Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis
Abstract
:1. Introduction
2. Results
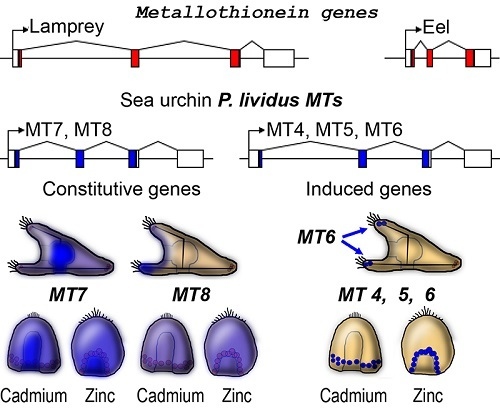
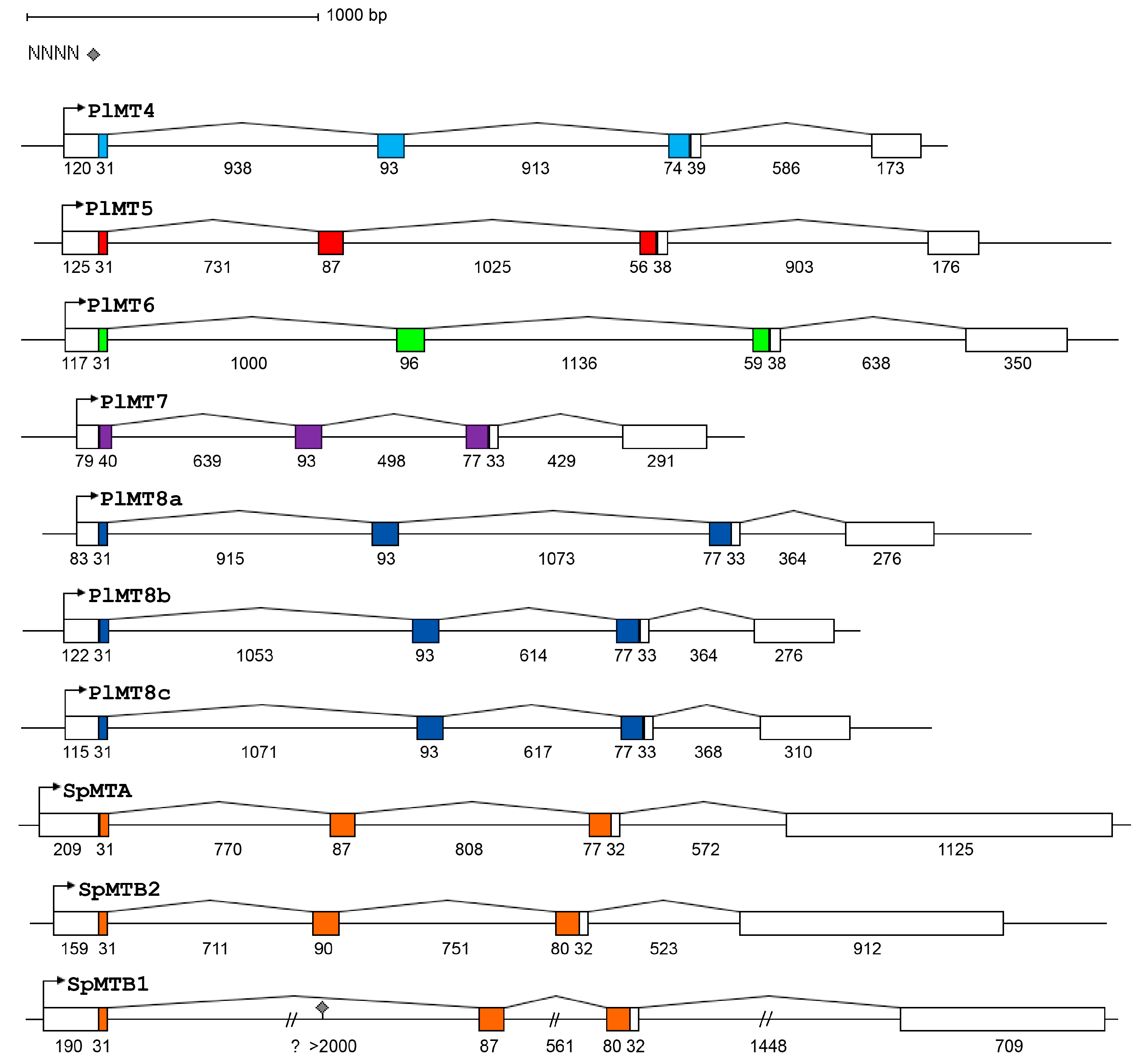
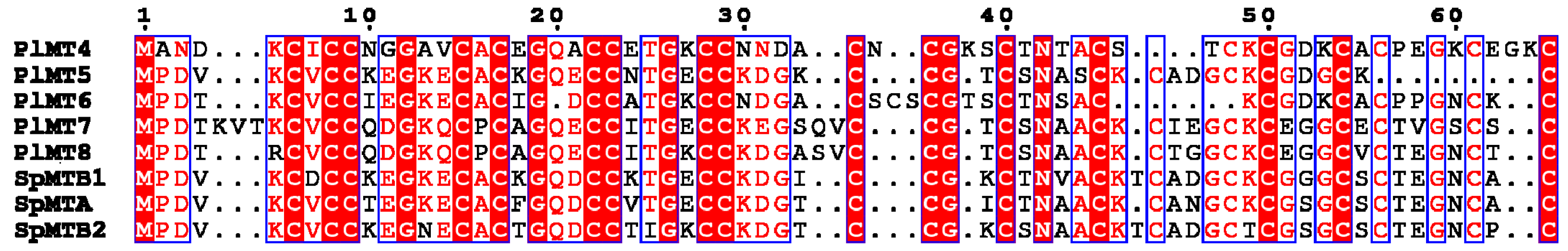
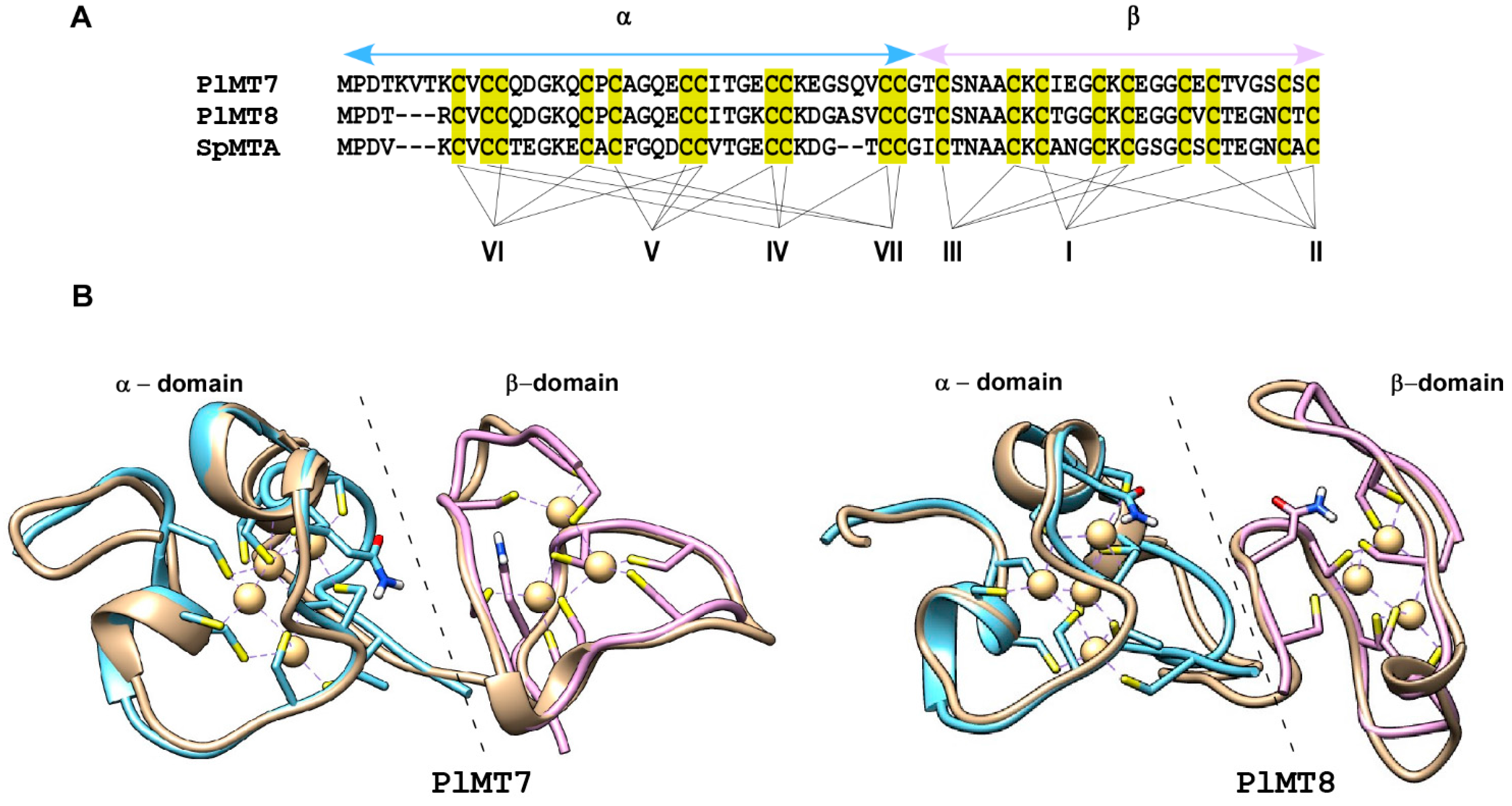
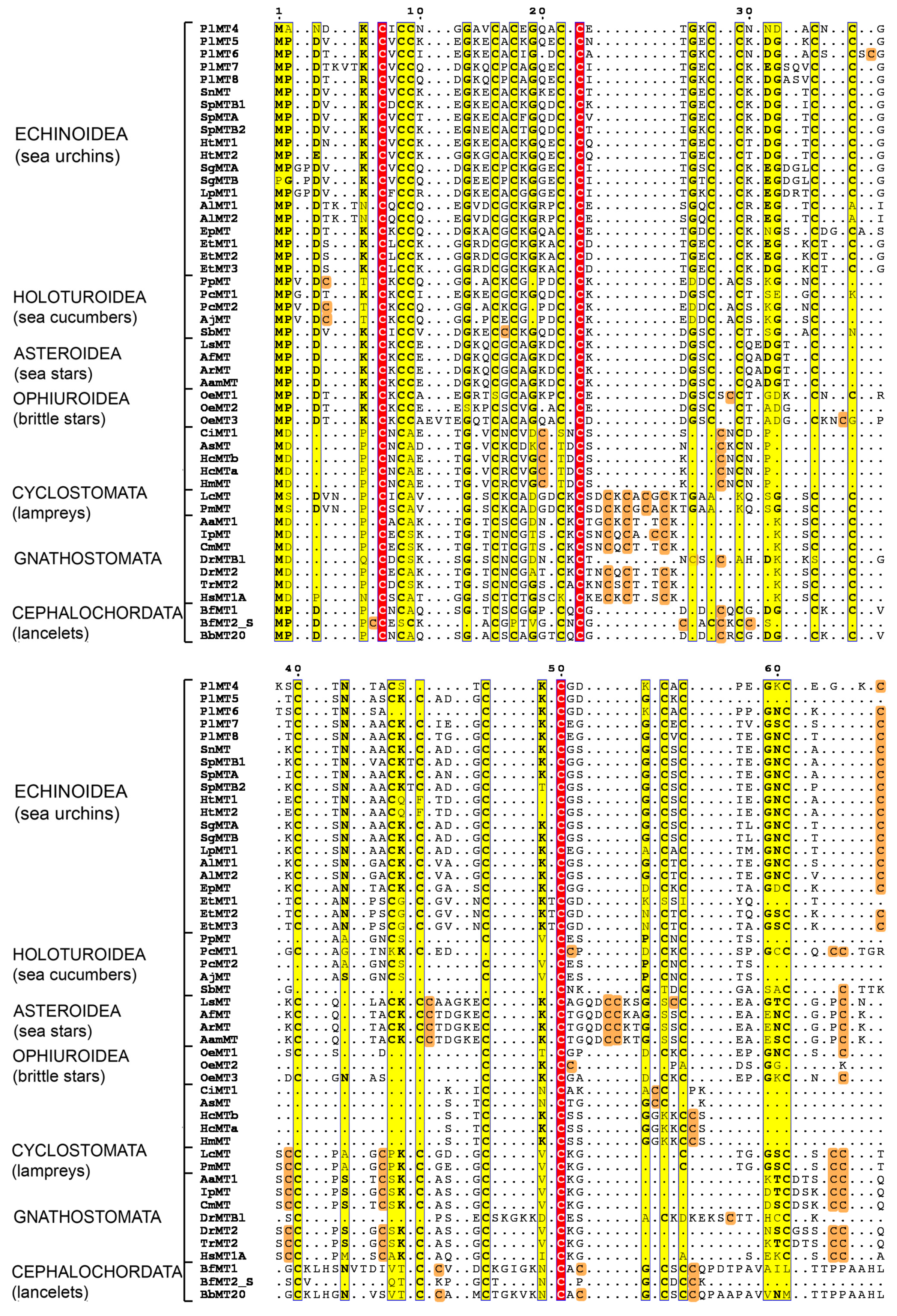
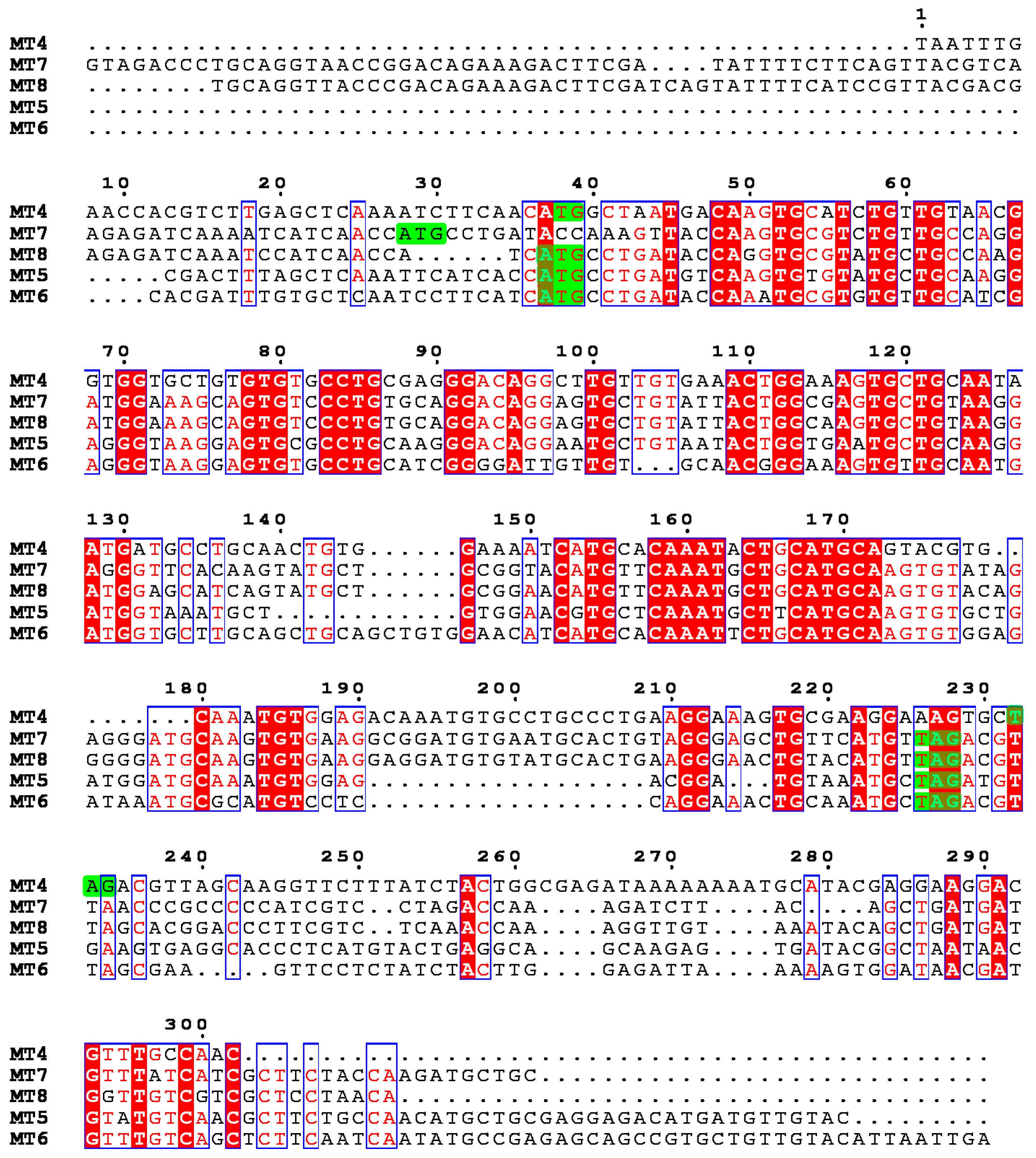
2.1. The Metallothionein Genes of P. lividus
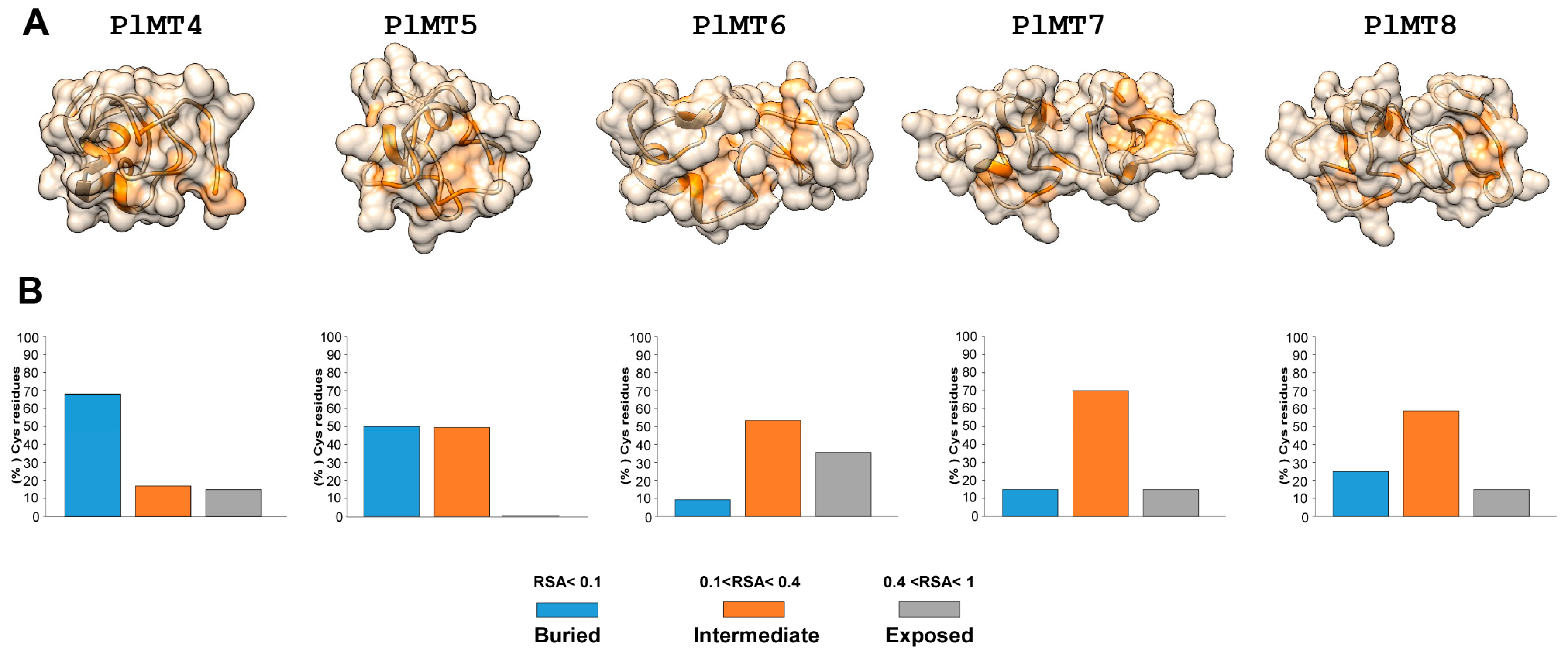
2.2. Predicted 3D Structural Model of P. lividus MTs
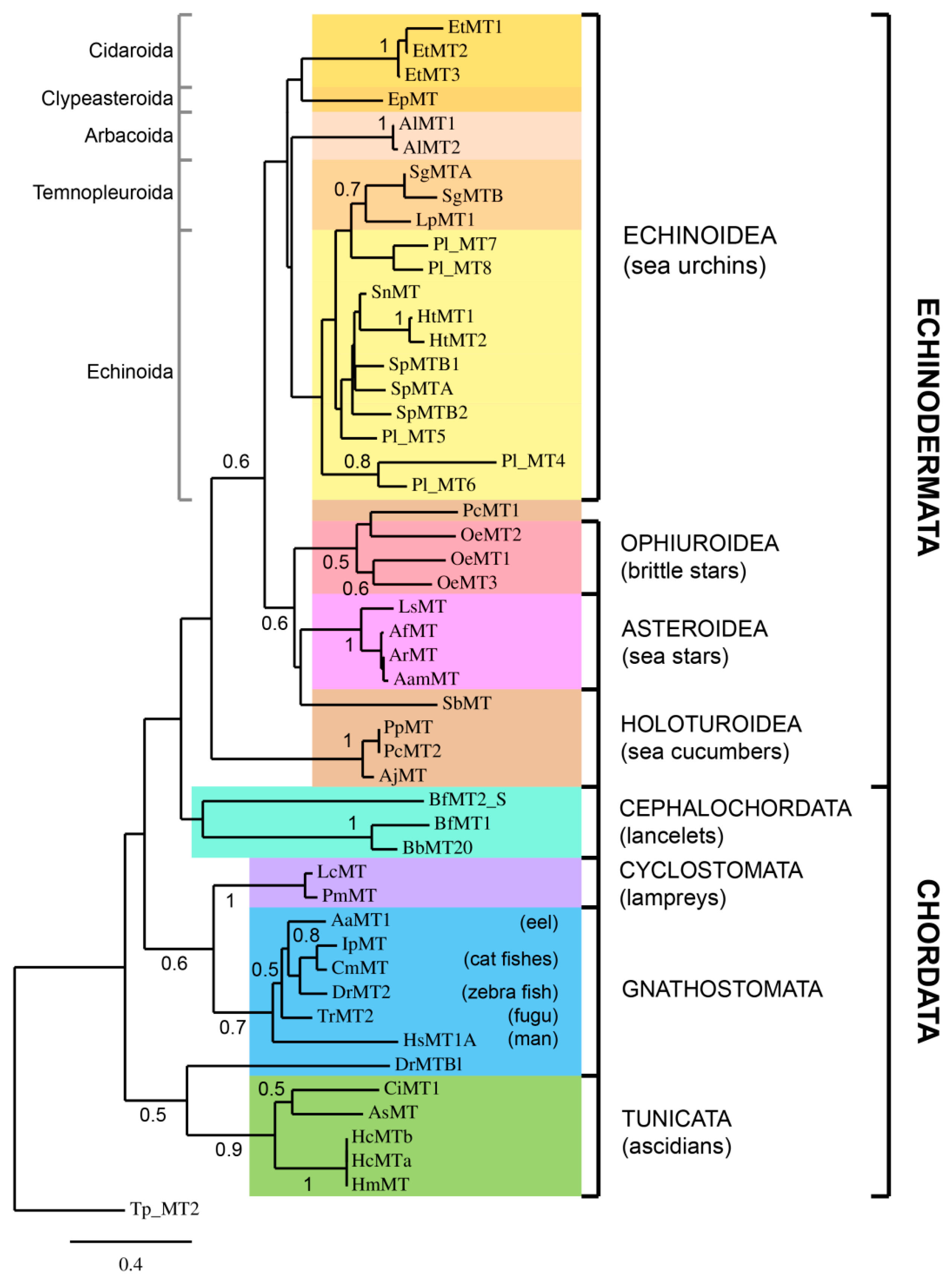
2.3. Phylogenetic Analysis of Deuterostome MTs
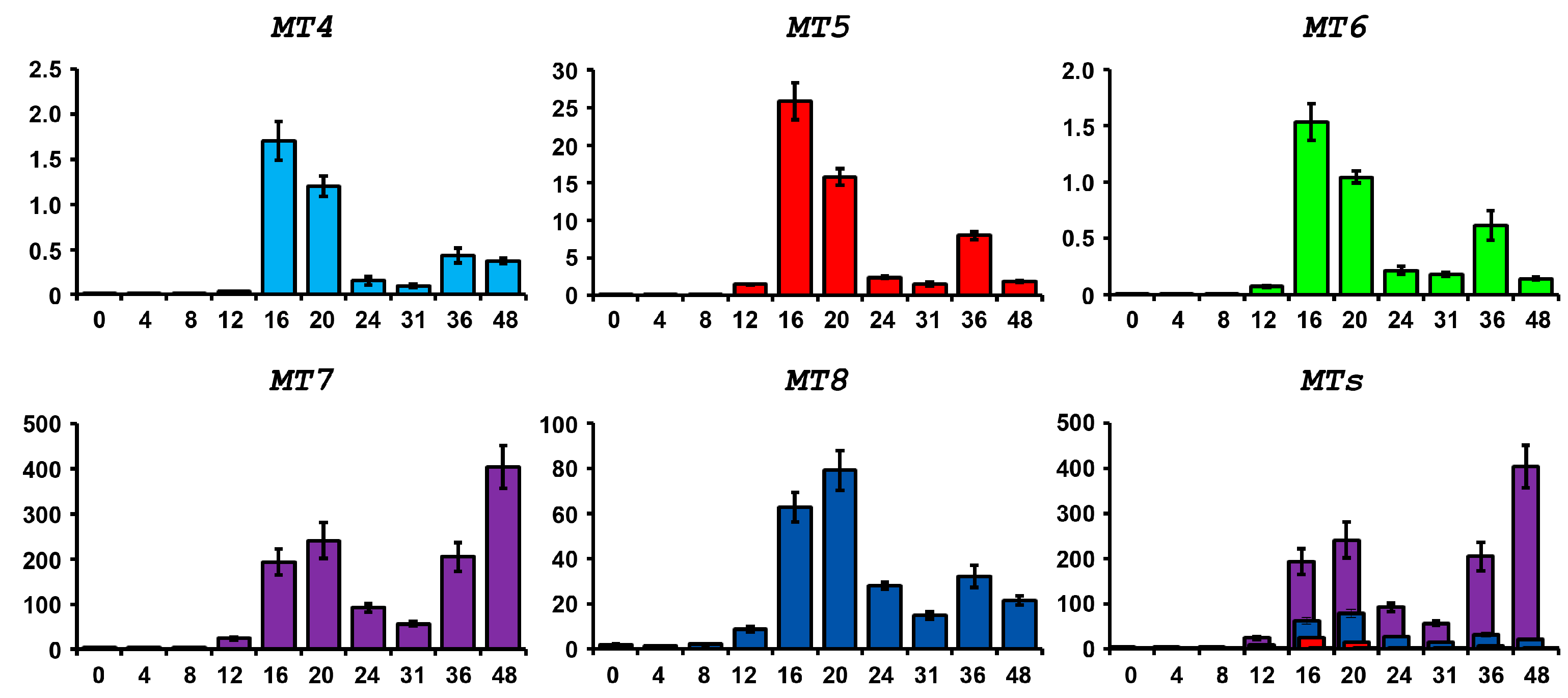
2.4. Expression of P. lividus MT Family Genes during Development and after Metal Treatments
2.4.1. Physiological Expression during Embryogenesis
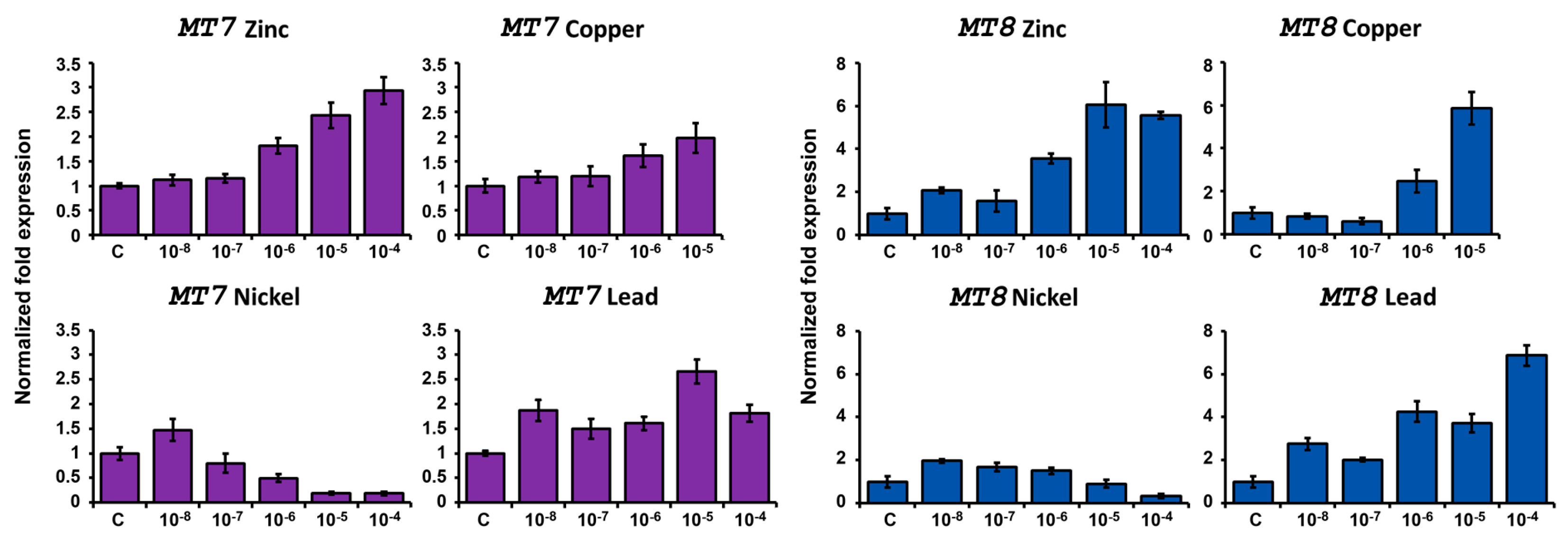
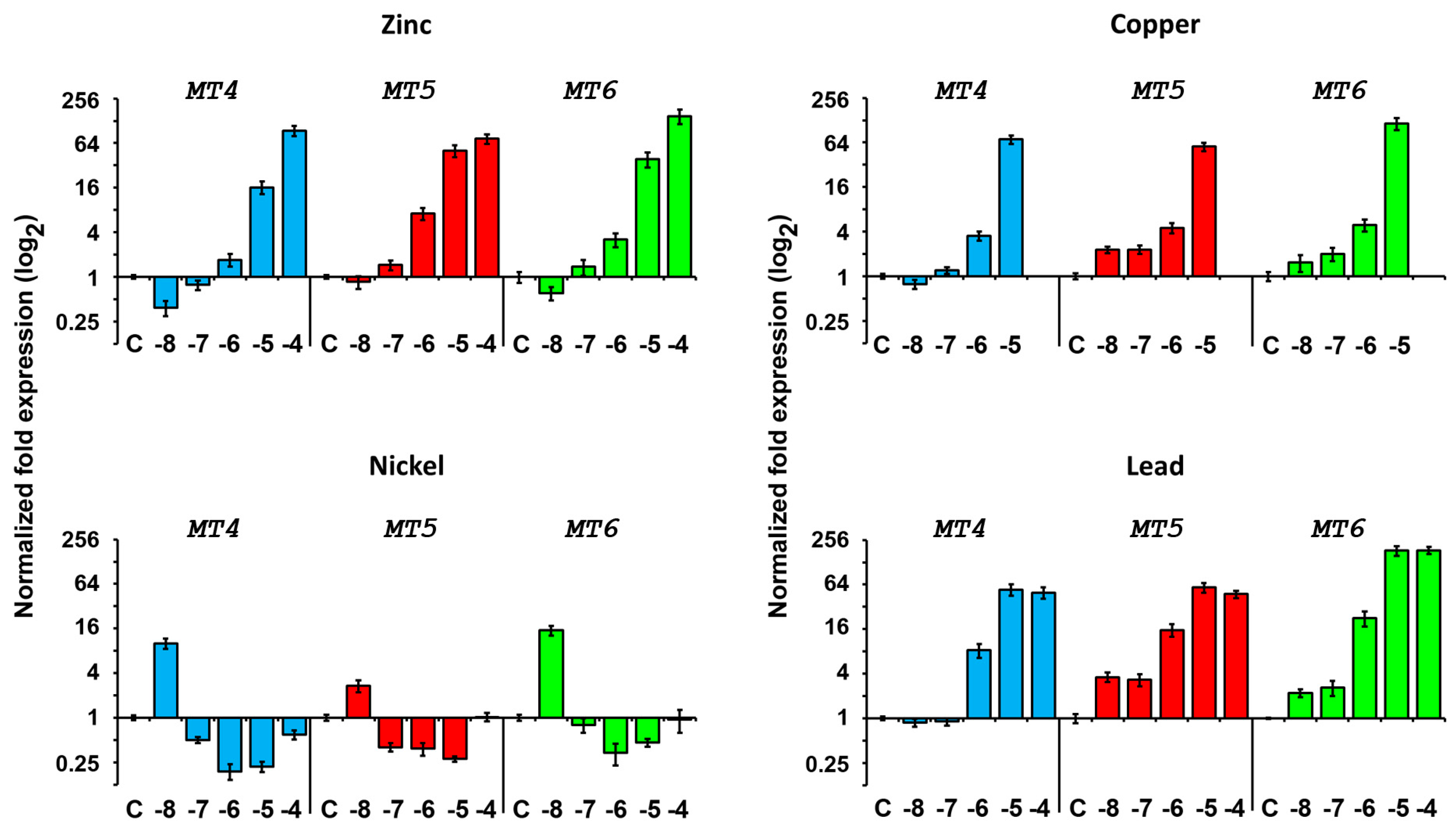
2.4.2. Expression after Metal Exposure
Zinc Treatment
Copper Treatment
Nickel Treatment
Lead Treatment
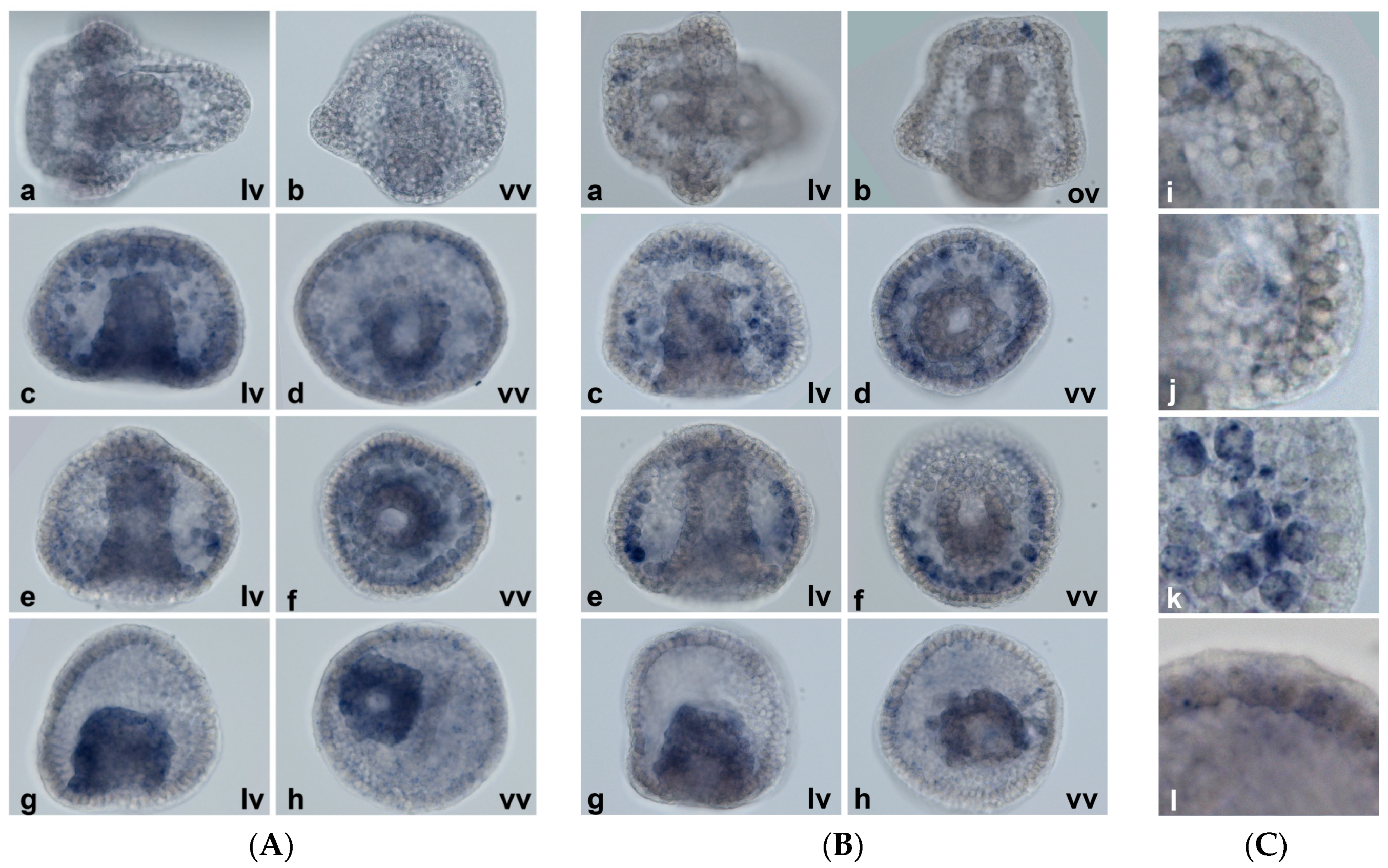
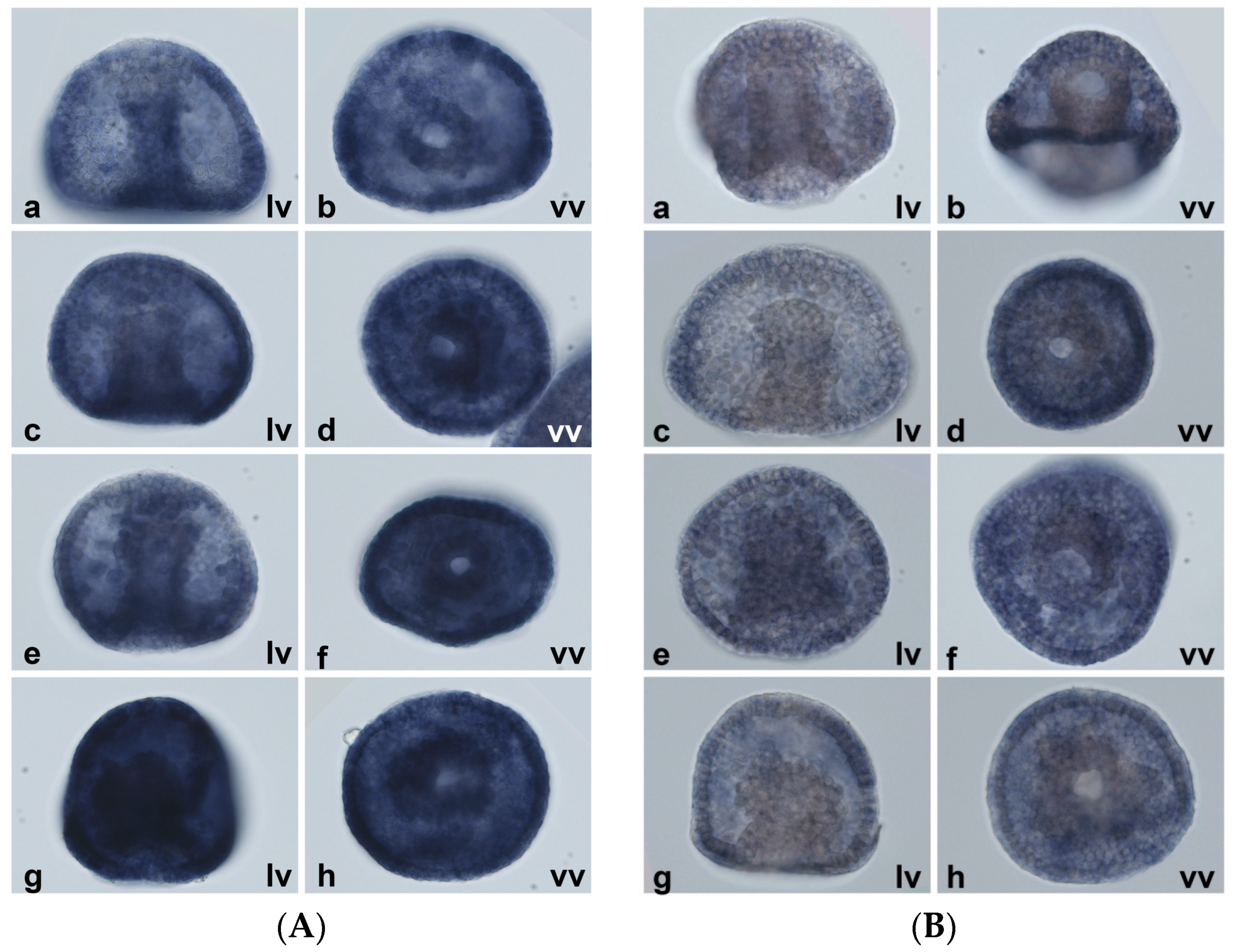
2.5. Spatial Patterns of MT Gene Expression
3. Discussion
4. Materials and Methods
4.1. Database Analysis, Cloning and Sequence Analysis of MT Genes
4.2. Sequences and Phylogenetic Analysis
4.3. Homology Modelling and Structural Characterization of PlMTs
4.4. Embryo Cultures, Metal Treatments and Morphological Analysis
4.5. RNA Extraction and RT-qPCR
4.6. Whole-Mount In Situ Hybridization
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| EST | Expressed sequence tags |
| MFSW | Millipore filtered sea water |
| MSA | Multiple sequence alignment |
| Myr | Million years ago |
| PlMTs | Paracentrotus lividus metallothioneins |
| PMCs | Primary mesenchyme cells |
| RSA | Relative solvent accessibility |
| RT-qPCR | Reverse transcription and quantitative (real time) PCR |
| SpMTs | Strongylocentrotus purpuratus metallothioneins |
| TSS | Transcription start site |
| WGS | Whole genome shotgun |
| WMISH | Whole-mount in situ hybridization |
References
- Braun, W.; Wagner, G.; Wörgötter, E.; Vasák, M.; Kägi, J.H.; Wüthrich, K. Polypeptide fold in the two metal clusters of metallothionein-2 by nuclear magnetic resonance in solution. J. Mol. Biol. 1986, 187, 125–129. [Google Scholar] [CrossRef]
- Kumari, M.V.; Hiramatsu, M.; Ebadi, M. Free radical scavenging actions of metallothionein isoforms I and II. Free Radic. Res. 1998, 29, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Macirella, R.; Guardia, A.; Pellegrino, D.; Bernabò, I.; Tronci, V.; Ebbesson, L.O.E.; Sesti, S.; Tripepi, S.; Brunelli, E. Effects of two sublethal concentrations of mercury chloride on the morphology and metallothionein activity in the liver of zebrafish (Danio rerio). Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.B.; Wei, H.W.; Wang, J.; Kong, Y.Q.; Wu, Y.Y.; Guo, J.L.; Li, T.F.; Li, J.K. Mammalian metallothionein-2A and oxidative stress. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, M.; Bofill, R.; Palacios, Ò.; Atrian, S. State-of-the-art of metallothioneins at the beginning of the 21st century. Coord. Chem. Rev. 2012, 256, 46–62. [Google Scholar] [CrossRef]
- Jin, R.; Chow, V.T.K.; Tan, P.H.; Dheen, S.T.; Duan, W.; Bay, B.H. Metallothionein 2A expression is associated with cell proliferation in breast cancer. Carcinogenesis 2002, 23, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, M.D.; Ivanova, I.A.; Cherian, M.G. Metallothionein and apoptosis during differentiation of myoblasts to myotubes: Protection against free radical toxicity. Toxicol. Appl. Pharmacol. 1999, 159, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Cherian, M.G.; Apostolova, M.D. Nuclear localization of metallothionein during cell proliferation and differentiation. Cell. Mol. Biol. 2000, 46, 347–356. [Google Scholar] [PubMed]
- Shimoda, R.; Achanzar, W.E.; Qu, W.; Nagamine, T.; Takagi, H.; Mori, M.; Waalkes, M.P. Metallothionein is a potential negative regulator of apoptosis. Toxicol. Sci. 2003, 73, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Pot, C.; Apetoh, L.; Thalhamer, T.; Zhu, B.; Murugaiyan, G.; Xiao, S.; Lee, Y.; Rangachari, M.; Yosef, N.; Kuchroo, V.K. Metallothioneins negatively regulate IL-27-induced type 1 regulatory T-cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 7802–7807. [Google Scholar] [CrossRef] [PubMed]
- Heger, Z.; Rodrigo, M.A.M.; Krizkova, S.; Ruttkay-Nedecky, B.; Zalewska, M.; del Pozo, E.M.P.; Pelfrene, A.; Pourrut, B.; Stiborova, M.; Eckschlager, T.; et al. Metallothionein as a scavenger of free radicals—New cardioprotective therapeutic agent or initiator of tumor chemoresistance? Curr. Drug Targets 2016, 17, 1438–1451. [Google Scholar] [CrossRef] [PubMed]
- Lynes, M.A.; Hidalgo, J.; Manso, Y.; Devisscher, L.; Laukens, D.; Lawrence, D.A. Metallothionein and stress combine to affect multiple organ systems. Cell Stress Chaperones 2014, 19, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Raudenska, M.; Gumulec, J.; Podlaha, O.; Sztalmachova, M.; Babula, P.; Eckschlager, T.; Adam, V.; Kizek, R.; Masarik, M. Metallothionein polymorphisms in pathological processes. Metallomics 2014, 6, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Werynska, B.; Pula, B.; Kobierzycki, C.; Dziegiel, P.; Podhorska-Okolow, M. Metallothioneins in the lung cancer. Folia Histochem. Cytobiol. 2015, 53, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.H.; Lukiw, W.J.; Bergeron, C.; Niznik, H.B.; Fraser, P.E. Metallothionein III is reduced in Alzheimer’s disease. Brain Res. 2001, 894, 37–45. [Google Scholar] [CrossRef]
- Binz, P.-A.; Kägi, J.H.R. Metallothionein: Molecular Evolution and Classification, In Metallothionein IV, Advances in Life Sciences; Klaassen, C.D., Ed.; Birkhäuser: Basel, Switzerland, 1999; pp. 7–13. [Google Scholar]
- Capdevila, M.; Atrian, S. Metallothionein protein evolution: A miniassay. J. Biol. Inorg. Chem. 2011, 16, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Palacios, O.; Atrian, S.; Capdevila, M. Zn- and Cu-thioneins: A functional classification for metallothioneins? J. Biol. Inorg. Chem. 2011, 16, 991–1009. [Google Scholar] [CrossRef] [PubMed]
- Palacios, Ò.; Pagani, A.; Pérez-Rafael, S.; Egg, M.; Höckner, M.; Brandstätter, A.; Capdevila, M.; Atrian, S.; Dallinger, R. Shaping mechanisms of metal specificity in a family of metazoan metallothioneins: Evolutionary differentiation of mollusc metallothioneins. BMC Biol. 2011, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Jenny, M.J.; Payton, S.L.; Baltzegar, D.A.; Lozier, J.D. Phylogenetic analysis of molluscan metallothioneins: Evolutionary insight from Crassostrea virginica. J. Mol. Evol. 2016, 83, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Nemer, M.; Wilkinson, D.G.; Travaglini, E.C.; Sternberg, E.J.; Butt, T.R. Sea urchin metallothionein sequence: Key to an evolutionary diversity. Proc. Natl. Acad. Sci. USA 1985, 82, 4992–4994. [Google Scholar] [CrossRef] [PubMed]
- Cserjesi, P.; Fang, H.; Brandhorst, B.P. Metallothionein gene expression in embryos of the sea urchin Lytechinus pictus. Mol. Reprod. Dev. 1997, 47, 39–46. [Google Scholar] [CrossRef]
- Scudiero, R.; Capasso, C.; Carginale, V.; Riggio, M.; Capasso, A.; Ciaramella, M.; Filosa, S.; Parisi, E. PCR amplification and cloning of metallothionein complementary DNAs in temperate and Antarctic sea urchin characterized by a large difference in egg metallothionein content. Cell. Mol. Life Sci. 1997, 53, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Prêcheur, B.; Wang, Y.; Mackay, E.A.; Wider, G.; Güntert, P.; Liu, A.; Kägi, J.H.; Wüthrich, K. NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA. J. Mol. Biol. 1999, 291, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.A.; Costa, S.; Gianguzza, M.; Roccheri, M.C.; Gianguzza, F. Effects of cadmium exposure on sea urchin development assessed by SSH and RT-qPCR: Metallothionein genes and their differential induction. Mol. Biol. Rep. 2013, 40, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B. Phylogenetic relationship, divergence times, and rates of molecular evolution for Camarodont sea urchin. Mol. Biol. Evol. 1988, 5, 345–365. [Google Scholar]
- Goldstone, J.V.; Hamdoun, A.; Cole, B.J.; Howard-Ashby, M.; Nebert, D.W.; Scally, M.; Dean, M.; Epel, D.; Hahn, M.E.; Stegeman, J.J. The chemical defensome: Environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev. Biol. 2006, 300, 366–384. [Google Scholar] [CrossRef] [PubMed]
- Franchi, N.; Boldrin, F.; Ballarin, L.; Piccinni, E. CiMT-1, an unusual chordate metallothionein gene in Ciona intestinalis genome: Structure and expression studies. J. Exp. Zool. A Ecol. Genet. Physiol. 2011, 315A, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Kamino, K.; Ueki, T.; Michibata, H. Expressed sequence tag analysis of vanadocytes in a vanadium-rich ascidian, Ascidia sydneiensis samea. Mar. Biotechnol. 2004, 6, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Guirola, M.; Pérez-Rafael, S.; Capdevila, M.; Palacios, O.; Atrian, S. Metal dealing at the origin of the Chordata phylum: The metallothionein system and metal overload response in amphioxus. PLoS ONE 2012, 7, e43299. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Wang, S.Y.; Kim, I.C.; Ki, J.S.; Raisuddin, S.; Lee, J.S.; Han, K.N. Cloning of a river pufferfish (Takifugu obscurus) metallothionein cDNA and study of its induction profile in cadmium-exposed fish. Chemosphere 2008, 71, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Zheng, Y.D.; Kuo, C.H. Expression of MT2 and SMT-B upon cadmium exposure and cold shock in zebrafish (Danio rerio). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008, 148, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Santovito, G.; Formigari, A.; Boldrin, F.; Piccinni, E. Molecular and functional evolution of Tetrahymena metallothioneins: New insights into the gene family of Tetrahymena thermophila. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007, 144, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Domènech, J.; Bofill, R.; Tinti, A.; Torreggiani, A.; Atrian, S.; Capdevila, M. Comparative insight into the Zn(II)-, Cd(II)- and Cu(I)-binding features of the protozoan Tetrahymena pyriformis MT1 metallothionein. Biochim. Biophys. Acta 2008, 1784, 693–704. [Google Scholar] [CrossRef] [PubMed]
- De Francisco, P.; Melgar, L.M.; Díaz, S.; Martín-González, A.; Gutiérrez, J.C. The Tetrahymena metallothionein gene family: Twenty-one new cDNAs, molecular characterization, phylogenetic study and comparative analysis of the gene expression under different abiotic stressors. BMC Genom. 2016, 17, 346. [Google Scholar] [CrossRef] [PubMed]
- Piccinni, E.; Staudenmann, W.; Albergoni, V.; de Gabrieli, R.; James, P. Purification and primary structure of metallothioneins induced by cadmium in the protists Tetrahymena pigmentosa and Tetrahymena pyriformis. Eur. J. Biochem. 1994, 226, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Piccinni, E.; Bertaggia, D.; Santovito, G.; Miceli, C.; Kraev, A. Cadmium metallothionein gene of Tetrahymena pyriformis. Gene 1999, 234, 51–59. [Google Scholar] [CrossRef]
- Fu, C.; Miao, W. Cloning and characterization of a new multi-stress inducible metallothionein gene in Tetrahymena pyriformis. Protist 2006, 157, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Pisani, D.; Mackenzie-Dodds, J.A.; Stockley, B.; Webster, B.L.; Littlewood, D.T.J. Testing the molecular clock: Molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata). Mol. Biol. Evol. 2006, 23, 1832–1851. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.J.; Eernisse, D.J. The phylogeny, evolutionary developmental biology, and paleobiology of the Deuterostomia: 25 years of new techniques, new discoveries, and new ideas. Org. Divers. Evol. 2016, 16, 401–418. [Google Scholar] [CrossRef]
- Quiniou, F.; Guillou, M.; Judas, A. Arrest and delay in embryonic development in sea urchin populations of the Bay of Brest (Brittany, France); link with environmental factors. Mar. Pollut. Bull. 1999, 38, 401–406. [Google Scholar] [CrossRef]
- Guillou, B.; Quiniou, E.; Huart, B.; Pagano, G. Comparison of embryonic development and metal contamination in several populations of the sea urchin Sphaerechinus granularis (Lamarck) exposed to anthropogenic pollution. Arch. Environ. Contam. Toxicol. 2000, 39, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Fernández, N.; Beiras, R. Combined toxicity of dissolved mercury with copper, lead and cadmium on embryogenesis and early larval growth of the Paracentrotus lividus sea-urchin. Ecotoxicology 2001, 10, 263–271. [Google Scholar] [CrossRef] [PubMed]
- King, C.K.; Riddle, M.J. Effects of metal contaminants on the development of the common Antarctic sea urchin Sterechinus neumayeri and comparisons of sensitivity with tropical and temperate echinoids. Mar. Ecol. Prog. Ser. 2001, 215, 143–154. [Google Scholar] [CrossRef]
- Kobayashi, N.; Okamura, H. Effects of heavy metals on sea urchin embryo development. 1. Tracing the cause by the effects. Chemosphere 2004, 55, 1403–1412. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Okamura, H. Effects of heavy metals on sea urchin embryo development. Part 2. Interactive toxic effects of heavy metals in synthetic mine effluents. Chemosphere 2005, 61, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Waterman, A.J. Effect of salts of heavy metals on development of the sea urchin, Arbacia punctulata. Biol. Bull. 1937, 73, 401–420. [Google Scholar] [CrossRef]
- Torres-Duarte, C.; Adeleye, A.S.; Pokhrel, S.; Mädler, L.; Keller, A.A.; Cherr, G.N. Developmental effects of two different copper oxide nanomaterials in sea urchin (Lytechinus pictus) embryos. Nanotoxicology 2016, 10, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Hardin, J.; Coffman, J.A.; Black, S.D.; McClay, D.R. Commitment along the dorsoventral axis of the sea urchin embryo is altered in response to NiCl2. Development 1992, 116, 671–685. [Google Scholar] [PubMed]
- Armstrong, N.; Hardin, J.; McClay, D.R. Cell–cell interactions regulate skeleton formation in the sea urchin embryo. Development 1993, 119, 833–840. [Google Scholar] [PubMed]
- Manzo, S.; Buono, S.; Cremisini, C. Cadmium, lead and their mixtures with copper: Paracentrotus lividus embryotoxicity assessment, prediction, and offspring quality evaluation. Ecotoxicology 2010, 19, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Radenac, G.; Fichet, D.; Miramand, P. Bioaccumulation and toxicity of four dissolved metals in Paracentrotus lividus sea-urchin embryo. Mar. Environ. Res. 2001, 51, 151–166. [Google Scholar] [CrossRef]
- Scudiero, R.; Capasso, C.; del Vecchio-Blanco, F.; Savino, G.; Capasso, A.; Parente, A.; Parisi, E. Isolation and primary structure determination of a metallothionein from Paracentrotus lividus (Echinodermata, Echinoidea). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 111, 329–336. [Google Scholar] [CrossRef]
- Russo, R.; Bonaventura, R.; Zito, F.; Schröder, H.C.; Müller, I.; Müller, W.E.G.; Matranga, V. Stress to cadmium monitored by metallothionein gene induction in Paracentrotus lividus embryos. Cell Stress Chaperones 2003, 8, 232–241. [Google Scholar] [CrossRef]
- Stetefeld, J.; Ruegg, M.A. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005, 30, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Peterson, K.J.; Wray, G.; Littlewood, D.T.J. From bilateral symmetry to pentaradiality: The phylogeny of hemichordates and echinoderms. In B Assembling the Tree of Life; Cracraft, J., Donoghue, M.J., Eds.; Oxford University Press: Oxford, UK, 2004; pp. 365–383. [Google Scholar]
- Russo, R.; Zito, F.; Matranga, V. Tissue-specificity and phylogenetics of Pl-MT mRNA during Paracentrotus lividus embryogenesis. Gene 2013, 519, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Harkey, M.A.; Whiteley, H.R.; Whiteley, A.H. Differential expression of the msp130 gene among skeletal lineage cells in the sea urchin embryo: A three dimensional in situ hybridization analysis. Mech. Dev. 1992, 37, 173–184. [Google Scholar] [CrossRef]
- Viarengo, A. Heavy metals in marine invertebrates: Mechanisms of regulation and toxicity at the cellular level. Aquat. Sci. 1989, 1, 295–317. [Google Scholar]
- Chiarelli, R.; Agnello, M.; Bosco, L.; Roccheri, M.C. Sea urchin embryos exposed to cadmium as an experimental model for studying the relationship between autophagy and apoptosis. Mar. Environ. Res. 2014, 93, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Tellis, M.S.; Lauer, M.M.; Nadella, S.; Bianchini, A.; Wood, C.M. Sublethal mechanisms of Pb and Zn toxicity to the purple sea urchin (Strongylocentrotus purpuratus) during early development. Aquat. Toxicol. 2014, 146, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Beniash, E.; Aizenberg, J.; Addadi, L.; Weiner, S. Amorphous calcium carbonate transforms into calcite during sea urchin larval spicule growth. Proc. Biol. Sci. 1997, 264, 461–465. [Google Scholar] [CrossRef]
- Vidavsky, N.; Addadi, S.; Mahamid, J.; Shimoni, E.; Ben-Ezra, D.; Shpigel, M.; Weiner, S.; Addadi, L. Initial stages of calcium uptake and mineral deposition in sea urchin embryos. Proc. Natl. Acad. Sci. USA 2014, 111, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.G.; Nemer, M. Metallothionein genes MTa and MTb expressed under distinct quantitative and tissue-specific regulation in sea urchin embryos. Mol. Cell. Biol. 1987, 7, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Angerer, L.M.; Kawczynski, G.; Wilkinson, D.G.; Nemer, M.; Angerer, R.C. Spatial patterns of metallothionein mRNA expression in the sea urchin embryo. Dev. Biol. 1986, 116, 543–547. [Google Scholar] [CrossRef]
- Varrella, S.; Romano, G.; Ianora, A.; Bentley, M.G.; Ruocco, N.; Costantini, M. Molecular response to toxic diatom-derived aldehydes in the sea urchin Paracentrotus lividus. Mar. Drugs 2014, 12, 2089–2113. [Google Scholar] [CrossRef] [PubMed]
- Varrella, S.; Romano, G.; Ruocco, N.; Ianora, A.; Bentley, M.G.; Costantini, M. First morphological and molecular evidence of the negative impact of diatom-derived hydroxyacids on the sea urchin Paracentrotus lividus. Toxicol. Sci. 2016, 151, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, O.; Castellano, I.; Romano, G.; Palumbo, A. Stress response to cadmium and manganese in Paracentrotus lividus developing embryos is mediated by nitric oxide. Aquat. Toxicol. 2014, 156, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Migliaccio, O.; Castellano, I.; Cirino, P.; Romano, G.; Palumbo, A. Maternal exposure to cadmium and manganese impairs reproduction and progeny fitness in the sea urchin Paracentrotus lividus. PLoS ONE 2015, 10, e0131815. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Chevenet, F.; Brun, C.; Bañuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Maggio, T.; Costa, S.; Salamone, M.; Tagliavia, M.; Mazzola, S.; Gianguzza, F.; Cuttitta, A. Maintenance of a protein structure in the dynamic evolution of TIMPs over 600 million years. Genome Biol. Evol. 2016, 8, 1056–1071. [Google Scholar] [CrossRef] [PubMed]
- Salamone, M.; Nicosia, A.; Bennici, C.; Quatrini, P.; Catania, V.; Mazzola, S.; Ghersi, G.; Cuttitta, A. Comprehensive analysis of a Vibrio parahaemolyticus strain extracellular serine protease VpSP37. PLoS ONE 2015, 10, e0126349. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Gianguzza, F.; Casano, C.; Ragusa, M. α-tubulin marker gene of neural territory of sea urchin embryos detected by whole-mount in situ hybridization. Int. J. Dev. Biol. 1995, 39, 477–483. [Google Scholar] [PubMed]
- Casano, C.; Ragusa, M.; Cutrera, M.; Costa, S.; Gianguzza, F. Spatial expression of α and β tubulin genes in the late embryogenesis of the sea urchin Paracentrotus lividus. Int. J. Dev. Biol. 1996, 40, 1033–1041. [Google Scholar] [PubMed]


| Taxonomy | Species | Name | ID Protein/Reference | ID mRNA | ID Gene/Scaffold # |
|---|---|---|---|---|---|
| Echinoidea Echinoida | Paracentrotus lividus | PlMT4/8 | Ragusa et al., 2013 | § | |
| Echinoidea Echinoida | Sterechinus neumayeri | SnMT | P55953 | ||
| Echinoidea Echinoida | Strongylocentrotus purpuratus | SpMTA | P04734.2 | SPU_017989.3a | Scaffold1168 v3.1 |
| SpMTB2 | Q27287 | SPU_017134.3c | Scaffold1906 v3.1 | ||
| SpMTB1 | SPU_001866.3a | Scaffold249 v3.1 | |||
| Echinoidea Echinoida | Heliocidaris tuberculata | HtMT1 | § | comp104953_c1_seq2 ◊ | |
| HtMT2 | comp104953_c1_seq1 ◊ | ||||
| Echinoidea Temnopleuroida | Sphaerechinus granularis | SgMTA | Q26497 | ||
| SgMTB | Q26496 | ||||
| Echinoidea Temnopleuroida | Lytechinus pictus | LpMT1 | O02033 | ||
| Echinoidea Arbacoida | Arbacia lixula | AlMT1 | § | c31462 _g1_i3 ¶ | |
| AlMT2 | c31386 _g2_i2 ¶ | ||||
| Echinoidea | Echinarachnius parma | EpMT | § | GAVF01002251 | |
| Clypeasteroida | |||||
| Echinoidea Cidaroida | Eucidaris tribuloides | EtMT1 | § | JI315060 | § JZLH010301553 |
| EtMT2 | GAZP01041405 | ||||
| EtMT3 | JI295076 | ||||
| Asteroidea | Asterias forbesi | AfMT | § | GAUS01062044.1 | |
| Asteroidea | Asterias rubens | ArMT | § | GAUU01048766.1 | |
| Asteroidea | Asterias amurensis | AamMT | § | GAVL01015559.1 | |
| Asteroidea | Leptasterias sp. AR-2014 | LsMT | § | GAVC01041863 | |
| Ophiuroidea | Ophiocoma echinata | OeMT1 | § | GAUQ01108229 | |
| OeMT2 | GAUQ01027563 | ||||
| OeMT3 | GAUQ01073893 | ||||
| Holothuroidea Aspidochirotida | Apostichopus japonicus | AjMT | § | GH551565 | § MODV01037111 +MODV01043468 |
| Holothuroidea Aspidochirotida | Parasthicopus parvimensis | PpMT | § | Locus_1_Transcript_31588/53328 | § Scaffold4005 KN886207.1 + Scaffold3285 KN885487.1 |
| Holothuroidea Aspidochirotida | Parastichopus californicus | PcMT1 | § | GAVO01014408.1 | |
| Holothuroidea Dendrochirotida | Sclerodactyla briareus | SbMT | § | GAUT01018048.1 | |
| Crinoidea | - | ||||
| Hemichordata | - | ||||
| Urochordata Enterogona | Ciona intestinalis | CiMT1 | ACN32211.2 Franchi et al., 2010 | BW491384 FJ217357 | Scaffold 186 (JGI database) |
| Urochordata Enterogona | Ascidia sydneiensis samea | AsMT | Yamaguchi et al., 2004 | Asy-sig-715 and Asy-sig-997 | |
| Urochordata Stolidobranchia | Herdmania curvata | HcMTA | § | AY314949 | |
| HcMTb | AY314939.1 | ||||
| Urochordata Stolidobranchia | Herdmania momus | HmMT | § | EL733027.1 | |
| Cephalochordata Amphioxiformes | Branchiostoma floridae | BfMT1 | Guirola et al., 2012 | BW764364 | Guirola et al., 2012 |
| BfMT2_S | FE561990.1 | ||||
| Cephalochordata Amphioxiformes | Branchiostoma belcheri | BbMT20 | XP_019631158.1 | XM_019775599.1 | NW_017803933 (AYSS01018500) |
| Craniata Cyclostomata | Lethenteron camtschaticum | LcMT | § | DC612982.1 | § APJL01076593 + APJL01076594 + APJL01076595 |
| Craniata Cyclostomata | Petromyzon marinus | PmMT | § | CO548937.1 | |
| Craniata Gnathostomata Elopomorpha | Anguilla anguilla | AaMT | ABF50549.1 | DQ493910.1 | § AZBK01727549 |
| Craniata Gnathostomata Otomorpha | Ictalurus punctatus | IpMT | O93571 | NM_001200077.1 or AF087935 and JT349175.1 | NC_030419 |
| Craniata Gnathostomata Otomorpha | Clarias macrocephalus | CmMT | AGC79138.1 | JX312865.1 | |
| Craniata Gnathostomata Otomorpha | Danio rerio | DrMTBl (like) | ENSDARP00000131449 (CAA65933.1) | ENSDART00000170342 | ENSDARG00000102051 |
| DrMT2 | ENSDARP00000061006 (AAH49475.1) | ENSDART00000061007 | ENSDARG00000041623 | ||
| Craniata Gnathostomata Euteleosteo-morpha | Takifugu rubripes | TrMT2 | ENSTRUP00000022394 | ENSTRUT00000022487 | ENSTRUG00000008907.1 |
| Craniata Gnathostomata Primates | Homo sapiens | HsMT1A | NP_005937.2 | NM_005946 | Gene ID: 4489 |
| Name | Forward Primer | Reverse Primer | Ta |
|---|---|---|---|
| MT4 | TTGGTGATAATAAATGAACCTTGGAG | AGACTGGACAGTATCTAATTGGACAG | 55 |
| MT5 | CAGACAGGTCTCCGTCTCGC | CGCTTCAAGCTCGCCACATT | 57 |
| MT6_UP | CACGGTGTTTTGGTTTGATGTC | TTTGTGCATGATGTTCCACAGC | 57 |
| MT6_DW | CACGATTTGTGCTCAATCCTTCAT | CAAGTGCTGGTTGCTATCCTG | 57 |
| MT7_UP | GCATGAGCAAAACCGTAGTCAG | ACATTCTGGATTCTTCTGCGTCG | 53 |
| MT7_DW | TAATGAAACCAGCCCACGATCA | TCACTCCGTCTTTGCAATCTT | 57 |
| MT8a | ATCAGTTACGACGAGAGATCAAATC | TTGGCTATCGCAAGACGTTC | 57 |
| MT8b | AAATGTAAATCAGTGACAGGACG | GAAACAATAAGTCATCAAATAACAAAAC | 53 |
| MT8c | CTTCTGAGTCTAGCGTTTCCTTGAAG | CTATCGCAAGACGTTCGTGAGC | 55 |
| Probe | Forward (Sense) Primer | Reverse (Antisense) Primer |
|---|---|---|
| MT4 | TAATTTGAACCACGTCTTGAGC | GTTGGCAAACGTCCTTCCTC |
| MT5 | CGACTTTAGCTCAAATTCATCACCATG | GTACAACATCATGTCTCCTCGC |
| MT6 | CACGATTTGTGCTCAATCCTTCAT | TCAATTAATGTAAAACAGCACGGC |
| MT7 | GTAGACCCTGCAGGTAACCG | GCAGCATCTTGGTAGAAGCG |
| MT8 | TGCAGGTTACCCGACAGAAAG | TGTTAGGAGCGACGACAACC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragusa, M.A.; Nicosia, A.; Costa, S.; Cuttitta, A.; Gianguzza, F. Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis. Int. J. Mol. Sci. 2017, 18, 812. https://doi.org/10.3390/ijms18040812
Ragusa MA, Nicosia A, Costa S, Cuttitta A, Gianguzza F. Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis. International Journal of Molecular Sciences. 2017; 18(4):812. https://doi.org/10.3390/ijms18040812
Chicago/Turabian StyleRagusa, Maria Antonietta, Aldo Nicosia, Salvatore Costa, Angela Cuttitta, and Fabrizio Gianguzza. 2017. "Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis" International Journal of Molecular Sciences 18, no. 4: 812. https://doi.org/10.3390/ijms18040812
APA StyleRagusa, M. A., Nicosia, A., Costa, S., Cuttitta, A., & Gianguzza, F. (2017). Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis. International Journal of Molecular Sciences, 18(4), 812. https://doi.org/10.3390/ijms18040812