Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology
Abstract
:1. Introduction
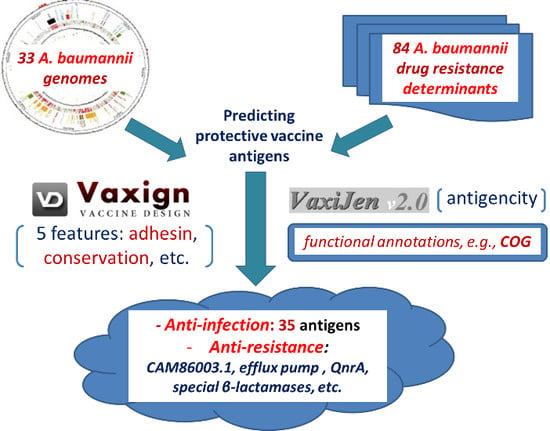
2. Results
2.1. Collection of A. baumannii Genome Sequences and Resistance Genes
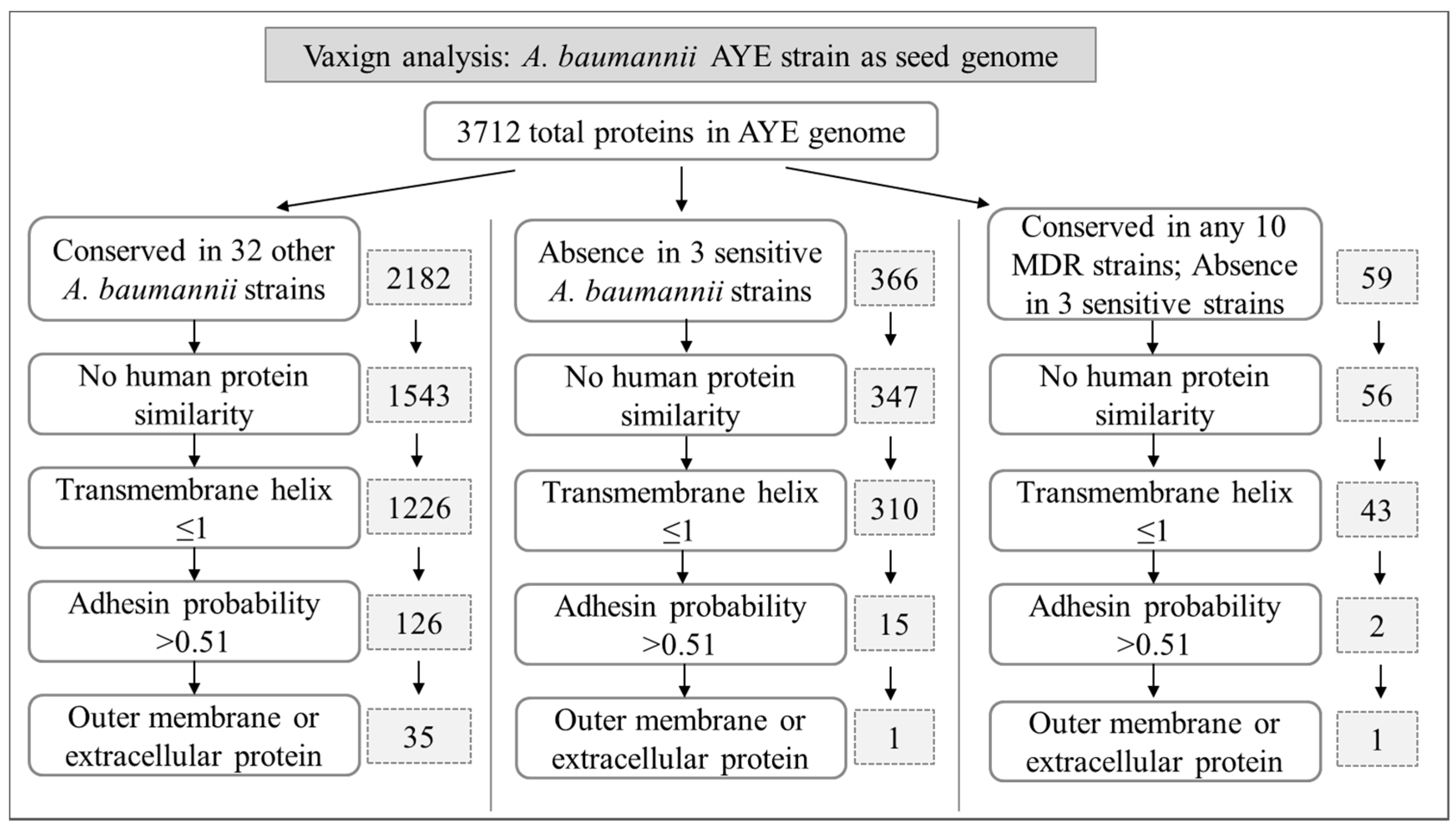
2.2. Predicted A. baumannii Vaccine Targets Based on RV Analysis of 33 Genomes
2.2.1. Predicted A. baumannii Vaccine Targets Conserved in All 33 Genomes
2.2.2. Predicted A. baumannii Vaccine Targets Absent from 3 Antibiotic-Sensitive Strains
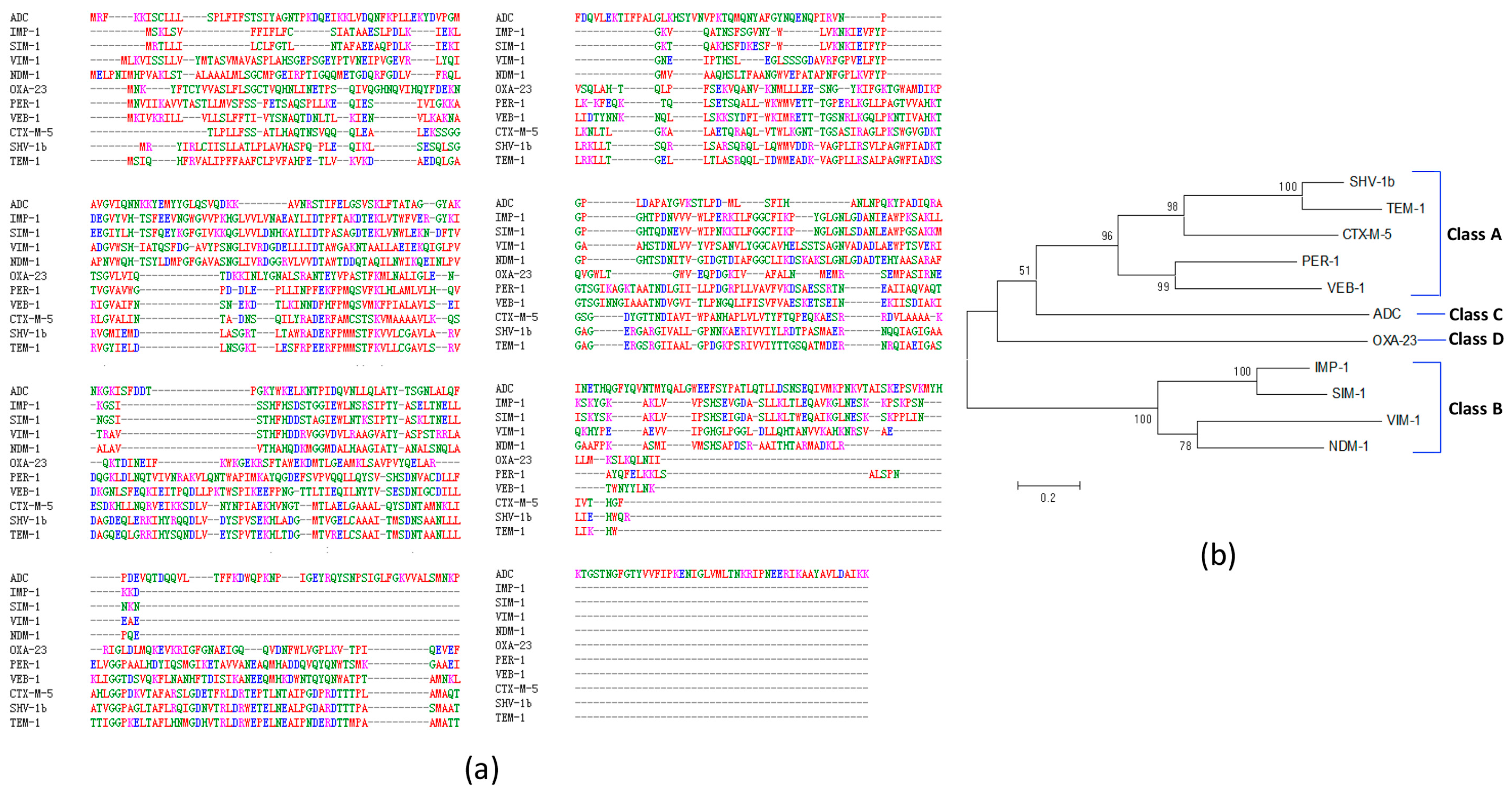
2.3. Predicted A. baumannii Vaccine Targets Based on RV Analysis of 84 Antibiotic Resistance Determinants
3. Discussion
4. Materials and Methods
4.1. Collection of A. baumannii Genome Sequences and Antibiotic Resistance Determinants
4.2. Vaxign Calculation of Sequence-Derived Features
4.3. Vaxign Results Analysis
4.4. Antigenicity Prediction
4.5. BLAST Analysis and COG Functional Annotation of Predicted Vaccine Candidates
4.6. PFam Conserved Domain Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Doyle, J.S.; Buising, K.L.; Thursky, K.A.; Worth, L.J.; Richards, M.J. Epidemiology of infections acquired in intensive care units. Semin. Respir. Crit. Care Med. 2011, 32, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Karveli, E.A.; Kelesidis, I.; Kelesidis, T. Community-acquired Acinetobacter infections. Eur. J. Clin. Microbiol. Infect. Dis. 2007, 26, 857–868. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Actis, L.; Pachon, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Pachon, J. Active and passive immunization against Acinetobacter baumannii using an inactivated whole cell vaccine. Vaccine 2010, 29, 1–5. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Dominguez-Herrera, J.; Smani, Y.; Lopez-Rojas, R.; Docobo-Perez, F.; Pachon, J. Vaccination with outer membrane complexes elicits rapid protective immunity to multidrug-resistant Acinetobacter baumannii. Infect. Immun. 2011, 79, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yao, Y.; Long, Q.; Yang, X.; Sun, W.; Liu, C.; Jin, X.; Li, Y.; Chu, X.; Chen, B.; et al. Immunization against multidrug-resistant Acinetobacter baumannii effectively protects mice in both pneumonia and sepsis models. PLoS ONE 2014, 9, e100727. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Lin, L.; Ibrahim, A.S.; Baquir, B.; Pantapalangkoor, P.; Bonomo, R.A.; Doi, Y.; Adams, M.D.; Russo, T.A.; Spellberg, B. Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS ONE 2012, 7, e29446. [Google Scholar] [CrossRef] [PubMed]
- Bentancor, L.V.; Routray, A.; Bozkurt-Guzel, C.; Camacho-Peiro, A.; Pier, G.B.; Maira-Litran, T. Evaluation of the trimeric autotransporter ata as a vaccine candidate against Acinetobacter baumannii infections. Infect. Immun. 2012, 80, 3381–3388. [Google Scholar] [CrossRef] [PubMed]
- Fattahian, Y.; Rasooli, I.; Mousavi Gargari, S.L.; Rahbar, M.R.; Darvish Alipour Astaneh, S.; Amani, J. Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (BAP). Microb. Pathog. 2011, 51, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Beanan, J.M.; Olson, R.; MacDonald, U.; Cox, A.D.; St Michael, F.; Vinogradov, E.V.; Spellberg, B.; Luke-Marshall, N.R.; Campagnari, A.A. The k1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect. Immun. 2013, 81, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Bentancor, L.V.; O’Malley, J.M.; Bozkurt-Guzel, C.; Pier, G.B.; Maira-Litran, T. Poly-n-acetyl-β-(1–6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect. Immun. 2012, 80, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, W. Current advances and challenges in the development of Acinetobacter vaccines. Hum. Vaccines Immunother. 2015, 11, 2495–2500. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Bagge, N.; Hoiby, N. Antibodies against β-lactamase can improve ceftazidime treatment of lung infection with β-lactam-resistant pseudomonas aeruginosa in a rat model of chronic lung infection. Acta Pathol. Microbiol. Immunol. Scand. 2002, 110, 881–891. [Google Scholar] [CrossRef]
- Ciofu, O. Pseudomonas aeruginosa chromosomal β-lactamase in patients with cystic fibrosis and chronic lung infection. Mechanism of antibiotic resistance and target of the humoral immune response. APMIS Suppl. 2003, 116, 1–47. [Google Scholar]
- Rappuoli, R. Reverse vaccinology. Curr. Opin. Microbiol. 2000, 3, 445–450. [Google Scholar] [CrossRef]
- Pizza, M.; Scarlato, V.; Masignani, V.; Giuliani, M.M.; Arico, B.; Comanducci, M.; Jennings, G.T.; Baldi, L.; Bartolini, E.; Capecchi, B.; et al. Identification of vaccine candidates against serogroup b meningococcus by whole-genome sequencing. Science 2000, 287, 1816–1820. [Google Scholar] [CrossRef] [PubMed]
- Ariel, N.; Zvi, A.; Grosfeld, H.; Gat, O.; Inbar, Y.; Velan, B.; Cohen, S.; Shafferman, A. Search for potential vaccine candidate open reading frames in the bacillus anthracis virulence plasmid pxo1: In silico and in vitro screening. Infect. Immun. 2002, 70, 6817–6827. [Google Scholar] [CrossRef] [PubMed]
- Wizemann, T.M.; Heinrichs, J.H.; Adamou, J.E.; Erwin, A.L.; Kunsch, C.; Choi, G.H.; Barash, S.C.; Rosen, C.A.; Masure, H.R.; Tuomanen, E.; et al. Use of a whole genome approach to identify vaccine molecules affording protection against streptococcus pneumoniae infection. Infect. Immun. 2001, 69, 1593–1598. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.C. Transcriptomics and proteomics: Tools for the identification of novel drug targets and vaccine candidates for tuberculosis. IUBMB Life 2002, 53, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Manque, P.A.; Tenjo, F.; Woehlbier, U.; Lara, A.M.; Serrano, M.G.; Xu, P.; Alves, J.M.; Smeltz, R.B.; Conrad, D.H.; Buck, G.A. Identification and immunological characterization of three potential vaccinogens against cryptosporidium species. Clin. Vaccine Immunol. 2011, 18, 1796–1802. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Garg, N.; Shukla, G.; Capalash, N.; Sharma, P. Immunoprotective efficacy of Acinetobacter baumannii outer membrane protein, FilF, predicted in silico as a potential vaccine candidate. Front. Microbiol. 2016, 7, 158. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.H.; Sung, W.C.; Lien, S.P.; Chen, Y.Z.; Lo, A.F.; Huang, J.H.; Kuo, S.C.; Chong, P. Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum. Vaccines Immunother. 2015, 11, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Naz, A.; Obaid, A.; Paracha, R.Z.; Naz, K.; Awan, F.M.; Muhmmad, S.A.; Janjua, H.A.; Ahmad, J.; Ali, A. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genom. 2016, 17, 732. [Google Scholar] [CrossRef] [PubMed]
- Moriel, D.G.; Beatson, S.A.; Wurpel, D.J.; Lipman, J.; Nimmo, G.R.; Paterson, D.L.; Schembri, M.A. Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS ONE 2013, 8, e77631. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiang, Z.; Mobley, H.L. Vaxign: The first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J. Biomed. Biotechnol. 2010, 2010, 297505. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; He, Y. Genome-wide prediction of vaccine targets for human herpes simplex viruses using vaxign reverse vaccinology. BMC Bioinform. 2013, 14, S2. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiang, Z. Bioinformatics analysis of brucella vaccines and vaccine targets using violin. Immunome Res. 2010, 6, S5. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.; Pei, J.; Mwangi, W.; Adams, L.G.; Rice-Ficht, A.; Ficht, T.A. Immunogenic and invasive properties of brucella melitensis 16 m outer membrane protein vaccine candidates identified via a reverse vaccinology approach. PLoS ONE 2013, 8, e59751. [Google Scholar] [CrossRef] [PubMed]
- Caro-Gomez, E.; Gazi, M.; Goez, Y.; Valbuena, G. Discovery of novel cross-protective rickettsia prowazekii t-cell antigens using a combined reverse vaccinology and in vivo screening approach. Vaccine 2014, 32, 4968–4976. [Google Scholar] [CrossRef] [PubMed]
- Pereira, U.P.; Soares, S.C.; Blom, J.; Leal, C.A.; Ramos, R.T.; Guimaraes, L.C.; Oliveira, L.C.; Almeida, S.S.; Hassan, S.S.; Santos, A.R.; et al. In silico prediction of conserved vaccine targets in streptococcus agalactiae strains isolated from fish, cattle, and human samples. Genet. Mol. Res. 2013, 12, 2902–2912. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.C.; Trost, E.; Ramos, R.T.; Carneiro, A.R.; Santos, A.R.; Pinto, A.C.; Barbosa, E.; Aburjaile, F.; Ali, A.; Diniz, C.A.; et al. Genome sequence of corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. J. Biotechnol. 2013, 167, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Meunier, M.; Guyard-Nicodeme, M.; Hirchaud, E.; Parra, A.; Chemaly, M.; Dory, D. Identification of novel vaccine candidates against campylobacter through reverse vaccinology. J. Immunol. Res. 2016, 2016, 5715790. [Google Scholar] [CrossRef] [PubMed]
- Valentine, S.C.; Contreras, D.; Tan, S.; Real, L.J.; Chu, S.; Xu, H.H. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in los angeles county, california. J. Clin. Microbiol. 2008, 46, 2499–2507. [Google Scholar] [CrossRef] [PubMed]
- Hood, M.I.; Jacobs, A.C.; Sayood, K.; Dunman, P.M.; Skaar, E.P. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob. Agents Chemother. 2010, 54, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Vallenet, D.; Barbe, V.; Audic, S.; Ogata, H.; Poirel, L.; Richet, H.; Robert, C.; Mangenot, S.; Abergel, C.; et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006, 2, e7. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative analysis of acinetobacters: Three genomes for three lifestyles. PLoS ONE 2008, 3, e1805. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Rappuoli, R.; De Groot, A.S.; Chen, R.T. Emerging vaccine informatics. J. Biomed. Biotechnol. 2010, 2010, 218590. [Google Scholar] [CrossRef] [PubMed]
- Vivona, S.; Bernante, F.; Filippini, F. Nerve: New enhanced reverse vaccinology environment. BMC Biotechnol. 2006, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Sayers, S.; Xiang, Z.; He, Y. Protegen: A web-based protective antigen database and analysis system. Nucleic Acids Res. 2011, 39, D1073–D1078. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiang, Z. Bioinformatics analysis of bacterial protective antigens in manually curated protegen database. Procedia Vaccinol. 2012, 6, 3–9. [Google Scholar] [CrossRef]
- Gardy, J.L.; Spencer, C.; Wang, K.; Ester, M.; Tusnady, G.E.; Simon, I.; Hua, S.; deFays, K.; Lambert, C.; Nakai, K.; et al. Psort-b: Improving protein subcellular localization prediction for gram-negative bacteria. Nucleic Acids Res. 2003, 31, 3613–3617. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, G.; Kumar, K.; Jain, P.; Ramachandran, S. Spaan: A software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 2005, 21, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. Vaxijen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. Cdd: Specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009, 37, D205–D210. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilkhani, H.; Rasooli, I.; Nazarian, S.; Sefid, F. In vivo validation of the immunogenicity of recombinant baumannii acinetobactin utilization a protein (rBauA). Microb. Pathog. 2016, 98, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.T.; Kuo, S.C.; Chiang, M.H.; Lee, Y.T.; Sung, W.C.; Chen, Y.H.; Chen, T.L.; Fung, C.P. Acinetobacter baumannii extracellular oxa-58 is primarily and selectively released via outer membrane vesicles after sec-dependent periplasmic translocation. Antimicrob. Agents Chemother. 2015, 59, 7346–7354. [Google Scholar] [CrossRef] [PubMed]
- Schaar, V.; Paulsson, M.; Morgelin, M.; Riesbeck, K. Outer membrane vesicles shield moraxella catarrhalis β-lactamase from neutralization by serum igg. J. Antimicrob. Chemother. 2013, 68, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Schauer, K.; Rodionov, D.A.; de Reuse, H. New substrates for tonb-dependent transport: Do we only see the tip of the iceberg? Trends Biochem. Sci. 2008, 33, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; Raffatellu, M. Metals in infectious diseases and nutritional immunity. Metallomics 2015, 7, 926–928. [Google Scholar] [CrossRef] [PubMed]
- Zimbler, D.L.; Penwell, W.F.; Gaddy, J.A.; Menke, S.M.; Tomaras, A.P.; Connerly, P.L.; Actis, L.A. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 2009, 22, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Sefid, F.; Rasooli, I.; Jahangiri, A. In silico determination and validation of baumannii acinetobactin utilization a structure and ligand binding site. BioMed Res. Int. 2013, 2013, 172784. [Google Scholar] [CrossRef] [PubMed]
- Mike, L.A.; Smith, S.N.; Sumner, C.A.; Eaton, K.A.; Mobley, H.L. Siderophore vaccine conjugates protect against uropathogenic escherichia coli urinary tract infection. Proc. Natl. Acad. Sci. USA 2016, 113, 13468–13473. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Marti, S.; Sanchez-Cespedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef] [PubMed]
- Fajardo Bonin, R.; Chapeaurouge, A.; Perales, J.; da Silva, J.G., Jr.; do Nascimento, H.J.; D’Alincourt Carvalho Assef, A.P.; Moreno Senna, J.P. Identification of immunogenic proteins of the bacterium Acinetobacter baumannii using a proteomic approach. Proteomics. Clin. Appl. 2014, 8, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Lee, J.S.; Lee, Y.C.; Park, T.I.; Lee, J.C. Acinetobacter baumannii invades epithelial cells and outer membrane protein a mediates interactions with epithelial cells. BMC Microbiol. 2008, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Duan, R.; Li, X.; Li, K.; Liang, J.; Liu, C.; Qiu, H.; Xiao, Y.; Jing, H.; Wang, X. Homology analysis and cross-immunogenicity of ompa from pathogenic Yersinia enterocolitica, Yersinia pseudotuberculosis and Yersinia pestis. Mol. Immunol. 2015, 68, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Piddock, L.J. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Coyne, S.; Courvalin, P.; Perichon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Chabane, Y.N.; Goussard, S.; Snesrud, E.; Courvalin, P.; de, E.; Grillot-Courvalin, C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 2015, 6, e00309-15. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.M. Relationships between the regulatory systems of quorum sensing and multidrug resistance. Front. Microbiol. 2016, 7, 958. [Google Scholar] [CrossRef] [PubMed]
- Reading, N.C.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Mata, A.; Fernandez-Dominguez, I.J.; Nunez-Reza, K.J.; Xiqui-Vazquez, M.L.; Baca, B.E. Networks involving quorum sensing, cyclic-di-gmp and nitric oxide on biofilm production in bacteria. Rev. Argent. Microbiol. 2014, 46, 242–255. [Google Scholar] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martinez, J.L. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Greco, V.; Martino, P.A.; Del Chierico, F.; Putignani, L.; Urbani, A.; Nally, J.E.; Bonizzi, L.; Roncada, P. Mechanisms of antibiotic resistance to enrofloxacin in uropathogenic escherichia coli in dog. J. Proteom. 2015, 127, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Gaviraghi, A.; Deriu, F.; De Martino, L.; Iovane, G.; Amoresano, A.; Roncada, P. Comparative proteomics to evaluate multi drug resistance in escherichia coli. Mol. BioSyst. 2012, 8, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, N.; Sharma, P.; Capalash, N. Quorum sensing in acinetobacter: An emerging pathogen. Crit. Rev. Microbiol. 2010, 36, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.Y.; Yang, Y.; Tay, S.B.; Chua, K.L.; Yew, W.S. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob. Agents Chemother. 2014, 58, 1802–1805. [Google Scholar] [CrossRef] [PubMed]
- Stacy, D.M.; Welsh, M.A.; Rather, P.N.; Blackwell, H.E. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native n-acyl homoserine lactones. ACS Chem. Biol. 2012, 7, 1719–1728. [Google Scholar] [CrossRef] [PubMed]
- Joice, R.; Lipsitch, M. Targeting imperfect vaccines against drug-resistance determinants: A strategy for countering the rise of drug resistance. PLoS ONE 2013, 8, e68940. [Google Scholar] [CrossRef] [PubMed]
- Tekle, Y.I.; Nielsen, K.M.; Liu, J.; Pettigrew, M.M.; Meyers, L.A.; Galvani, A.P.; Townsend, J.P. Controlling antimicrobial resistance through targeted, vaccine-induced replacement of strains. PLoS ONE 2012, 7, e50688. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xu, F.; Lin, J. Development and evaluation of cmec subunit vaccine against campylobacter jejuni. J. Vaccines Vaccin. 2010, 1, 112. [Google Scholar] [PubMed]
- Pannek, S.; Higgins, P.G.; Steinke, P.; Jonas, D.; Akova, M.; Bohnert, J.A.; Seifert, H.; Kern, W.V. Multidrug efflux inhibition in Acinetobacter baumannii: Comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J. Antimicrob. Chemother. 2006, 57, 970–974. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Hancock, R.E. Genetic definition of the substrate selectivity of outer membrane porin protein oprd of pseudomonas aeruginosa. J. Bacteriol. 1993, 175, 7793–7800. [Google Scholar] [CrossRef] [PubMed]
- Epp, S.F.; Kohler, T.; Plesiat, P.; Michea-Hamzehpour, M.; Frey, J.; Pechere, J.C. C-terminal region of pseudomonas aeruginosa outer membrane porin oprd modulates susceptibility to meropenem. Antimicrob. Agents Chemother. 2001, 45, 1780–1787. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Chen, S.S.; Hung, K.H.; Wu, H.M.; Hsueh, P.R.; Yan, J.J.; Wu, J.J. Overproduction of active efflux pump and variations of oprd dominate in imipenem-resistant pseudomonas aeruginosa isolated from patients with bloodstream infections in taiwan. BMC Microbiol. 2016, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Catel-Ferreira, M.; Nehme, R.; Molle, V.; Aranda, J.; Bouffartigues, E.; Chevalier, S.; Bou, G.; Jouenne, T.; De, E. Deciphering the function of the outer membrane protein oprd homologue of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2012, 56, 3826–3832. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martinez, L.; Pascual, A.; Jacoby, G.A. Quinolone resistance from a transferable plasmid. Lancet 1998, 351, 797–799. [Google Scholar] [CrossRef]
- Cattoir, V.; Nordmann, P. Plasmid-mediated quinolone resistance in gram-negative bacterial species: An update. Curr. Med. Chem. 2009, 16, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Zervosen, A.; Saegerman, C.; Antoniotti, I.; Robert, B.; Ruth, N.; Collard, A.; Schynts, F.; Galleni, M.; Mercuri, P.S. Characterization of the cattle serum antibody responses against TEM β-lactamase and the nonimmunogenic Escherichia coli heat-stable enterotoxin (STai). FEMS Immunol. Med. Microbiol. 2008, 54, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.S.; Karthika, R.U.; Singh, S.P.; Shashikala, P.; Kanungo, R.; Jayachandran, S.; Prashanth, K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 2008, 26, 333–337. [Google Scholar] [PubMed]
- Lipsitch, M.; Siber, G.R. How can vaccines contribute to solving the antimicrobial resistance problem? mBio 2016, 7, e00428-16. [Google Scholar] [CrossRef] [PubMed]
- Gardy, J.L.; Laird, M.R.; Chen, F.; Rey, S.; Walsh, C.J.; Ester, M.; Brinkman, F.S. Psortb v.2.0: Expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 2005, 21, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. Orthomcl: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. Eggnog 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
| Strain | NCBI BioProject No. | MDR | Year | Country | Disease/Source | Proteins |
|---|---|---|---|---|---|---|
| ATCC17978 | 17,477 | No | 1951 | USA | Meningitis | 3803 |
| AB307-0294 | 30,993 | No | 1994 | USA | Bloodstream infection | 3363 |
| D1279779 | 61,919 | No | 2009 | Australia | Bacteremia | 3371 |
| A1 | 269,083 | Yes | 1982 | UK | - | 3626 |
| LAC-4 | 242,902 | Yes | 1997 | USA | Nosocomial outbreak | 3633 |
| AYE | 28,921 | Yes | 2001 | France | Urinary tract infection | 3725 |
| AB0057 | 21,111 | Yes | 2004 | USA | Bloodstream infection | 3669 |
| 1656-2 | 42,153 | Yes | 2004–2005 | Korea | Outbreak strain | 3733 |
| ACICU | 17827 | Yes | 2005 | Italy | Outbreak strain causing meningitis | 3613 |
| MDR-ZJ06 | 28,333 | Yes | 2006 | China | Ventilator-associated pneumonia | 3688 |
| AbH12O-A2 | 261,783 | Yes | 2006–2008 | Spain | Nosocomial outbreak | 3542 |
| AB5075-UW | 243,297 | Yes | 2008 | USA | Combatant wound infection | 3819 |
| 3207 | 311,558 | Yes | 2008 | Mexico | Bronchoalveolar lavage fluid | 3790 |
| D36 | 294,725 | Yes | 2008 | Australia | - | 3848 |
| TCDC-AB0715 | 62,279 | Yes | 2007–2009 | Taiwan | Bacteremia | 3956 |
| AC29 | 238,628 | Yes | 2011 | Malaysia | Wounds | 3555 |
| AC30 | 173,033 | Yes | 2011 | Malaysia | Wounds | 3660 |
| MDR-TJ | 52,959 | Yes | 2012 | China | - | 3872 |
| TYTH-1 | 74,551 | Yes | 2012 | Taiwan | Bacteremia | 3628 |
| KBN10P02143 | 291,316 | Yes | 2012 | Korea | Surgical patient pus | 3871 |
| BJAB07104 | 74,421 | Yes | 2013 | China | Bloodstream infection | 3754 |
| BJAB0715 | 74,423 | Yes | 2013 | China | Cerebrospinal fluid | 3756 |
| BJAB0868 | 74,425 | Yes | 2013 | China | Ascites | 3689 |
| AB031 | 256,158 | Yes | 2014 | Canada | Bloodstream infection | 3472 |
| AB030 | 256,157 | Yes | 2014 | Canada | Bloodstream infection | 4155 |
| ZW85-1 | 219,230 | Yes | 2014 | China | Diarrheal patient feces | 3544 |
| NCGM237 | 1466 | Yes | 2014 | Japan | - | 3741 |
| XH386 | 273,343 | Yes | 2016 | China | Pediatric hospital | 3942 |
| YU-R612 | 309,091 | Yes | 2016 | Korea | Sepsis | 3900 |
| IOMTU433 | 3154 | Yes | - | Nepal | - | 3868 |
| CIP70.10 | 9585 | Yes | - | - | - | 3551 |
| R2090 | 9721 | Yes | - | - | - | 3526 |
| R2091 | 12,156 | Yes | - | - | - | 3567 |
| Drug Class | Resistance Mechanism | Protein Category |
|---|---|---|
| β-lactams | β-Lactamases | |
| Ambler Class A | TEM-1,-19,-116,-128,-193,-194,-195; SHV-1b,-2,-5,-12,-18,-48,-56,-71,-96; CTX-M-2,-5,-15,-43; PER-1; VEB-1,-7; | |
| Ambler Class B | IMP-1,-4,-5,-8,-19;VIM-1,-2,-11; SIM;NDM-1; | |
| Ambler Class C | ADC; | |
| Ambler Class D | OXA-2,-10,-23,-24,-58; | |
| OMPs | CarO; | |
| Efflux pumps | RND family: adeC/adeK/oprM, AdeA/AdeI, AdeB, hemolysin D; MATE family efflux transporter: AbeM; MFS family drug transporter: Bcr/CflA | |
| Aminoglycoside | Aminoglycoside-modifying enzymes | Aac(6')-Ib, AacC2, Aac (3)-Ia, Aac(6′)-Ih, Aac(6')-Ik, Aac6-II, Aac(3)-Ia, Aad(2′)-1a, SPH, AphA1-IAB, APH(3′)-VIa; |
| Quinolones | Quinolone resistance protein | QnrA1; |
| Tetracyclines | Tetracycline-specific efflux | TETA(A), TETA(G); |
| Chloramphenicol | chloramphenicol acetyltransferase | chloramphenicol acetyltransferase; chloramphenicol resistance protein; |
| Sulfanilamide | Sul1; | |
| Tremethoprim | dihydrofolate reductase | DfrA1, DfrA10, DHFRX; |
| Polymyxin | Polymyxin resistance protein ArnT; |
| Protein Accession | Protein Name | Localization | Adhesin Probability | Length | Antigenecity Score | COG Group | Pfam Domains | Functional Description |
|---|---|---|---|---|---|---|---|---|
| TonB Dependent Receptor Proteins | ||||||||
| CAM86801.1 | putative ferric siderophore receptor | OM | 0.639 | 734 | 0.60 | P | PF00593 | TonB dependent receptor |
| CAM88090.1 | putative ferric siderophore receptor | OM | 0.612 | 743 | 0.67 | P | PF00593 | TonB dependent receptor |
| CAM86392.1 | putative siderophore receptor | OM | 0.558 | 756 | 0.59 | P | PF00593 | TonB dependent receptor |
| CAM86878.1 | putative ferric siderophore receptor | OM | 0.600 | 772 | 0.61 | P | PF00593 | TonB dependent receptor |
| CAM85131.1 | putative ferric siderophore receptor | OM | 0.595 | 736 | 0.67 | P | PF00593 | TonB dependent receptor |
| CAM86022.1 | putative ferric acinetobactin receptor (bauA) | OM | 0.627 | 767 | 0.59 | P | PF00593 | TonB dependent receptor |
| CAM86399.1 | putative OM porin, receptor for Fe(III)-coprogen, Fe(III)-ferrioxamine B and Fe(III)-rhodotrulic acid uptake (FhuE) | OM | 0.574 | 718 | 0.58 | P | PF00593 | TonB dependent receptor |
| CAM87481.1 | putative TonB-dependent siderophore receptor precursor | OM | 0.531 | 705 | 0.70 | P | PF00593 | TonB dependent receptor |
| CAM86048.1 | putative TonB-dependent receptor | OM | 0.566 | 924 | 0.59 | P | PF00593 | TonB dependent receptor |
| CAM86923.1 | putative OM TonB-dependent receptor | OM | 0.544 | 904 | 0.62 | P | PF00593 | TonB dependent receptor |
| CAM85573.1 | putative TonB-dependent OM receptor for vitamin B12/cobalamin transport (Btub) | OM | 0.644 | 638 | 0.67 | P | PF00593 | TonB dependent receptor |
| Fimbria or Pilus Related Proteins | ||||||||
| CAM87009.1 | putative Fimbria adhesin protein | EC | 0.710 | 341 | 0.68 | NU | PF00419 | Fimbrial protein |
| CAM87933.1 | putative pilus assembly protein (FilF) | OM | 0.677 | 641 | 0.73 | |||
| Porin Proteins | ||||||||
| CAM87753.1 | putative OMP | OM | 0.606 | 217 | 0.75 | M | PF00691 | OmpA family lipoprotein |
| CAM88440.1 | putative OMP | OM | 0.549 | 438 | 0.67 | M | PF03573 | OprD |
| CAM85599.1 | putative glucose-sensitive porin (OprB-like) | OM | 0.655 | 417 | 0.55 | M | PF04966 | OprB |
| CAM86576.1 | porin | OM | 0.605 | 439 | 0.61 | M | PF04966 | Carbohydrate-selective porin, OprB family |
| CAM85154.1 | conserved, putative exported protein | OM | 0.580 | 255 | 0.82 | S | PF16956 | Porin_7 superfamily |
| CAM85174.1 | conserved, putative exported protein | OM | 0.599 | 300 | 0.72 | S | PF16596 | Porin_7 Superfamily |
| CAM85116.1 | conserved, putative exported protein | OM | 0.584 | 241 | 0.80 | M | PF03502 | Nucleoside-specific channel-forming, Tsx |
| CAM87023.1 | conserved, putative exported protein | OM | 0.659 | 443 | 0.67 | S | PF 02530 | OM_channels |
| Efflux Related Proteins | ||||||||
| CAM85249.1 | cation efflux system protein (CzcC) | OM | 0.518 | 471 | 0.64 | M | PF02321 | OM efflux protein |
| CAM85825.1 | conserved, putative exported protein | OM | 0.625 | 499 | 0.51 | M | PF02321 | OM efflux protein |
| CAM88576.1 | polysaccharide export protein | OM | 0.530 | 366 | 0.46 | M | PF02563 | Polysaccharide biosynthesis/export protein |
| CAM87663.1 | conserved, putative exported protein | OM | 0.608 | 398 | 0.71 | S | ||
| CAM85361.1 | toluene tolerance efflux transporter (ABC superfamily, peri-bind) | EC | 0.662 | 226 | 0.74 | Q | PF02470 | MlaD protein |
| CAM86485.1 | conserved, putative exported protein | EC | 0.533 | 294 | 0.69 | S | PF 16331 | TolA binding protein trimerization |
| CAM87843.1 | conserved, putative exported protein | EC | 0.744 | 715 | 0.75 | S | ||
| Other Putative OM Proteins Or Lipoproteins | ||||||||
| CAM86480.1 | putative OM protein | OM | 0.541 | 841 | 0.63 | M | PF01103 | Bac_surface_Ag |
| CAM87743.1 | OM lipoprotein | OM | 0.536 | 132 | 0.42 | M | PF04355 | SmpA_OmlA |
| CAM87612.1 | putative lipoprotein | OM | 0.513 | 159 | 0.68 | MP | PF04170 | copper resistance protein NlpE |
| Other Putative Extracellular Proteins | ||||||||
| CAM85672.1 | putative fatty acid transport protein | EC | 0.660 | 476 | 0.64 | M | PF03349 | |
| CAM88107.1 | putative phosphatase; alkaline phosphatase | EC | 0.561 | 726 | 0.49 | S | PF05787 | unknown function |
| CAM85335.1 | putative alkaline protease | EC | 0.551 | 461 | 0.52 | O | PF00082 | Peptidase_S8 |
| CAM85336.1 | conserved, putative signal peptide | EC | 0.570 | 143 | 0.44 | |||
| Accession Number | Resistance Protein Name | Protein Length | Localization | Adhesion Probability | Antigenicity Score | COG Group | Pfam Domains | Functional Description |
|---|---|---|---|---|---|---|---|---|
| ADB64519.1 | QnrA1 | 218 | EC | 0.330 | 0.58 | S | PF00805 PF13599 | Pentapeptide repeat protein |
| WP_000045119.1 | adeC/adeK/oprM family multidrug efflux complex outer membrane factor | 465 | OM | 0.339 | 0.62 | M | PF02321 | RND efflux system, outer membrane lipoprotein |
| WP_000018327.1 | OprD-like protein | 469 | OM | 0.533 | 0.70 | M | PF03573 | OprD super family Outer membrane porin |
| AAL68825.1 | CTX-M-5 | 276 | PP | 0.598 | 0.47 | V | PF13354 | β-lactamase |
| AAZ14955.1 | CTX-M-43 | 291 | PP | 0.548 | 0.43 | V | PF13354 | β-lactamase |
| BAD34451.1 | CTX-M-2 | 291 | PP | 0.537 | 0.42 | V | PF13354 | β-lactamase |
| AEQ20897.1 | CTX-M15 | 291 | PP | 0.427 | 0.44 | V | PF13354 | β-lactamase |
| ACJ61335.1 | RTG-4 | 298 | PP | 0.384 | 0.42 | V | PF13354 | β-lactamase |
| AFA35105.1 | ADC | 383 | PP | 0.367 | 0.34 | V | PF00144 | β-lactamase |
| ACO56763.1 | VEB-7 | 299 | PP | 0.354 | 0.54 | V | PF13354 | β-lactamase |
| AMB18971.1 | TEM-116 | 291 | PP | 0.251 | 0.50 | V | PF13354 | β-lactamase |
| AFN21551.1 | TEM-19 | 286 | PP | 0.234 | 0.47 | V | PF13354 | β-lactamase |
| AAQ57123.1 | TEM-128 | 286 | PP | 0.233 | 0.47 | V | PF13354 | β-lactamase |
| AGW28875.1 | TEM-1 | 286 | PP | 0.226 | 0.48 | V | PF13354 | β-lactamase |
| AFC75524.1 | TEM 194 | 286 | PP | 0.212 | 0.49 | V | PF13354 | β-lactamase |
| AFC75525.1 | TEM 195 | 286 | PP | 0.212 | 0.48 | V | PF13354 | β-lactamase |
| AAQ55480.1 | SHV-56 | 286 | PP | 0.210 | 0.46 | V | PF13354 | β-lactamase |
| AFC75523.1 | TEM 193 | 286 | PP | 0.207 | 0.47 | V | PF13354 | β-lactamase |
| ACG63555.1 | SHV-18 | 286 | PP | 0.203 | 0.43 | V | PF13354 | β-lactamase |
| AAV38100.1 | SHV-1b | 286 | PP | 0.194 | 0.46 | V | PF13354 | β-lactamase |
| AAP20889.1 | SHV-12 | 286 | PP | 0.185 | 0.45 | V | PF13354 | β-lactamase |
| ABC25482.1 | SHV-71 | 286 | PP | 0.177 | 0.45 | V | PF13354 | β-lactamase |
| AAP20890.1 | SHV-48 | 286 | PP | 0.176 | 0.46 | V | PF13354 | β-lactamase |
| ABN49112.1 | SHV-96 | 286 | PP | 0.165 | 0.48 | V | PF13354 | β-lactamase |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Z.; Chen, Y.; Ong, E.; He, Y. Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology. Int. J. Mol. Sci. 2017, 18, 458. https://doi.org/10.3390/ijms18020458
Ni Z, Chen Y, Ong E, He Y. Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology. International Journal of Molecular Sciences. 2017; 18(2):458. https://doi.org/10.3390/ijms18020458
Chicago/Turabian StyleNi, Zhaohui, Yan Chen, Edison Ong, and Yongqun He. 2017. "Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology" International Journal of Molecular Sciences 18, no. 2: 458. https://doi.org/10.3390/ijms18020458
APA StyleNi, Z., Chen, Y., Ong, E., & He, Y. (2017). Antibiotic Resistance Determinant-Focused Acinetobacter baumannii Vaccine Designed Using Reverse Vaccinology. International Journal of Molecular Sciences, 18(2), 458. https://doi.org/10.3390/ijms18020458