De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Quality Control and De Novo Transcriptome Assembly
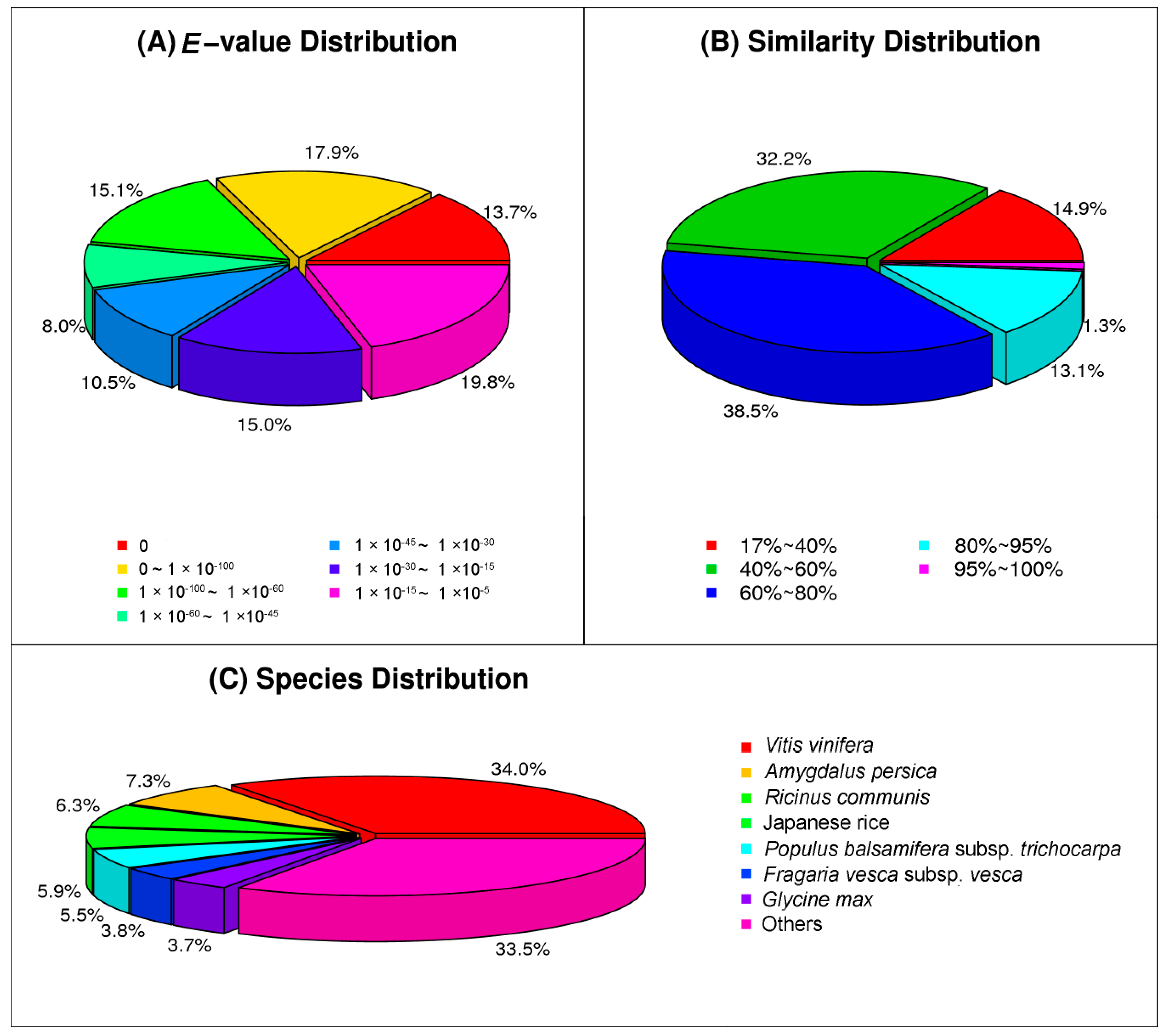
2.2. Functional Annotation of Assembled Unigenes and Coding Sequence (CDS) Prediction
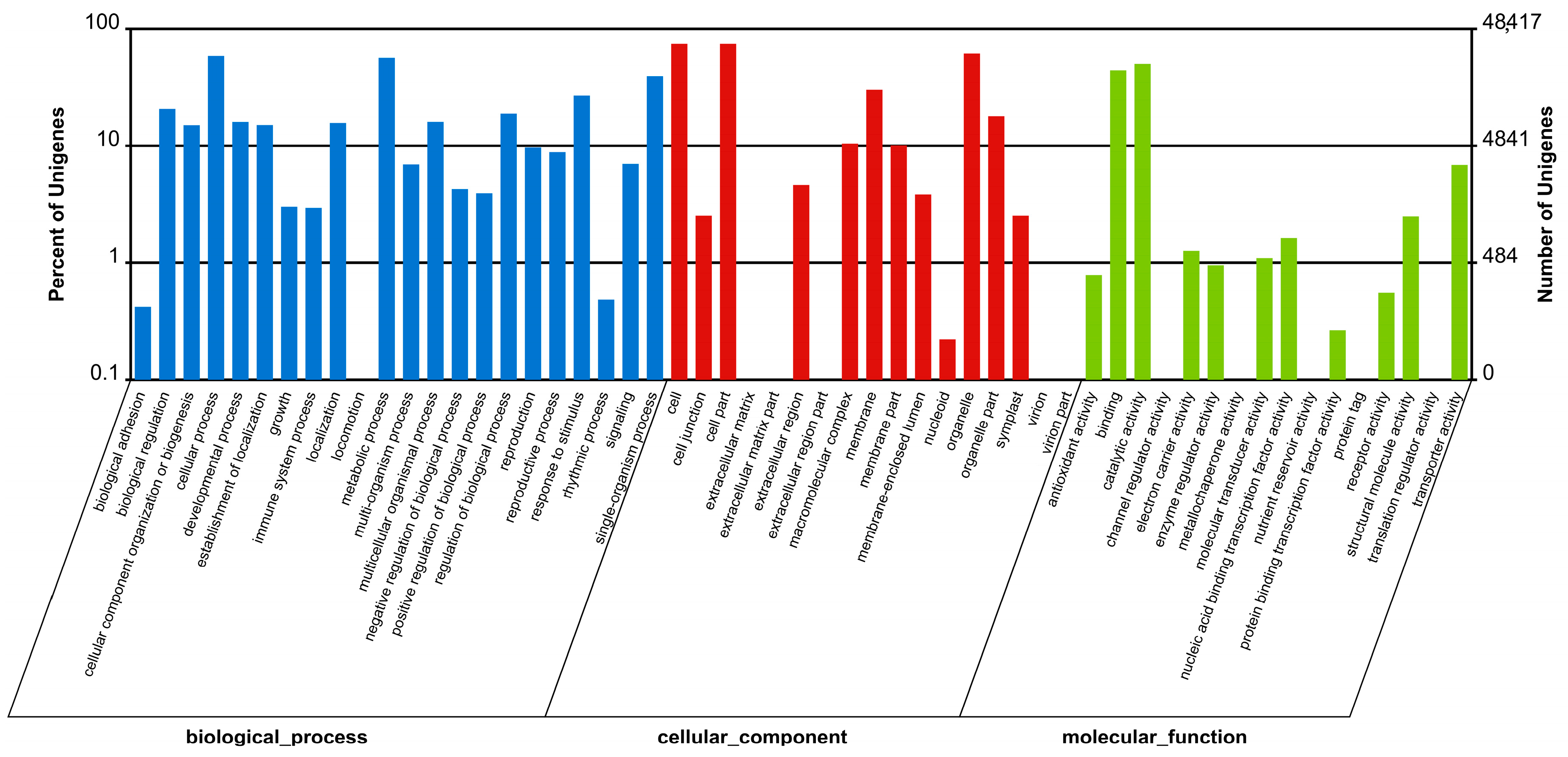
2.2.1. Gene Ontology (GO) Classification
2.2.2. Clusters of Orthologous Groups (COG) Classification
2.2.3. Kyoto Encyclopedia of Genes and Genomes (KEGG) Classification
2.2.4. CDS Prediction
2.2.5. Comparison with Reported Transcriptome Assemblies
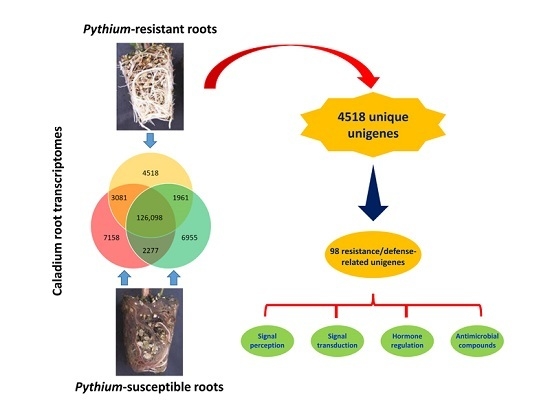
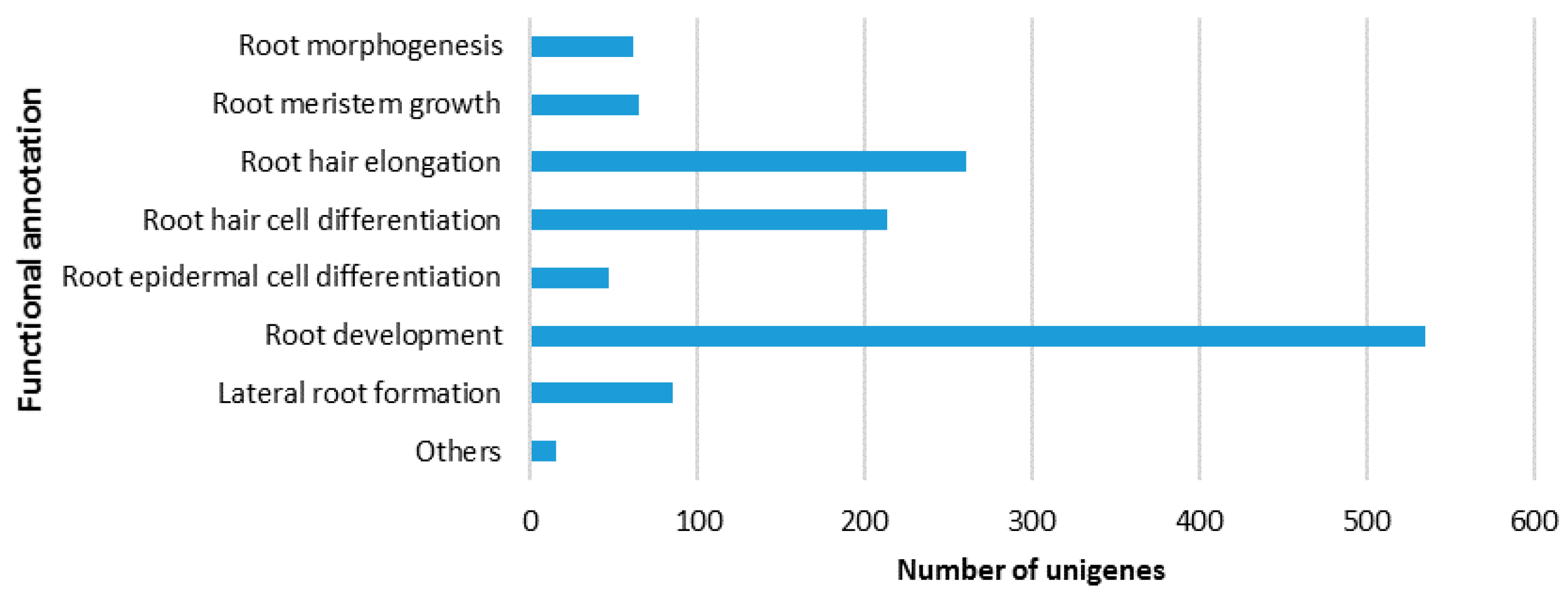
2.3. Identification of Putative Unigenes Involved in Resistance and Defense against Necrotrophic Pathogens
2.3.1. Early Signal Perception and Transduction in Plant-Pathogen Interactions
2.3.2. Hormone Regulation during Plant-Pathogen Interactions
2.3.3. Secondary Metabolites with Antimicrobial Activities
2.4. Simple Sequence Repeat (SSR) and Single Nucleotide Polymorphism (SNP) Discovery
3. Material and Methods
3.1. Plant Material
3.2. RNA Extraction and Quality Control
3.3. Illumina cDNA Library Preparation and Transcriptome Sequencing
3.4. Raw Sequencing Data Trimming and De Novo Transcriptome Assembly
3.5. Unigenes Annotation and Coding DNA Sequence (CDS) Prediction
3.6. Detection of SSR and SNP Sites
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CDS | Coding DNA sequence |
| COG | Clusters of Orthologous Groups |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NCBI | National Center for Biotechnology Information |
| NR | Non-redundant protein database (NCBI) |
| NT | Nucleotide sequence database (NCBI) |
| PAMP | Pathogen-associated molecular pattern |
| PTI | PAMP-triggered immunity |
| SNP | Single nucleotide polymorphism |
| SSR | Simple sequence repeat |
| USDA | United States Department of Agriculture |
References
- Cao, Z.; Mclaughlin, M.; Deng, Z. Interspecific genome size and chromosome number variation shed new light on species classification and evolution in caladium. J. Am. Soc. Hortic. Sci. 2014, 139, 449–459. [Google Scholar]
- Gong, L.; Deng, Z. Development and characterization of microsatellite markers for caladiums (Caladium Vent.). Plant Breed. 2011, 130, 591–595. [Google Scholar] [CrossRef]
- Deng, Z.; Goktepe, F.; Harbaugh, B.K.; Hu, J.G. Assessment of genetic diversity and relationships among caladium cultivars and species using molecular markers. J. Am. Soc. Hortic. Sci. 2007, 132, 219–229. [Google Scholar]
- Deng, Z.; Harbaugh, B.K.; Kelly, R.O.; Seijo, T.; McGovern, R.J. Pythium root rot resistance in caladium cultivars. HortScience 2005, 40, 549–552. [Google Scholar]
- Deng, Z.; Harbaugh, B.K.; Kelly, R.O.; Seijo, T.; McGovern, R.J. Screening for resistance to pythium root rot among twenty-three caladium cultivars. HortTechnology 2005, 15, 631–634. [Google Scholar]
- Ridings, W.H.; Hartman, R.D. Pathogenicity of Pythium myriotylum and other species of Pythium to caladium derived from shoot-tip culture. Phytopathology 1976, 66, 704–709. [Google Scholar] [CrossRef]
- Bruehl, G.W. Nonspecific genetic resistance to soil-borne fungi. Phytopathology 1983, 73, 948–951. [Google Scholar] [CrossRef]
- Goktepe, F.; Seijo, T.; Deng, Z.; Harbaugh, B.K.; Peres, N.A. Toward breeding for resistance to fusarium tuber rot in caladium: Inoculation technique and sources of resistance. HortScience 2007, 42, 1135–1139. [Google Scholar]
- Deng, Z.; Harbaugh, B.K.; Kelly, R.; Seijo, T.; McGovern, R.J. Evaluation of caladium cultivars for resistance to pythium root rot. HortScience 2004, 39, 772–773. [Google Scholar]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Sela-Buurlage, M.B.; Budai-Hadrian, O.; Pan, Q.; Carmel-Goren, L.; Vunsch, R.; Zamir, D.; Fluhr, R. Genome-wide dissection of Fusarium resistance in tomato reveals multiple complex loci. Mol. Genet. Genom. 2001, 265, 1104–1111. [Google Scholar]
- Komatsu, S.; Yang, G.; Hayashi, N.; Kaku, H.; Umemura, K.; Iwasaki, Y. Alterations by a defect in a rice G protein α subunit in probenazole and pathogen-induced responses. Plant Cell Environ. 2004, 27, 947–957. [Google Scholar] [CrossRef]
- Wang, X.L.; Jiang, N.; Liu, J.L.; Liu, W.D.; Wang, G.L. The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 2014, 5, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Diener, A.C.; Ausubel, F.M. Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef] [PubMed]
- De Vleesschauwer, D.; Xu, J.; Hofte, M. Making sense of hormone-mediated defense networking: From rice to Arabidopsis. Front. Plant Sci. 2014, 5, 611. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Prasad, V.; Sanjaya; Chen, K.H.; Liu, P.C.; Chan, M.T.; Chiu-Ping, C. Transgenic tomato plants expressing an Arabidopsis thionin (thi2.1) driven by fruit-inactive promoter battle against phytopathogenic attack. Planta 2005, 221, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.A.; Ponnala, L.; Weber, C.A. Strategies for transcriptome analysis in non-model plants. Am. J. Bot. 2012, 99, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Zhang, Y.; Chen, X.; Lee, E.J.; Barber, C.J.S.; Chakrabarty, R.; Desgagne-Penix, I.; Haslam, T.M.; Kim, Y.B.; Liu, E.W.; et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 2013, 166, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Xia, E.H.; Huang, H.; Jiang, J.J.; Liu, B.Y.; Gao, L.Z. De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genom. 2015, 16, 298. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Gu, Y.H.; Tao, X.; Cheng, X.J.; Wei, C.H.; Fu, J.; Cheng, Z.Q.; Zhang, Y.Z. De novo transcriptome sequencing of Oryza officinalis Wall ex Watt to identify disease-resistance genes. Int. J. Mol. Sci. 2015, 16, 29482–29495. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Q.; Esselink, G.D.; Visser, R.G.F.; van Tuyl, J.M.; Arens, P. Transcriptome analysis of Gerbera hybrida including in silico confirmation of defense genes found. Front. Plant Sci. 2016, 7, 247. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Oh, S.K.; Lee, J.H.; Lee, B.M.; Jo, S.H. Genome-wide SNP calling using next generation sequencing data in tomato. Mol. Cells 2014, 37, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.F.; Wiegert-Rininger, K.E.; Vallejo, V.A.; Barry, C.S.; Warner, R.M. Transcriptome-enabled marker discovery and mapping of plastochron-related genes in Petunia spp. BMC Genom. 2015, 16, 726. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.F.; Pan, C.; Diao, Y.; You, Y.N.; Yang, C.Z.; Hu, Z.L. Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC Genom. 2013, 14, 490. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, J.; Zhang, X.; Xu, L. Comparative transcriptome analysis of Anthurium “Albama” and its anthocyanin-loss mutant. PLoS ONE 2015, 10, e0119027. [Google Scholar] [CrossRef] [PubMed]
- Schliesky, S.; Gowik, U.; Weber, A.P.; Brautigam, A. RNA-Seq assembly—Are we there yet? Front. Plant Sci. 2012, 3, 220. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, H.; Ramagoni, R.K.; Anchoju, V.C.; Vankudavath, R.N.; Syed, A.U. De novo transcriptome analysis of Allium cepa L. (onion) bulb to identify allergens and epitopes. PLoS ONE 2015, 10, e0135387. [Google Scholar] [CrossRef] [PubMed]
- Cherukupalli, N.; Divate, M.; Mittapelli, S.R.; Khareedu, V.R.; Vudem, D.R. De novo assembly of leaf transcriptome in the medicinal plant Andrographis paniculata. Front. Plant Sci. 2016, 7, 1203. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, P.; Zhou, S.; Sun, Y.; Liu, N.; Li, X.; Hou, Y. De novo transcriptome sequencing and comprehensive analysis of the drought-responsive genes in the desert plant Cynanchum komarovii. BMC Genom. 2015, 16, 753. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Zhou, W.; Li, T.; Tian, C. De novo assembly and transcriptome characterization of spruce dwarf mistletoe Arceuthobium sichuanense uncovers gene expression profiling associated with plant development. BMC Genom. 2016, 17, 771. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, V.A.; Zhernakov, A.I.; Kulaeva, O.A.; Ershov, N.I.; Borisov, A.Y.; Tikhonovich, I.A. De novo assembly of the pea (Pisum sativum L.) nodule transcriptome. Int. J. Genom. 2015, 2015, 695947. [Google Scholar]
- Xu, L.; Wang, J.; Lei, M.; Li, L.; Fu, Y.; Wang, Z.; Ao, M.; Li, Z. Transcriptome analysis of storage roots and fibrous roots of the traditional medicinal herb Callerya speciosa (Champ.) schot. PLoS ONE 2016, 11, e0160338. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Gu, C.; Liu, L.; Zhu, X.; Zhao, Y.; Huang, S. Transcriptome profiling of Louisiana iris root and identification of genes involved in lead-stress response. Int. J. Mol. Sci. 2015, 16, 28087–28097. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.R.; Walker, J.C. Receptor-like protein kinases: The keys to response. Curr. Opin. Plant Biol. 2003, 6, 339–342. [Google Scholar] [CrossRef]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.S.; Tzeng, Y.H.; Mayer, K.F.X.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Gish, L.A.; Clark, S.E. The RLK/Pelle family of kinases. Plant J. 2011, 66, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Govers, F. Arabidopsis l-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J. Exp. Bot. 2009, 60, 4383–4396. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cordewener, J.H.G.; America, A.H.P.; Shan, W.X.; Bouwmeester, K.; Govers, F. Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol. Plant Microbe Int. 2015, 28, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.G.; Du, L.Q.; Chen, Z.X. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojarvi, J.; Rayapuram, C.; Idanheimo, N.; Hunter, K.; Kimura, S.; Merilo, E.; et al. Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 2015, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Rong, W.; Qi, L.; Li, J.; Wei, X.; Zhang, Z. Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis. Sci. Rep. UK 2013, 3, 3021. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.H.; Xing, L.P.; Wang, X.Y.; Yang, X.M.; Wang, W.; Sun, Y.L.; Qian, C.; Ni, J.L.; Chen, Y.P.; Liu, D.J.; et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Martin, G.B. Signal recognition and transduction mediated by the tomato Pto kinase: A paradigm of innate immunity in plants. Microbes Infect. 2000, 2, 1591–1597. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Z.C.; Lu, H.J.; Lu, S.W.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed]
- Haffani, Y.Z.; Silva-Gagliardi, N.F.; Sewter, S.K.; Grace Aldea, M.; Zhao, Z.; Nakhamchik, A.; Cameron, R.K.; Goring, D.R. Altered expression of PERK receptor kinases in Arabidopsis leads to changes in growth and floral organ formation. Plant Signal. Behav. 2006, 1, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.; Goring, D.R. The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 2002, 50, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Lecourieux, D.; Raneva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yoshioka, M.; Asai, S.; Nomura, H.; Kuchimura, K.; Mori, H.; Doke, N.; Yoshioka, H. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012, 196, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Takabatake, R.; Karita, E.; Seo, S.; Mitsuhara, I.; Kuchitsu, K.; Ohashi, Y. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2007, 48, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Lehmann, S.; Serrano, M.; L’Haridon, F.; Tjamos, S.E.; Metraux, J.P. Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 2015, 112, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Bhar, A.; Chatterjee, M.; Das, S. Fusarium oxysporum f.sp ciceri race 1 induced redox state alterations are coupled to downstream defense signaling in root tissues of chickpea (Cicer arietinum L.). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Parkhi, V.; Kenerley, C.M.; Rathore, K.S. Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 2009, 230, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.Q.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Qu, N.; Gao, M.; Zhang, Z.; Ding, X.; Yang, F.; Li, Y.; Dong, O.X.; Chen, S.; Li, X.; et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 2012, 24, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, S131–S151. [Google Scholar] [PubMed]
- Cho, Y.H.; Yoo, S.D. Novel connections and gaps in ethylene signaling from the ER membrane to the nucleus. Front. Plant Sci. 2015, 5, 733. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Eggermont, K.; Tierens, K.F.M.J.; Broekaert, W.F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999, 121, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, T.; Schmidt, J.S.; Zheng, X.Y.; Bent, A.F. Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 1999, 119, 935–949. [Google Scholar] [CrossRef] [PubMed]
- AbuQamar, S.; Chai, M.F.; Luo, H.L.; Song, F.M.; Mengiste, T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008, 20, 1964–1983. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, P.; Shockey, J.; Levesque, C.A.; Cook, R.J.; Browse, J. A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7209–7214. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Gisi, U.; Niderman, T. Local and systemic protection against Phytophthora infestans induced in potato and tomato plants by jasmonic acid and jasmonic methyl-ester. Phytopathology 1993, 83, 1054–1062. [Google Scholar] [CrossRef]
- Baetz, U.; Martinoia, E. Root exudates: The hidden part of plant defense. Trends Plant Sci. 2014, 19, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Lanoue, A.; Burlat, V.; Henkes, G.J.; Koch, I.; Schurr, U.; Rose, U.S.R. De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol. 2010, 185, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Arfaoui, A.; El Hadrami, A.; Mabrouk, Y.; Sifi, B.; Boudabous, A.; El Hadrami, I.; Daayf, F.; Cherif, M. Treatment of chickpea with Rhizobium isolates enhances the expression of phenylpropanoid defense-related genes in response to infection by Fusarium oxysporum F. sp. ciceris. Plant Physiol. Biochem. 2007, 45, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Balmer, D.; de Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.M.; Wang, Q.; Webster, F.X.; Kiemle, D.; Hong, Y.J.; Tantillo, D.J.; Coates, R.M.; Wray, A.T.; Askew, W.; O’Donnell, C.; et al. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 2013, 25, 1108–1125. [Google Scholar] [CrossRef] [PubMed]
- Steeghs, M.; Bais, H.P.; de Gouw, J.; Goldan, P.; Kuster, W.; Northway, M.; Fall, R.; Vivanco, J.M. Proton-transfer-reaction mass spectrometry as a new tool for real time analysis of root-secreted volatile organic compounds in Arabidopsis. Plant Physiol. 2004, 135, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Delaux, P.M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Sejalon-Delmas, N.; Combier, J.P.; Becard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Sawada, Y.; Shimada, Y.; Hirai, M.Y.; Sasaki, E.; Krischke, M.; Brown, P.D.; Saito, K.; Kamiya, Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 2011, 67, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Ching, A.; Caldwell, K.S.; Jung, M.; Dolan, M.; Smith, O.S.; Tingey, S.; Morgante, M.; Rafalski, A.J. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet. 2002, 3, 19. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.; Oliver, R. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large est datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]

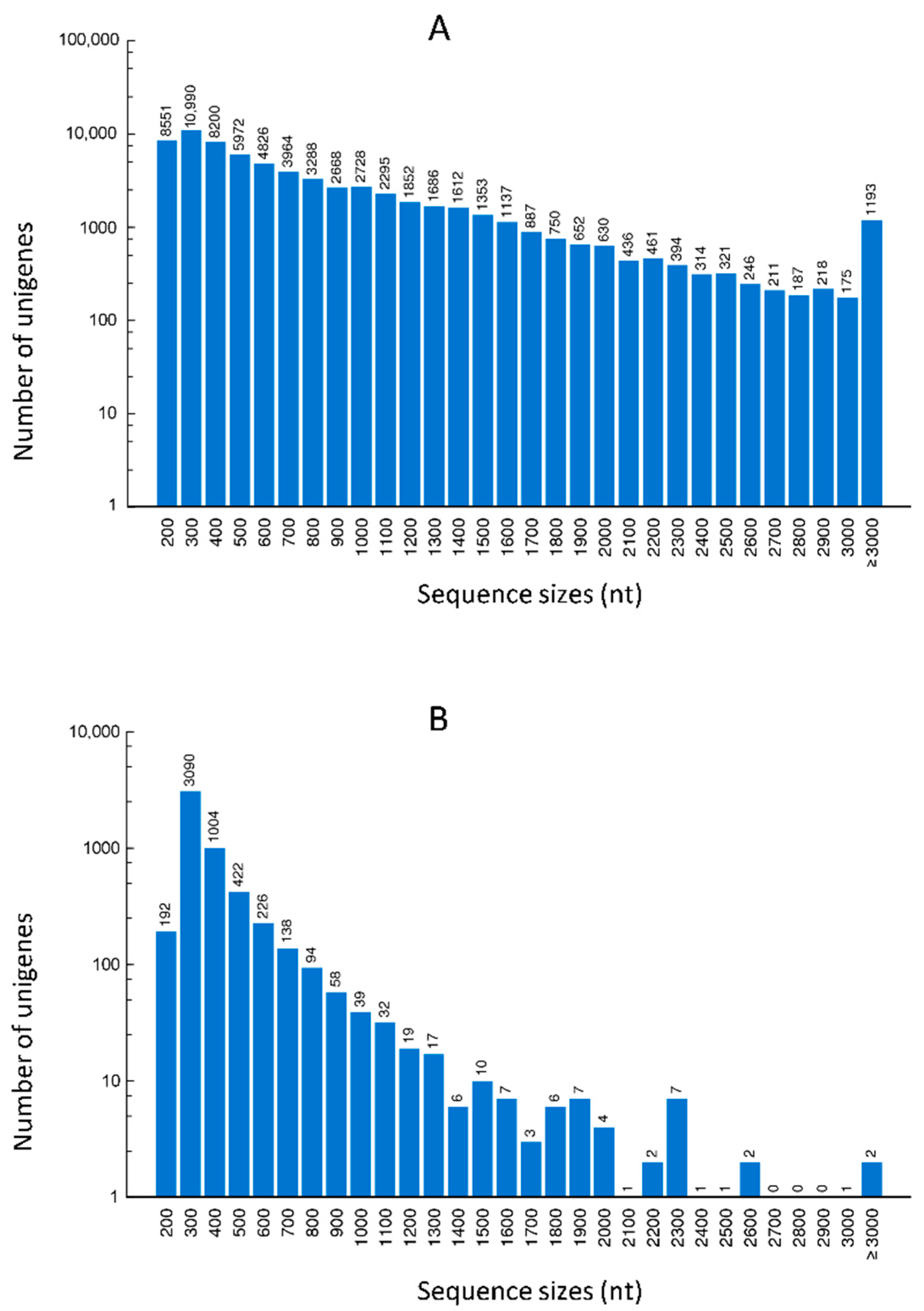
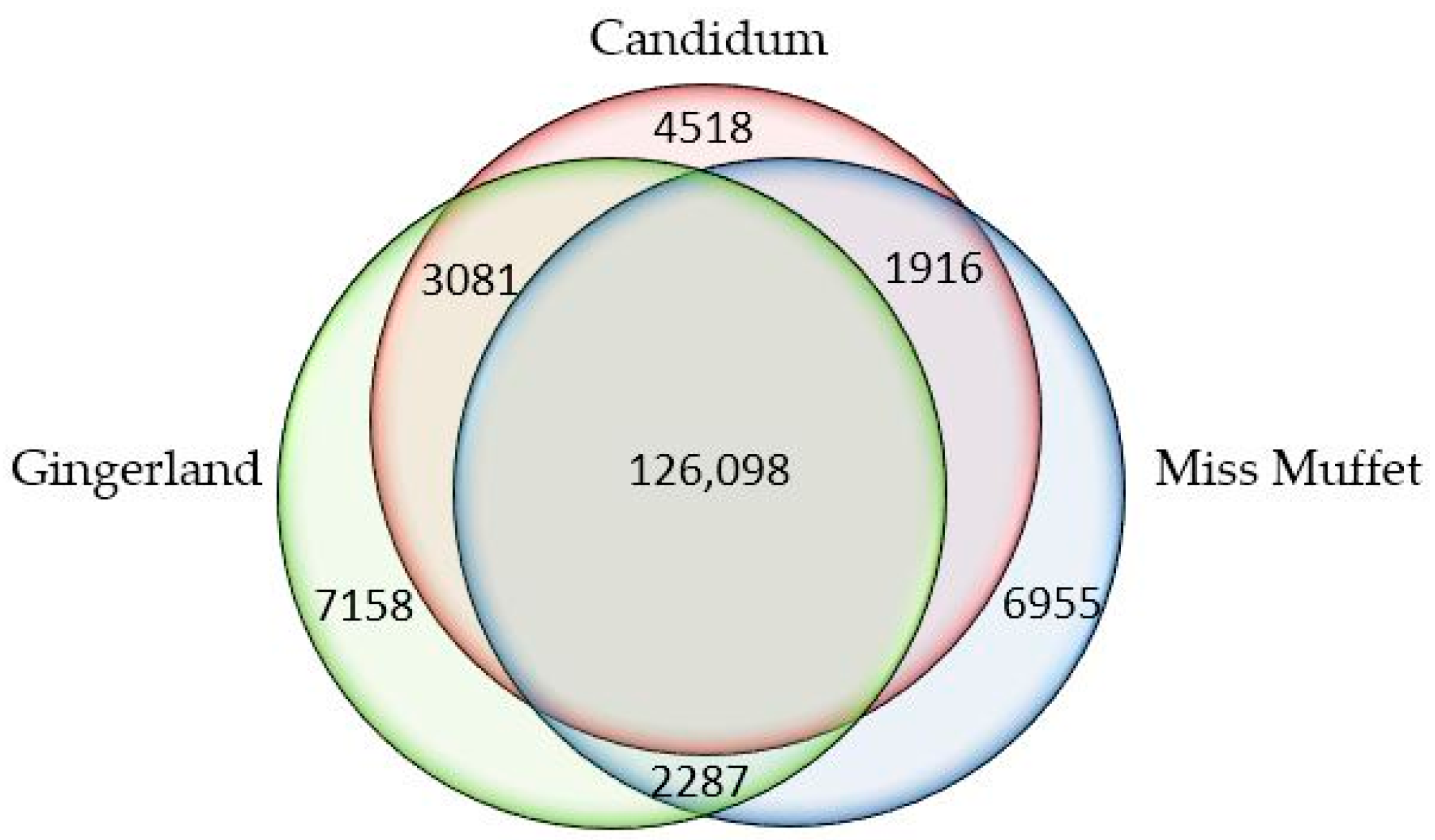
| Cultivars | Raw Reads | Clean Reads | Total Clean Nucleotides (nt) | Q20 Percentage (%) | N Percentage (%) | GC Percentage (%) |
|---|---|---|---|---|---|---|
| “Candidum” | 68,000,078 | 61,439,682 | 6,143,968,200 | 96.84 | 0.01 | 49.20 |
| “Gingerland” | 50,980,474 | 46,148,492 | 4,614,849,200 | 96.97 | 0.01 | 49.47 |
| “Miss Muffet” | 67,583,936 | 61,034,284 | 6,103,428,400 | 96.92 | 0.01 | 49.84 |
| Sample | Total Number | Total Length (nt) | Mean Length (nt) | N50 Length (nt) | Total Consensus Sequences | Distinct Clusters | Distinct Singletons | |
|---|---|---|---|---|---|---|---|---|
| Contig | “Candidum” | 255,215 | 74,985,719 | 294 | 471 | - | - | - |
| “Gingerland” | 232,333 | 69,427,118 | 299 | 492 | - | - | - | |
| “Miss Muffet” | 258,116 | 75,467,248 | 292 | 463 | - | - | - | |
| Unigene | “Candidum” | 133,737 | 97,471,052 | 729 | 1525 | 133,737 | 46,962 | 86,775 |
| “Gingerland” | 122,994 | 90,548,700 | 736 | 1514 | 122,994 | 43,721 | 79,273 | |
| “Miss Muffet” | 135,589 | 97,184,027 | 717 | 1487 | 135,589 | 47,844 | 87,745 | |
| All | 137,354 | 137,253,956 | 999 | 1755 | 137,354 | 62,353 | 75,001 |
| Databases | Number of Caladium Unigenes with Hits in Databases | Percentage of All Unigenes (%) |
|---|---|---|
| NR | 68,827 | 50.11 |
| NT | 52,233 | 38.03 |
| Swiss-Prot | 47,907 | 34.88 |
| KEGG | 46,406 | 33.79 |
| COG | 31,417 | 22.87 |
| GO | 48,417 | 35.25 |
| All | 71,825 | 52.29 |
| Rank | Pathway | Count (46,406) | Pathway ID | Function |
|---|---|---|---|---|
| 1 | Metabolic pathways | 12,474 | ko01100 | Metabolism |
| 2 | RNA transport | 5390 | ko03013 | Genetic Information Processing |
| 3 | Biosynthesis of secondary metabolites | 4780 | ko01110 | Metabolism |
| 4 | mRNA surveillance pathway | 4198 | ko03015 | Genetic Information Processing |
| 5 | Glycerophospholipid metabolism | 3613 | ko00564 | Metabolism |
| 6 | Endocytosis | 3563 | ko04144 | Cellular Processes |
| 7 | Ether lipid metabolism | 3169 | ko00565 | Metabolism |
| 8 | Plant-pathogen interaction | 2415 | ko04626 | Organismal Systems |
| 9 | Spliceosome | 2024 | ko03040 | Genetic Information Processing |
| 10 | RNA degradation | 1823 | ko03018 | Genetic Information Processing |
| Functional Pathway | Putative Annotation | Number of Unigenes | Root-Specific |
|---|---|---|---|
| Signal perception | l-type lection receptor kinase (LecRK) | 1 | No |
| Cysteine-rich receptor-like kinase (CRK) | 1 | No | |
| Receptor-like serine/threonine-protein kinases (STKs) | 12 | No | |
| Proline-rich receptor-like protein kinases (PERKs) | 31 | No | |
| Receptor-like protein kinases (RLKs) | 5 | No | |
| Signal transduction | Calcium-dependent protein kinases (CPKs) | 8 | No |
| Calmodulins (CaM) or calmodulins-like (CaM-like) | 4 | No | |
| Superoxide dismutase | 5 | No | |
| Mitogen-activated protein kinases (MAPKs) | 8 | No | |
| Hormone regulation | Ethylene receptors | 1 | No |
| S-adenosylmethionine (S-AdoMet) | 1 | No | |
| 1-aminocyclopropane-1-carboxylic acid (ACC) | 1 | No | |
| Acyl-CoA oxidase (ACX1A) | 1 | No | |
| Antimicrobial compounds | Phenylpropanoid biosythetic process | 2 | No |
| Flavonoid | 1 | No | |
| Flavonoid 3 | 1 | No | |
| Flavonoid 6-hydroxylase | 1 | No | |
| Flavonoid 3-monooxygenase | 1 | No | |
| Terpene synthase | 4 | No | |
| Tryptophan biosynthetic process | 1 | No | |
| Tryptophan 5-monooxygenase | 5 | No | |
| Tryptophan catabolic process | 1 | Yes | |
| Tryptophan 5-monooxygenase | 1 | No | |
| Tryptophan biosynthetic process | 1 | No |
| Number of Repeats | Mono-Nucleotide Repeats | Di-Nucleotide Repeats | Tri-Nucleotide Repeats | Quad-Nucleotide Repeat | Penta-Nucleotide Repeats | Hexa-Nucleotide Repeats |
|---|---|---|---|---|---|---|
| 4 | 0 | 0 | 0 | 0 | 293 | 362 |
| 5 | 0 | 0 | 5183 | 356 | 53 | 4 |
| 6 | 0 | 3489 | 2857 | 87 | 0 | 1 |
| 7 | 0 | 2659 | 1431 | 0 | 0 | 0 |
| 8 | 0 | 2879 | 102 | 0 | 0 | 0 |
| 9 | 0 | 3501 | 7 | 0 | 0 | 0 |
| 10 | 0 | 2397 | 1 | 0 | 0 | 0 |
| 11 | 0 | 553 | 2 | 0 | 0 | 0 |
| 12 | 1086 | 18 | 0 | 0 | 0 | 0 |
| 13 | 543 | 4 | 0 | 0 | 0 | 0 |
| 14 | 336 | 4 | 0 | 0 | 0 | 0 |
| 15 | 178 | 0 | 0 | 0 | 0 | 0 |
| 16 | 109 | 0 | 0 | 0 | 0 | 0 |
| 17 | 66 | 0 | 0 | 0 | 0 | 0 |
| 18 | 52 | 0 | 0 | 0 | 0 | 0 |
| 19 | 37 | 0 | 0 | 0 | 0 | 0 |
| 20 | 41 | 0 | 0 | 0 | 0 | 0 |
| 21 | 54 | 0 | 0 | 0 | 0 | 0 |
| 22 | 38 | 0 | 0 | 0 | 0 | 0 |
| 23 | 53 | 0 | 0 | 0 | 0 | 0 |
| 24 | 1 | 0 | 0 | 0 | 0 | 0 |
| SubTotal | 2594 | 15,504 | 9583 | 443 | 346 | 367 |
| SNP Type | “Candidum” | “Gingerland” | “Miss Muffet” |
|---|---|---|---|
| Transition | 169,687 | 147,579 | 162,919 |
| A (adenine) ↔ G (guanine) | 85,666 | 74,538 | 82,157 |
| C (cytosine) ↔ T (thymine) | 84,021 | 73,041 | 80,762 |
| Transversion | 112,041 | 97,106 | 107,298 |
| A (adenine) ↔ C (cytosine) | 27,274 | 23,705 | 25,952 |
| A (adenine) ↔ T (thymine) | 23,246 | 19,717 | 21,984 |
| C (cytosine) ↔ G (guanine) | 34,446 | 30,372 | 33,440 |
| G (guanine) ↔ T (thymine) | 27,075 | 23,312 | 25,922 |
| Total | 281,728 | 244,685 | 270,217 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Z.; Deng, Z. De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes. Int. J. Mol. Sci. 2017, 18, 712. https://doi.org/10.3390/ijms18040712
Cao Z, Deng Z. De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes. International Journal of Molecular Sciences. 2017; 18(4):712. https://doi.org/10.3390/ijms18040712
Chicago/Turabian StyleCao, Zhe, and Zhanao Deng. 2017. "De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes" International Journal of Molecular Sciences 18, no. 4: 712. https://doi.org/10.3390/ijms18040712
APA StyleCao, Z., & Deng, Z. (2017). De Novo Assembly, Annotation, and Characterization of Root Transcriptomes of Three Caladium Cultivars with a Focus on Necrotrophic Pathogen Resistance/Defense-Related Genes. International Journal of Molecular Sciences, 18(4), 712. https://doi.org/10.3390/ijms18040712