [PubMed]
- Antony, P.; Rose, M.; Heidenreich, A.; Knuchel, R.; Gaisa, N.T.; Dahl, E. Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer 2014, 14, 901. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, T.; Edvardsen, H.; Solvang, H.K.; Daviaud, C.; Naume, B.; Borresen-Dale, A.L.; Kristensen, V.N.; Tost, J. Integrated analysis of high-resolution DNA methylation profiles, gene expression, germline genotypes and clinical end points in breast cancer patients. Int. J. Cancer 2014, 134, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Kroes, R.A.; Moskal, J.R. The Role of DNA Methylation in ST6Gal1 Expression in Gliomas. Glycobiology 2016, 26, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Shimono, Y.; Mizutani, K.; Nobutani, K.; Momose, K.; Azuma, T.; Takai, Y. Reduction of the ST6 β-Galactosamide α-2,6-Sialyltransferase 1 (ST6GAL1)-catalyzed Sialylation of Nectin-like Molecule 2/Cell Adhesion Molecule 1 and Enhancement of ErbB2/ErbB3 Signaling by MicroRNA-199a. J. Biol. Chem. 2013, 288, 11845–11853. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dong, W.; Su, Z.; Zhao, L.; Miao, Y.; Li, N.; Zhou, H.; Jia, L. Functional roles of sialylation in breast cancer progression through miR-26a/26b targeting ST8SIA4. Cell Death Dis. 2016, 7, e2561. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhao, X.; Liang, L.; Pan, X.; Lv, H.; Zhao, Y. Sialyltransferase ST3GAL6 mediates the effect of microRNA-26a on cell growth, migration, and invasion in hepatocellular carcinoma through the protein kinase B/mammalian target of rapamycin pathway. Cancer Sci. 2017, 108, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, S.; Dong, W.; Song, X.; Zhou, H.; Zhao, L.; Jia, L. Α-2, 3-sialyltransferases regulate the multidrug resistance of chronic myeloid leukemia through miR-4701-5p targeting ST3GAL1. Lab. Investig. 2016, 96, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Jia, L.; Zhou, H.; Song, X.; Zhou, M.; Xu, J.; Zhao, L.; Feng, X.; Zhao, Y. miR-4299 mediates the invasive properties and tumorigenicity of human follicular thyroid carcinoma by targeting ST6GALNAC4. IUBMB Life 2016, 68, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Denomme, G.A. Molecular basis of blood group expression. Transfus. Apher. Sci. 2011, 44, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Dotz, V.; Wuhrer, M. Histo-blood group glycans in the context of personalized medicine. Biochim. Biophys. Acta 2016, 1860, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Chihara, Y.; Sugano, K.; Kobayashi, A.; Kanai, Y.; Yamamoto, H.; Nakazono, M.; Fujimoto, H.; Kakizoe, T.; Fujimoto, K.; Hirohashi, S.; et al. Loss of blood group A antigen expression in bladder cancer caused by allelic loss and/or methylation of the ABO gene. Lab. Investig. 2005, 85, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Worm, J.; Guldberg, P.; Eiberg, H.; Krogdahl, A.; Liu, C.J.; Reibel, J.; Dabelsteen, E. Genetic and epigenetic alterations of the blood group ABO gene in oral squamous cell carcinoma. Int. J. Cancer 2004, 109, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Dabelsteen, E.; Gao, S. ABO blood-group antigens in oral cancer. J. Dent. Res. 2005, 84, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Yen, C.J.; Chou, R.H.; Chen, J.N.; Huang, W.C.; Wu, C.Y.; Yu, Y.L. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int. J. Cancer 2014, 134, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- Bui, C.; Ouzzine, M.; Talhaoui, I.; Sharp, S.; Prydz, K.; Coughtrie, M.W.; Fournel-Gigleux, S. Epigenetics: Methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 2010, 24, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Karibe, T.; Fukui, H.; Sekikawa, A.; Shiratori, K.; Fujimori, T. EXTL3 promoter methylation down-regulates EXTL3 and heparan sulphate expression in mucinous colorectal cancers. J. Pathol. 2008, 216, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Rabinovich, G.A. Galectins as modulators of tumour progression. Nat. Rev. Cancer 2005, 5, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Rao, U.S. Galectin-1 is silenced by promoter hypermethylation and its re-expression induces apoptosis in human colorectal cancer cells. Cancer Lett. 2011, 301, 38–46. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Tan, J.X.; Dai, H.S.; Chen, H.W.; Xu, X.J.; Yang, A.G.; Zhang, Y.J.; Bai, L.H.; Bie, P. MiRNA-22 inhibits oncogene galectin-1 in hepatocellular carcinoma. Oncotarget 2016, 7, 57099–57116. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Banerjee, P.P.; Vasta, G.R. Differential expression of galectins in normal, benign and malignant prostate epithelial cells: Silencing of galectin-3 expression in prostate cancer by its promoter methylation. Biochem. Biophys. Res. Commun. 2007, 358, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Cappello, F.; Rodolico, V.; Vasta, G.R. Evidence of heavy methylation in the galectin 3 promoter in early stages of prostate adenocarcinoma: Development and validation of a methylated marker for early diagnosis of prostate cancer. Transl. Oncol. 2009, 2, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Ben Mahmoud, L.K.; Arfaoui, A.; Khiari, M.; Chaar, I.; el Amine, O.; Ben Hmida, A.M.; Gharbi, L.; Mzabi, S.R.; Bouraoui, S. Loss of Galectin-3 Expression in Mucinous Colorectal Carcinomas is Associated With 5′CpG Island Methylation in Tunisian Patients. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Ruebel, K.H.; Jin, L.; Qian, X.; Scheithauer, B.W.; Kovacs, K.; Nakamura, N.; Zhang, H.; Raz, A.; Lloyd, R.V. Effects of DNA methylation on galectin-3 expression in pituitary tumors. Cancer Res. 2005, 65, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Angrisano, T.; Florio, E.; Pero, R.; Decaussin-Petrucci, M.; Troncone, G.; Capasso, M.; Lembo, F.; Fusco, A.; Chiariotti, L. DNA methylation state of the galectin-3 gene represents a potential new marker of thyroid malignancy. Oncol. Lett. 2013, 6, 86–90. [Google Scholar] [PubMed]
- Lu, W.; Wang, J.; Yang, G.; Yu, N.; Huang, Z.; Xu, H.; Li, J.; Qiu, J.; Zeng, X.; Chen, S.; et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget 2017, 8, 15242. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Duraisamy, S.; Barbashov, S.; Kawano, T.; Kharbanda, S.; Kufe, D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol. Cell 2007, 27, 992–1004. [Google Scholar] [CrossRef] [PubMed]
- Demers, M.; Couillard, J.; Giglia-Mari, G.; Magnaldo, T.; St Pierre, Y. Increased galectin-7 gene expression in lymphoma cells is under the control of DNA methylation. Biochem. Biophys. Res. Commun. 2009, 387, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Hwang, J.A.; Ro, J.Y.; Lee, Y.S.; Chun, K.H. Galectin-7 is epigenetically-regulated tumor suppressor in gastric cancer. Oncotarget 2013, 4, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, T.; Morishita, A.; Fujihara, S.; Iwama, H.; Niki, T.; Fujita, K.; Akashi, E.; Mimura, S.; Oura, K.; Sakamoto, T.; et al. Galectin-9: An anticancer molecule for gallbladder carcinoma. Int. J. Oncol. 2016, 48, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, W.; Zhuang, C.; Geng, Z.; Hou, C.; Huang, D.; Hu, L.; Wang, X. microRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol. Rep. 2015, 34, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Love, D.C.; Krause, M.W.; Hanover, J.A. O-GlcNAc cycling: Emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 2010, 21, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, K.; Wang, Z.; Hart, G.W. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, B.; Nairn, A.V.; Nagy, T.; Moremen, K.W.; Buckhaults, P.; Pierce, M. O-Linked N-Acetylglucosamine (O-GlcNAc) Expression Levels Epigenetically Regulate Colon Cancer Tumorigenesis by Affecting the Cancer Stem Cell Compartment via Modulating Expression of Transcriptional Factor MYBL1. J. Biol. Chem. 2017, 292, 4123–4137. [Google Scholar] [CrossRef] [PubMed]
| Target | Epigenetic Mechanism | Effect | Tissues/Cells Involved | Reference |
|---|---|---|---|---|
| Galactosyltransferases | ||||
| A3GALT1/A3GALNT | Promoter methylation | Down-regulation | Bladder and oral cancer | [107,108] |
| B3GALT5 native | Promoter methylation | Down-regulation | Colon cancer | [53,54] |
| B3GALT5 LTR | Distant sequence methylation | Up-regulation | Colon cancer | [54,56] |
| B4GALT1 | Promoter methylation | Down-regulation | Colon cancer | [57] |
| B4GALT3 | miR-27a | Up-regulation | Cervical cancer | [58] |
| C1GALT1 chaperone Cosmc | Promoter methylation | Down-regulation | B lymphocyte and other model cell lines | [43,44] |
| N-acetyl-galactosaminyl transferases | ||||
| GALNT1 | miR-129 | Down-regulation | Bladder cancer | [34] |
| GALNT4 | miR-9 | Down-regulation | Liver and cervical cancer | [35,36] |
| GALNT7 | miR-34° -34c | Down-regulation | Laryngeal cancer | [37] |
| miR-214 | Down-regulation | Esophageal cancer | [38] | |
| miR-17-3p | Down-regulation | Liver cell lines and mouse model | [40] | |
| GALNT10 | miR-122 | Down-regulation | Liver cancer | [41] |
| GALNT13 | miR-424 | Down-regulation | Breast and HEK293 cell lines | [33] |
| GALNT14 | miR-125a | Down-regulation | Ovarian cancer | [42] |
| A3GALNT/A3GALT1 | Promoter methylation | Down-regulation | Bladder and oral cancer | [107,108] |
| B4GALNT2 | Promoter methylation | Down-regulation | Gastrointestinal cancer | [80,81] |
| N-acetyl-glucosaminyl transferases | ||||
| MGAT3 | Promoter methylation | Down-regulation | Mammary model and ovarian and liver cell lines | [25,26,27,28,29] |
| MGAT4 | miR-424 | Down-regulation | Breast and HEK293 cell lines | [33] |
| MGAT5 | Distant methylation | Down-regulation | Ovarian cell line | [28] |
| MGAT5 | Distant methylation | Up-regulation | Melanoma hybrid cell line | [32] |
| B3GLCT | miR-200 | Down-regulation | Breast cell line | [50] |
| B3GNT7 | Promoter methylation | Down-regulation | Colon cancer | [59] |
| OGT | miR-424 | unclear | Breast and HEK293 cell lines | [33] |
| Sialyltransferases | ||||
| ST3GAL1 | miR-4701-5p | Down-regulation | Chronic myeloid leukemia | [103] |
| ST3GAL2 | Promoter methylation | Down-regulation | Prostate cell lines | [48] |
| ST3GAL3 | Promoter methylation | unknown | Whole methylome | [70] |
| ST3GAL5 | miR-200 | Down-regulation | Breast cell line | [50] |
| miR-26a, miR-548l, miR-34a | Down-regulation | Liver cancer | [51] | |
| ST3GAL6 | Promoter methylation | Down-regulation | Colon cancer cell line | [71] |
| miR-26a | Down-regulation | Liver cancer | [102] | |
| ST6GAL1 | Promoter methylation | Down-regulation | Glioma, bladder and breast cancer | [97,98,99] |
| miR-199a | Down-regulation | Lung and HEK293 cell lines | [100] | |
| ST6GALNAC1 | Promoter methylation | Down-regulation | Esophageal cancer | [46] |
| ST6GALNAC4 | miR-4299 | Down-regulation | Thyroid cancer | [104] |
| ST6GALNAC5 | Promoter methylation | No regulation | Colon cancer | [49] |
| miR-200 | Down-regulation | Breast cell line | [50] | |
| ST6GALNAC6 | Putative promoter methylation/ histone deacetylation | Down-regulation | Colon cancer | [67] |
| ST8SIA4 | miR26a-26b | Down-regulation | Breast cancer | [101] |
| Fucosyltransferases | ||||
| FUT2 | miR-15b | Down-regulation | Liver cancer | [110] |
| FUT3 | Promoter methylation | Down-regulation | Gastric and tongue cell lines | [68,69] |
| FUT4 | Promoter methylation | Down-regulation | Skin cell lines | [84] |
| miR-224-3p | Down-regulation | Breast cancer | [85] | |
| miR-493-5p | Down-regulation | Breast cancer | [86] | |
| FUT6 | miR-106b | Down-regulation | Breast cancer | [87] |
| FUT8 | miR-198 | Up-regulation | Colon cancer | [19] |
| miR-122 -34a 26a -146a | Down-regulation | Liver cancer | [20,21] | |
| Sulfotransferases | ||||
| HS3STs | Promoter methylation | Down-regulation | Chondrosarcoma cell line | [111] |
| Nucleotide donor transporters | ||||
| DTDST | Histone deacetylation | Down-regulation | Colon cancer | [72] |
| Galectins | ||||
| LGALS3 | Promoter methylation | Down-regulation | Colon, prostate and pituitary cancer | [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119] |
| Promoter hypomethylation | Up-regulation | Thyroid cancer | [120] | |
| miR-322 | Down-regulation | Breast, lung, prostate and HEK293 cell lines | [122] | |
| miR-128 | Down-regulation | Colon cancer | [121] | |
| LGALS7 | Promoter methylation | Down-regulation | Gastric cancer and lymphomas | [123,124] |
| LGALS9 | miR-22 | Down-regulation | Liver cancer | [126] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dall’Olio, F.; Trinchera, M. Epigenetic Bases of Aberrant Glycosylation in Cancer. Int. J. Mol. Sci. 2017, 18, 998. https://doi.org/10.3390/ijms18050998
Dall’Olio F, Trinchera M. Epigenetic Bases of Aberrant Glycosylation in Cancer. International Journal of Molecular Sciences. 2017; 18(5):998. https://doi.org/10.3390/ijms18050998
Chicago/Turabian StyleDall’Olio, Fabio, and Marco Trinchera. 2017. "Epigenetic Bases of Aberrant Glycosylation in Cancer" International Journal of Molecular Sciences 18, no. 5: 998. https://doi.org/10.3390/ijms18050998
APA StyleDall’Olio, F., & Trinchera, M. (2017). Epigenetic Bases of Aberrant Glycosylation in Cancer. International Journal of Molecular Sciences, 18(5), 998. https://doi.org/10.3390/ijms18050998







