Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer
Abstract
:1. Background
2. Long Non-Coding RNAs
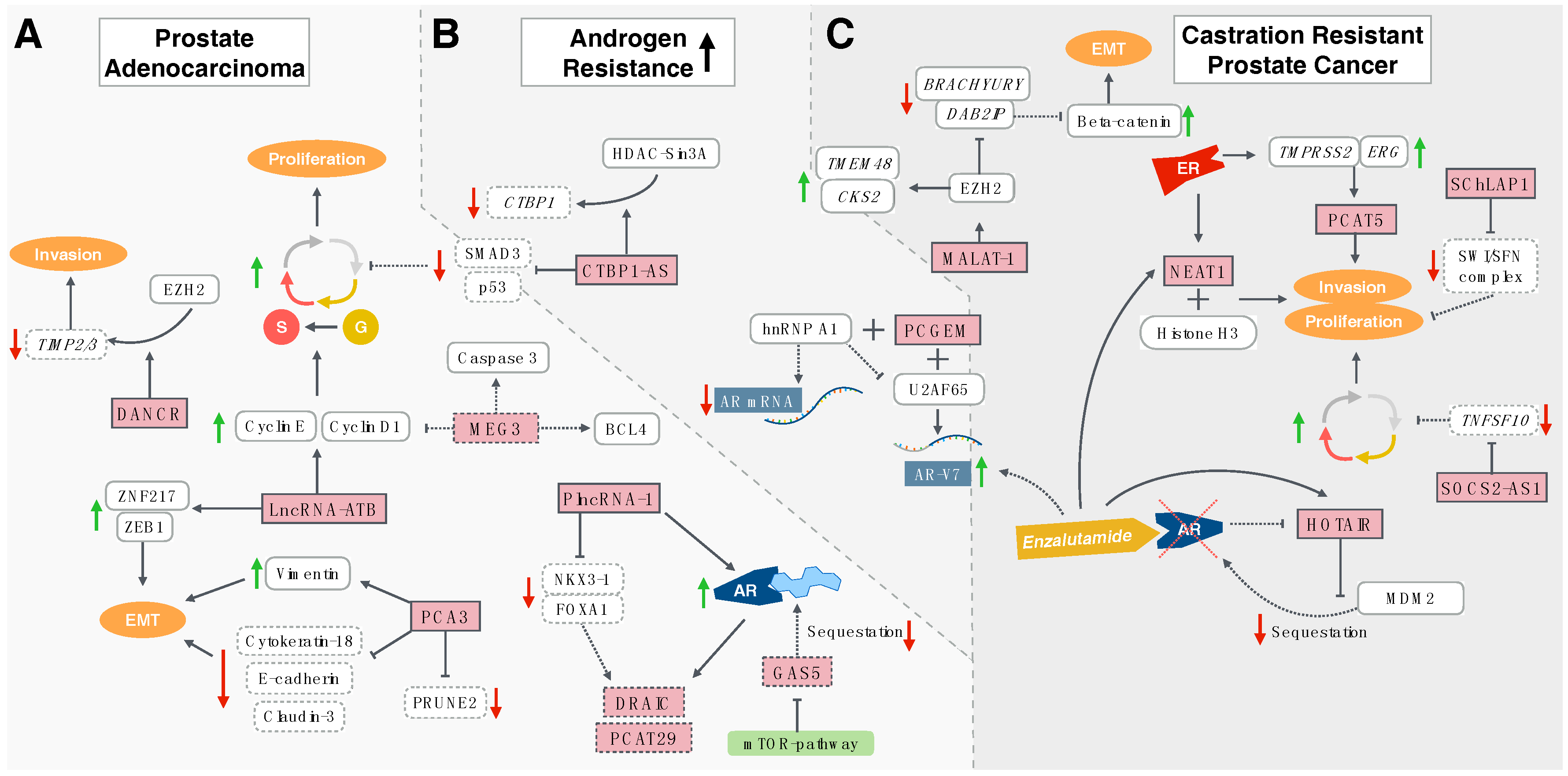
3. Long Non-Coding RNAs and Prostate Cancer
3.1. Hormone-Sensitive Prostate Cancer
3.1.1. Long Non-Coding RNA Activated by Transforming Growth Factor-β
3.1.2. Prostate Cancer Antigen 3
3.1.3. Maternally Expressed Gene 3
3.1.4. Differentiation Antagonising Non-Protein Coding RNA
3.1.5. Prostate Cancer-Associated Transcript 29 and Downregulated RNA in Cancer
3.1.6. PlncRNA-1
3.1.7. Growth Arrest-Specific 5
3.2. Promoters of Castration Resistance
3.2.1. Prostate Cancer Gene Expression Marker 1
3.2.2. C-Terminal Binding Protein 1-Antisense
3.3. Castration-Resistant Prostate Cancer
3.3.1. HOX Transcript Antisense RNA
3.3.2. Metastasis-Associated Lung Adenocarcinoma Transcript-1
3.3.3. Nuclear-Enriched Abundant Transcript 1
3.3.4. Prostate Cancer-Associated Transcript 5
3.3.5. Second Chromosome Locus Associated with Prostate-1
3.3.6. Suppressor of Cytokine Signalling 2-Antisense Transcript 1
4. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| ADT | Androgen deprivation therapy |
| AR | Androgen receptor |
| BCL4 | B-cell lymphoma like-2 like protein 4 |
| BPH | Benign prostatic hyperplasia |
| CKS2 | Cyclin-dependent kinases regulatory subunit 2 |
| CTBP1 | C-terminal binding protein 1-antisense |
| CTBP1-AS | C-terminal binding protein 1-antisense |
| DAB2IP | Disabled homolog 2-interacting protein |
| DANCR | Differentiation antagonising non-protein coding RNA |
| DRAIC | Downregulated RNA in cancer |
| EMT | Epithelial- to -mesenchymal transition |
| ERG | ETS-(E-twenty-six) related gene |
| ERα | Oestrogen receptor αalpha |
| EZH2 | Enhancer of zeste homolog |
| FOXA1 | Forkhead box protein A1 |
| GAS5 | Growth arrest-specific 5 |
| HDAC-Sin3A | Histone decarboxylase paired amphipathic helix protein Sin3a |
| hnRNP A1 | Heterogeneous nuclear ribonucleoprotein A1 |
| HOTAIR | HOX transcript antisense RNA |
| lncRNA | Long non-coding RNA |
| MALAT-1 | Metastasis-associated lung adenocarcinoma transcript-1 |
| mCRPC | Metastatic castration-resistant prostate cancer |
| MDM2 | Mouse double minute 2 homolog. |
| MEG3 | LncRNA Maternally expressed gene 3 |
| NEAT-1 | Nuclear-enriched abundant transcript 1 |
| NKX3-1 | Homeobox protein Nkx-3.1 |
| PCA | Prostate adenocarcinoma |
| PCA3 | Prostate cancer antigen 3 |
| PCAT29 | Prostate cancer-associated transcript 29 |
| PCAT5 | Prostate cancer-associated transcript 5 |
| PCCGEM | Prostate cancer gene expression marker 1 |
| PRUNE2 | Prune homolog 2 |
| PSA | Prostate Prostate-specific antigen |
| SChLAP1 | Second chromosome locus associated with prostate-1 |
| SMAD3 | Mothers against decapentaplegic homolog 3 |
| SOCS2-AS1 | Cytokine signalling 2-antisense transcript 1 |
| SWI/SFN | SWItch Sucrose Non-Fermentable complex |
| TIMP 2/3 | Tissue inhibitor of metalloproteinase |
| TMEM48 | Transmembrane Protein 48 |
| TMPRSS2 | Transmembrane Protease, Serine 2 |
| TNSF10 | Tumour necrosis factor superfamily member 10 |
| U2AF65 | U2 Small Nuclear RNA Auxillary Factor 2 |
| ZEB1 | Zinc-finger E-box binding homeobox 1 |
| ZNF217 | Zinc finger protein 217 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Goldgar, D.E.; Easton, D.F.; Cannon-Albright, L.A.; Skolnick, M.H. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J. Natl. Cancer Inst. 1994, 86, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.K.; Lavrovsky, Y.; Song, C.S.; Chen, S.; Jung, M.H.; Velu, N.K.; Bi, B.Y.; Chatterjee, B. Regulation of androgen action. Vitam. Horm. 1999, 55, 309–352. [Google Scholar] [PubMed]
- Ruizeveld de Winter, J.A.; Janssen, P.J.; Sleddens, H.M.; Verleun-Mooijman, M.C.; Trapman, J.; Brinkmann, A.O.; Santerse, A.B.; Schröder, F.H.; van der Kwast, T.H. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am. J. Pathol. 1994, 144, 735–746. [Google Scholar] [PubMed]
- Koochekpour, S. Androgen receptor signaling and mutations in prostate cancer. Asian J. Androl. 2010, 12, 639–657. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.A.; Yen, A.E.; Weigel, N.L. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 2013, 140, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Taplin, M.E.; Bubley, G.J.; Ko, Y.J.; Small, E.J.; Upton, M.; Rajeshkumar, B.; Balk, S.P. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 1999, 59, 2511–2515. [Google Scholar] [PubMed]
- Yang, X.; Guo, Z.; Sun, F.; Li, W.; Alfano, A.; Shimelis, H.; Chen, M.; Brodie, A.M.; Chen, H.; Xiao, Z.; et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J. Biol. Chem. 2011, 286, 36152–36160. [Google Scholar] [CrossRef] [PubMed]
- Attard, G.; Parker, C.; Eeles, R.A.; Schröder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Bill-Axelson, A.; Holmberg, L.; Garmo, H.; Rider, J.R.; Taari, K.; Busch, C.; Nordling, S.; Häggman, M.; Andersson, S.O.; Spångberg, A.; et al. Radical prostatectomy or watchful waiting in early prostate cancer. N. Engl. J. Med. 2014, 370, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Widmark, A.; Klepp, O.; Solberg, A.; Damber, J.E.; Angelsen, A.; Fransson, P.; Lund, J.A.; Tasdemir, I.; Hoyer, M.; Wiklund, F.; et al. Endocrine treatment, with or without radiotherapy, in locally advanced prostate cancer (SPCG-7/SFUO-3): An open randomised phase III trial. Lancet 2009, 373, 301–308. [Google Scholar] [CrossRef]
- Chen, C.D.; Welsbie, D.S.; Tran, C.; Baek, S.H.; Chen, R.; Vessella, R.; Rosenfeld, M.G.; Sawyers, C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004, 10, 33–39. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.; Ouk, S.; Clegg, N.J.; Chen, Y.; Watson, P.A.; Arora, V.; Wongvipat, J.; Smith-Jones, P.M.; Yoo, D.; Kwon, A.; et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009, 324, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Beer, T.M.; Armstrong, A.J.; Rathkopf, D.E.; Loriot, Y.; Sternberg, C.N.; Higano, C.S.; Iversen, P.; Bhattacharya, S.; Carles, J.; Chowdhury, S.; et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014, 371, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.; Nilsson, S.; Heinrich, D.; Helle, S.I.; O'Sullivan, J.M.; Fosså, S.D.; Chodacki, A.; Wiechno, P.; Logue, J.; Seke, M.; et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N. Engl. J. Med. 2013, 369, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Rickman, D.S.; Park, K.; Chae, S.S.; Sboner, A.; MacDonald, T.Y.; Wang, Y.; Sheikh, K.L.; Terry, S.; Tagawa, S.T.; et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011, 1, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, M.B.; Stephenson, M.L.; Scott, J.F.; Hecht, L.I.; Zamecnik, P.C. A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958, 231, 241–257. [Google Scholar] [PubMed]
- Pichler, M.; Calin, G.A. MicroRNAs in cancer: From developmental genes in worms to their clinical application in patients. Br. J. Cancer 2015, 113, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Ravasi, T.; Suzuki, H.; Pang, K.C.; Katayama, S.; Furuno, M.; Okunishi, R.; Fukuda, S.; Ru, K.; Frith, M.C.; Gongora, M.M.; et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006, 16, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.; Uranitsch, S.; Gerger, A.; Pichler, M.; Haybaeck, J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int. J. Mol. Sci. 2014, 15, 13993–14013. [Google Scholar] [CrossRef] [PubMed]
- Seles, M.; Hutterer, G.C.; Kiesslich, T.; Pummer, K.; Berindan-Neagoe, I.; Perakis, S.; Schwarzenbacher, D.; Stotz, M.; Gerger, A.; Pichler, M. Current Insights into Long Non-Coding RNAs in Renal Cell Carcinoma. Int. J. Mol. Sci. 2016, 17, 573. [Google Scholar] [CrossRef] [PubMed]
- Cerk, S.; Schwarzenbacher, D.; Adiprasito, J.B.; Stotz, M.; Hutterer, G.C.; Gerger, A.; Ling, H.; Calin, G.A.; Pichler, M. Current Status of Long Non-Coding RNAs in Human Breast Cancer. Int. J. Mol. Sci. 2016, 17, 1485. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.A.; Bullock, M.D.; Ling, H.; Pichler, M.; Haybaeck, J. Long Non-Coding RNAs in Endometrial Carcinoma. Int. J. Mol. Sci. 2015, 16, 26463–26472. [Google Scholar] [CrossRef] [PubMed]
- Bezan, A.; Gerger, A.; Pichler, M. MicroRNAs in testicular cancer: Implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2014, 34, 2709–2713. [Google Scholar] [PubMed]
- Ling, H.; Krassnig, L.; Bullock, M.D.; Pichler, M. MicroRNAs in Testicular Cancer Diagnosis and Prognosis. Urol. Clin. N. Am. 2016, 43, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Zebisch, A.; Hatzl, S.; Pichler, M.; Wolfler, A.; Sill, H. Therapeutic Resistance in Acute Myeloid Leukemia: The Role of Non-Coding RNAs. Int. J. Mol. Sci. 2016, 17, 2080. [Google Scholar] [CrossRef] [PubMed]
- Troppan, K.; Wenzl, K.; Deutsch, A.; Ling, H.; Neumeister, P.; Pichler, M. MicroRNAs in diffuse large B-cell lymphoma: Implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2014, 34, 557–564. [Google Scholar] [PubMed]
- Li, P.; Yang, R.; Gao, W.Q. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Mol. Cancer 2014, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Ress, A.L.; Winter, E.; Stiegelbauer, V.; Karbiener, M.; Schwarzenbacher, D.; Scheideler, M.; Ivan, C.; Jahn, S.W.; Kiesslich, T.; et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br. J. Cancer 2014, 110, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Kiesslich, T.; Pichler, M.; Neureiter, D. Epigenetic control of epithelial-mesenchymal-transition in human cancer. Mol. Clin. Oncol. 2013, 1, 3–11. [Google Scholar] [PubMed]
- Sun, Y.; Wang, B.E.; Leong, K.G.; Yue, P.; Li, L.; Jhunjhunwala, S.; Chen, D.; Seo, K.; Modrusan, Z.; Gao, W.Q.; et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: Implications for androgen-deprivation therapy. Cancer Res. 2012, 72, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yi, X.M.; Tang, C.P.; Ge, J.P.; Zhang, Z.Y.; Zhou, W.Q. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol. Rep. 2016, 36, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Jennbacken, K.; Tesan, T.; Wang, W.; Gustavsson, H.; Damber, J.E.; Welen, K. N-cadherin increases after androgen deprivation and is associated with metastasis in prostate cancer. Endocr. Relat. Cancer 2010, 17, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Bussemakers, M.J.; van Bokhoven, A.; Verhaegh, G.W.; Smit, F.P.; Karthaus, H.F.; Schalken, J.A.; Debruyne, F.M.; Ru, N.; Isaacs, W.B. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999, 59, 5975–5979. [Google Scholar] [PubMed]
- Crawford, E.D.; Rove, K.O.; Trabulsi, E.J.; Qian, J.; Drewnowska, K.P.; Kaminetsky, J.C.; Huisman, T.K.; Bilowus, M.L.; Freedman, S.J.; Glover, W.L., Jr. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: A prospective study of 1,962 cases. J. Urol. 2012, 188, 1726–1731. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.; Auprich, M.; Ahyai, S.A.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.; Huland, H.; Fisch, M.; et al. Initial prostate biopsy: Development and internal validation of a biopsy-specific nomogram based on the prostate cancer antigen 3 assay. Eur. Urol. 2013, 63, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Auprich, M.; Haese, A.; Walz, J.; Pummer, K.; de la Taille, A.; Graefen, M.; de Reijke, T.; Fisch, M.; Kil, P.; Gontero, P.; et al. External validation of urinary PCA3-based nomograms to individually predict prostate biopsy outcome. Eur. Urol. 2010, 58, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Auprich, M.; Chun, F.K.; Ward, J.F.; Pummer, K.; Babaian, R.; Augustin, H.; Luger, F.; Gutschi, S.; Budäus, L.; Fisch, M.; et al. Critical assessment of preoperative urinary prostate cancer antigen 3 on the accuracy of prostate cancer staging. Eur. Urol. 2011, 59, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Ploussard, G.; Durand, X.; Xylinas, E.; Moutereau, S.; Radulescu, C.; Forgue, A.; Nicolaiew, N.; Terry, S.; Allory, Y.; Loric, S.; et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. Eur. Urol. 2011, 59, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Lemos, A.E.; Ferreira, L.B.; Batoreu, N.M.; de Freitas, P.P.; Bonamino, M.H.; Gimba, E.R. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumour Biol. 2016, 37, 11339–11348. [Google Scholar] [CrossRef] [PubMed]
- Salameh, A.; Lee, A.K.; Cardó-Vila, M.; Nunes, D.N.; Efstathiou, E.; Staquicini, F.I.; Dobroff, A.S.; Marchiò, S.; Navone, N.M.; Hosoya, H.; et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. USA 2015, 112, 8403–8408. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Wang, M.; Wu, X.; Tao, D.; Xiao, X.; Wang, L.; Min, F.; Zeng, F.; Jiang, G. Long Non-Coding RNA MEG3 Inhibits Cell Proliferation and Induces Apoptosis in Prostate Cancer. Cell. Physiol. Biochem. 2015, 37, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef] [PubMed]
- Mignard, V.; Lalier, L.; Paris, F.; Vallette, F.M. Bioactive lipids and the control of Bax pro-apoptotic activity. Cell Death Dis. 2014, 5, e1266. [Google Scholar] [CrossRef] [PubMed]
- Heer, R.; Robson, C.N.; Shenton, B.K.; Leung, H.Y. The role of androgen in determining differentiation and regulation of androgen receptor expression in the human prostatic epithelium transient amplifying population. J. Cell. Physiol. 2007, 212, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.L.; Jeng, L.B.; Lai, H.C.; Liao, P.Y.; Chang, C. Androgen receptor enhances cell adhesion and decreases cell migration via modulating beta1-integrin-AKT signaling in hepatocellular carcinoma cells. Cancer Lett. 2014, 351, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.H.; Izumi, K.; Lee, S.O.; Lin, W.J.; Yeh, S.; Chang, C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis. 2013, 4, e764. [Google Scholar] [CrossRef] [PubMed]
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Li, F.; Tang, X.S.; Xu, S.; Gao, Y.; Shi, Q.; Guo, W.; Wang, X.; He, D.; Guo, P. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget 2016, 7, 37868–37881. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, K.; Reon, B.J.; Anaya, J.; Dutta, A. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol. Cancer Res. 2015, 13, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Patel, L.; Prensner, J.R.; Shi, Y.; Iyer, M.K.; Subramaniyan, S.; Carley, A.; Niknafs, Y.S.; Sahu, A.; Han, S.; et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol. Cancer Res. 2014, 12, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Ren, S.; Lu, J.; Wang, F.; Xu, W.; Sun, Y.; Wei, M.; Chen, J.; Gao, X.; Xu, C.; et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol. Oncol. 2013, 31, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.; Gelmann, E.P. NKX3.1 activates cellular response to DNA damage. Cancer Res. 2010, 70, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- Nam, R.K.; Zhang, W.W.; Loblaw, D.A.; Klotz, L.H.; Trachtenberg, J.; Jewett, M.A.; Stanimirovic, A.; Davies, T.O.; Toi, A.; Venkateswaran, V.; et al. A genome-wide association screen identifies regions on chromosomes 1q25 and 7p21 as risk loci for sporadic prostate cancer. Prostate Cancer Prostatic Dis. 2008, 11, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res. Treat. 2014, 145, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Mourtada-Maarabouni, M.; Williams, G.T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta 2013, 1832, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Yacqub-Usman, K.; Pickard, M.R.; Williams, G.T. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate 2015, 75, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Lu, C.; Mostaghel, E.A.; Yegnasubramanian, S.; Gurel, M.; Tannahill, C.; Edwards, J.; Isaacs, W.B.; Nelson, P.S.; Bluemn, E.; et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012, 72, 3457–3462. [Google Scholar] [CrossRef] [PubMed]
- Petrovics, G.; Zhang, W.; Makarem, M.; Street, J.P.; Connelly, R.; Sun, L.; Sesterhenn, I.A.; Srikantan, V.; Moul, J.W.; Srivastava, S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene 2004, 23, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, N.; Huang, J.; Ho, T.T.; Zhu, Z.; Qiu, Z.; Zhou, X.; Bai, C.; Wu, F.; Xu, M.; et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 2016, 7, 15481–15491. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Horie-Inoue, K.; Katayama, S.; Suzuki, T.; Tsutsumi, S.; Ikeda, K.; Urano, T.; Fujimura, T.; Takagi, K.; Takahashi, S.; et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013, 32, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhao, J.C.; Kim, J.; Fong, K.W.; Yang, Y.A.; Chakravarti, D.; Mo, Y.Y.; Yu, J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015, 13, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X.; et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013, 190, 2278–2287. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045–41055. [Google Scholar] [PubMed]
- Saramaki, O.R.; Tammela, T.L.; Martikainen, P.M.; Vessella, R.L.; Visakorpi, T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer 2006, 45, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Gore, C.; Liu, J.; Pong, R.C.; Mason, R.; Hao, G.; Long, M.; Kabbani, W.; Yu, L.; Zhang, H.; et al. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc. Natl. Acad. Sci. USA 2010, 107, 2485–2490. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ren, S.; Chen, R.; Lu, J.; Shi, X.; Zhu, Y.; Zhang, W.; Jing, T.; Zhang, C.; Shen, J.; et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget 2014, 5, 11091–11102. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef] [PubMed]
- Setlur, S.R.; Mertz, K.D.; Hoshida, Y.; Demichelis, F.; Lupien, M.; Perner, S.; Sboner, A.; Pawitan, Y.; Andrén, O.; Johnson, L.A.; et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J. Natl. Cancer Inst. 2008, 100, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, J.; Mani, R.S.; Cao, Q.; Brenner, C.J.; Cao, X.; Wang, X.; Wu, L.; Li, J.; Hu, M.; et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010, 17, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Ylipaa, A.; Kivinummi, K.; Kohvakka, A.; Annala, M.; Latonen, L.; Scaravilli, M.; Kartasalo, K.; Leppänen, S.P.; Karakurt, S.; Seppälä, J.; et al. Transcriptome Sequencing Reveals PCAT5 as a Novel ERG-Regulated Long Noncoding RNA in Prostate Cancer. Cancer Res. 2015, 75, 4026–4031. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Reisman, D.; Glaros, S.; Thompson, E.A. The SWI/SNF complex and cancer. Oncogene 2009, 28, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R.; Shi, Y.; Udager, A.M.; Prensner, J.R.; Sahu, A.; Iyer, M.K.; Siddiqui, J.; Cao, X.; Wei, J.; Jiang, H.; et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia 2014, 16, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Mehra, R.; Udager, A.M.; Ahearn, T.U.; Cao, X.; Feng, F.Y.; Loda, M.; Petimar, J.S.; Kantoff, P.; Mucci, L.A.; Chinnaiyan, A.M. Overexpression of the Long Non-coding RNA SChLAP1 Independently Predicts Lethal Prostate Cancer. Eur. Urol. 2016, 70, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Misawa, A.; Takayama, K.; Urano, T.; Inoue, S. Androgen-induced Long Noncoding RNA (lncRNA) SOCS2-AS1 Promotes Cell Growth and Inhibits Apoptosis in Prostate Cancer Cells. J. Biol. Chem. 2016, 291, 17861–17880. [Google Scholar] [CrossRef] [PubMed]
- Starr, R.; Willson, T.A.; Viney, E.M.; Murray, L.J.; Rayner, J.R.; Jenkins, B.J.; Gonda, T.J.; Alexander, W.S.; Metcalf, D.; Nicola, N.A.; et al. A family of cytokine-inducible inhibitors of signalling. Nature 1997, 387, 917–921. [Google Scholar] [PubMed]
- Farooqi, A.A.; Bhatti, S.; Ismail, M. TRAIL and vitamins: Opting for keys to castle of cancer proteome instead of open sesame. Cancer Cell Int. 2012, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, Y.; He, A.; Li, J.; Chen, M.; Zhan, Y.; Lin, J.; Zhuang, C.; Liu, L.; Zhao, G.; et al. Theophylline controllable RNAi-based genetic switches regulate expression of lncRNA TINCR and malignant phenotypes in bladder cancer cells. Sci. Rep. 2016, 6, 30798. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Roychowdhury-Saha, M.; Black, C.; Watt, A.T.; Marcusson, E.G.; Freier, S.M.; Edgington, T.S. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011, 585, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Chandra Gupta, S.; Nandan Tripathi, Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int. J. Cancer 2016. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Spitale, R.C.; Chang, H.Y. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011, 71, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Gofrit, O.N.; Benjamin, S.; Halachmi, S.; Leibovitch, I.; Dotan, Z.; Lamm, D.L.; Ehrlich, N.; Yutkin, V.; Ben-Am, M.; Hochberg, A. DNA based therapy with diphtheria toxin-A BC-819: A phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urol. 2014, 191, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
| LncRNA | Expression Pattern | Relevance |
|---|---|---|
| Diagnostic Biomarker | ||
| PCA3 | Overexpression | Predicts prostate cancer in combination with PSA |
| MALAT-1 | Overexpression | More sensitive than PSA for initial diagnosis |
| Prognostic/Predictive Biomarker | ||
| HOTAIR | Overexpression | Associated with resistance to enzalutamide |
| MALAT-1 | Overexpression | Correlates with ADT-resistance |
| SChLAP1 | Overexpression | Predicts lethal mCRPC |
| LncRNA-ATB | Overexpression | Correlates with unfavourable clinical features |
| PCAT29 | Underexpression | Associated with early biochemical recurrence |
| DRAIC | Underexpression | |
| NEAT1 | Overexpression | Predicts early biochemical recurrence |
| Therapeutic Target | ||
| LncRNA-ATB | Overexpression | Blockage could slow down tumour progression |
| PCA3 | Overexpression | Inhibition may retard progression of disease |
| MEG3 | Underexpression | Induction of expression could decelerate tumour progression |
| DANCR | Overexpression | Metastatic spread prevented by blockage upon enzalutamide-treatment |
| PlncRNA-1 | Overexpression | Blockage could help to slow down cancer progression |
| GAS5 | Underexpression | Indirectly upregulated by mTOR (mechanistic target of rapamycin)-inhibitors |
| PCGEM1 | Overexpression | Efficacy of ADT enhanced by blockage |
| CTBP1-AS | Overexpression | Blockage could reduce proliferation rate |
| HOTAIR | Overexpression | Efficacy of enzalutamide enhanced upon blockage |
| PCAT5 | Overexpression | Inhibition could be effective, particularly in ERG-positive prostate cancers |
| SOCS2-AS1 | Overexpression | Blockage may reverse anti-apoptotic abilities |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolle, M.A.; Bauernhofer, T.; Pummer, K.; Calin, G.A.; Pichler, M. Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 473. https://doi.org/10.3390/ijms18020473
Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. International Journal of Molecular Sciences. 2017; 18(2):473. https://doi.org/10.3390/ijms18020473
Chicago/Turabian StyleSmolle, Maria A., Thomas Bauernhofer, Karl Pummer, George A. Calin, and Martin Pichler. 2017. "Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer" International Journal of Molecular Sciences 18, no. 2: 473. https://doi.org/10.3390/ijms18020473
APA StyleSmolle, M. A., Bauernhofer, T., Pummer, K., Calin, G. A., & Pichler, M. (2017). Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. International Journal of Molecular Sciences, 18(2), 473. https://doi.org/10.3390/ijms18020473







