The Cell Death Triggered by the Nuclear Localized RxLR Effector PITG_22798 from Phytophthora infestans Is Suppressed by the Effector AVR3b
Abstract
:1. Introduction
2. Results
2.1. PITG_22798 Is Induced Early during Infection of P. infestans and Promotes Pathogen Colonization
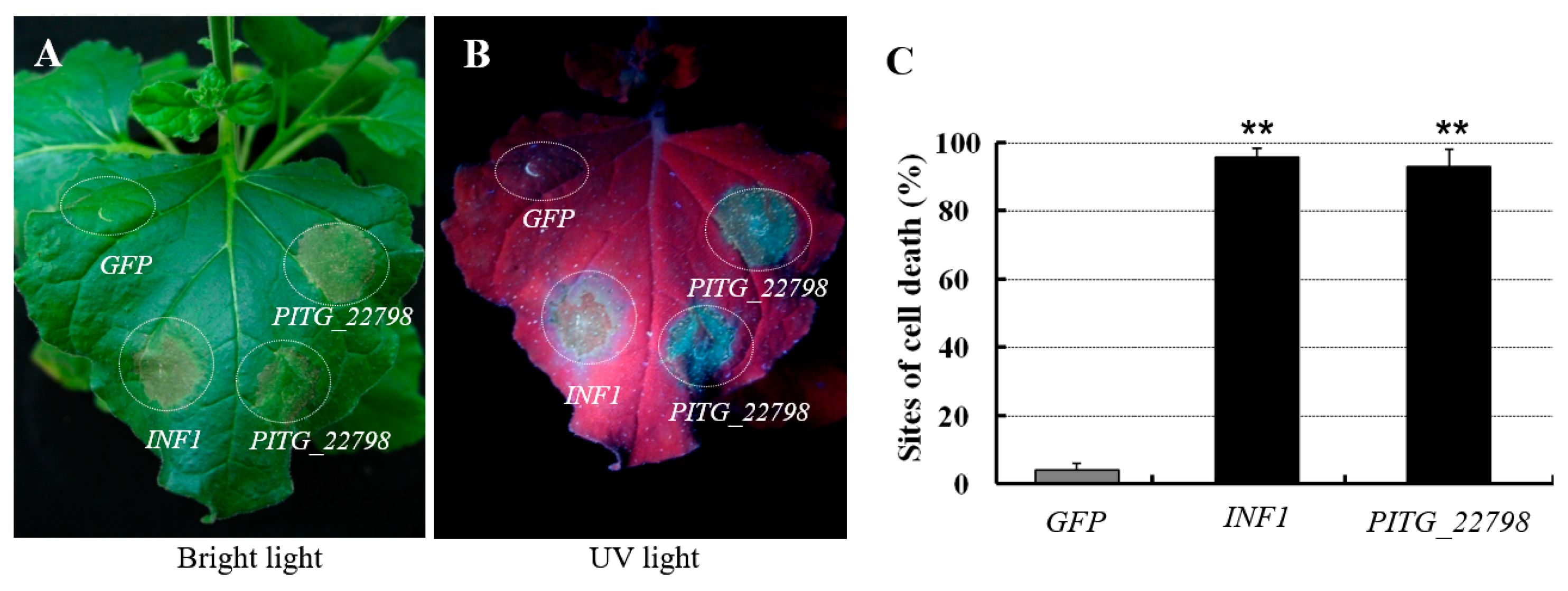
2.2. PITG_22798 Causes Cell Death in N. benthamiana
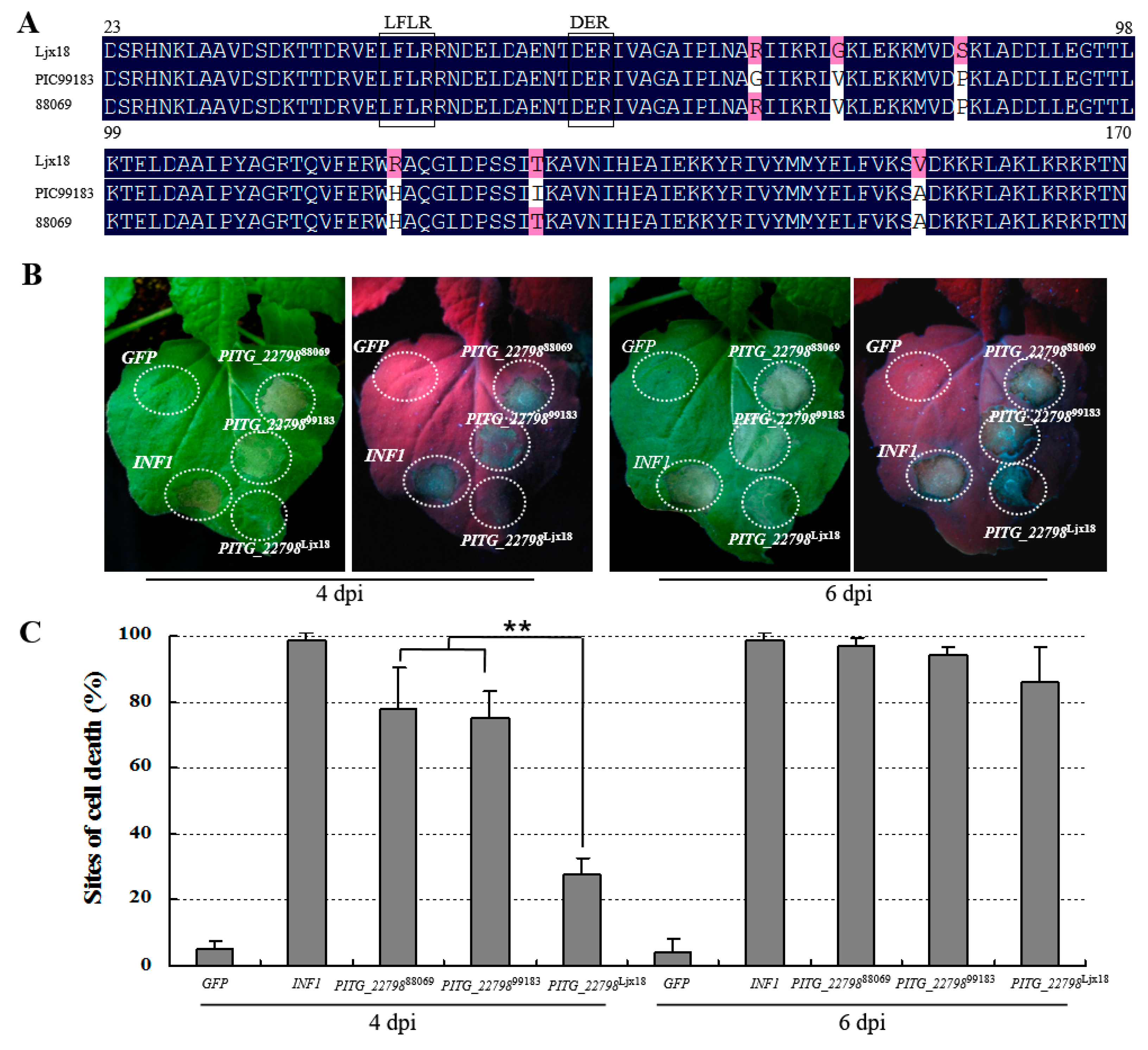
2.3. PITG_22798 Is Conserved in P. infestans Isolates
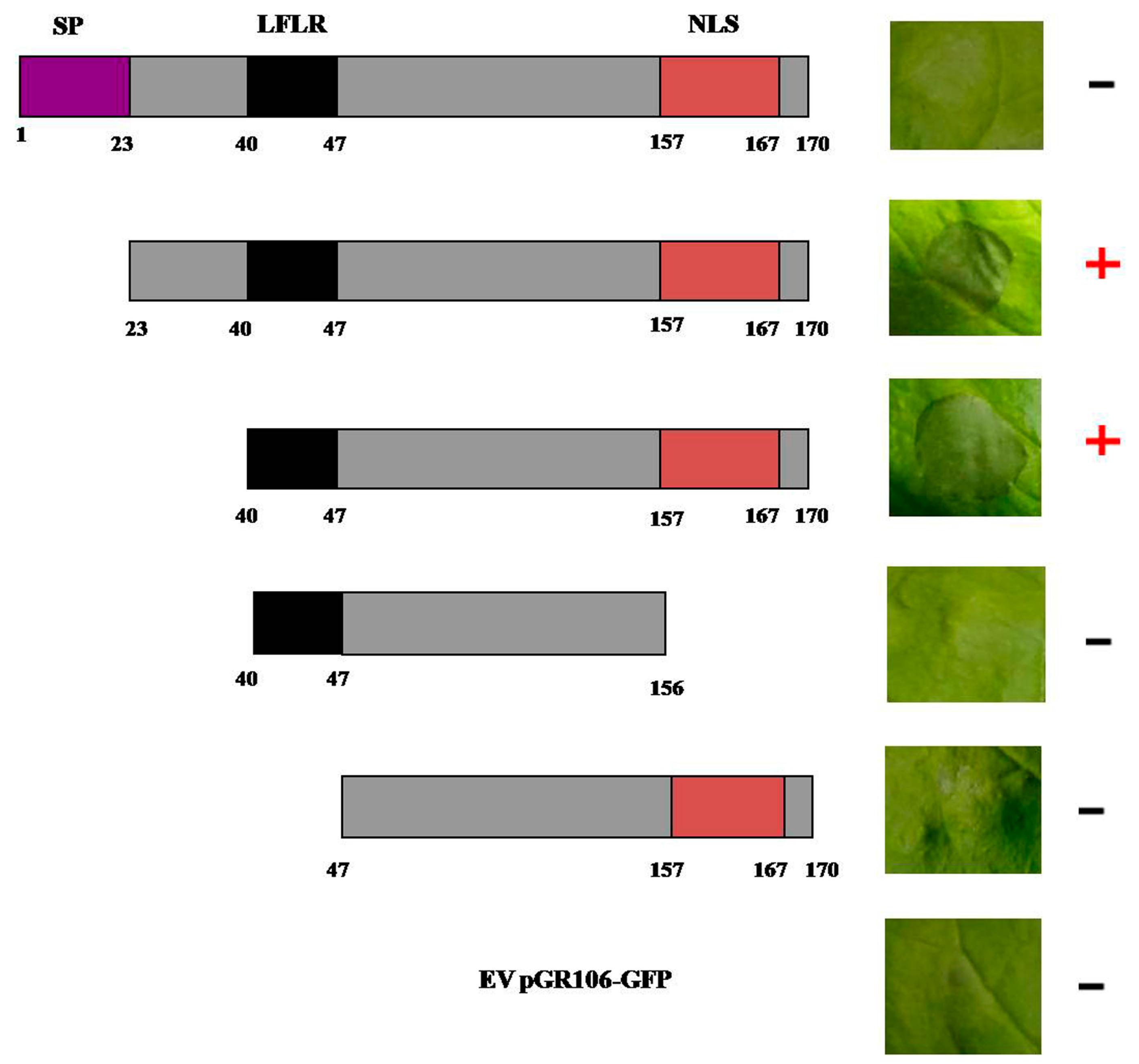
2.4. Effector PITG_22798 Functions in the Nucleus to Induce Cell Death
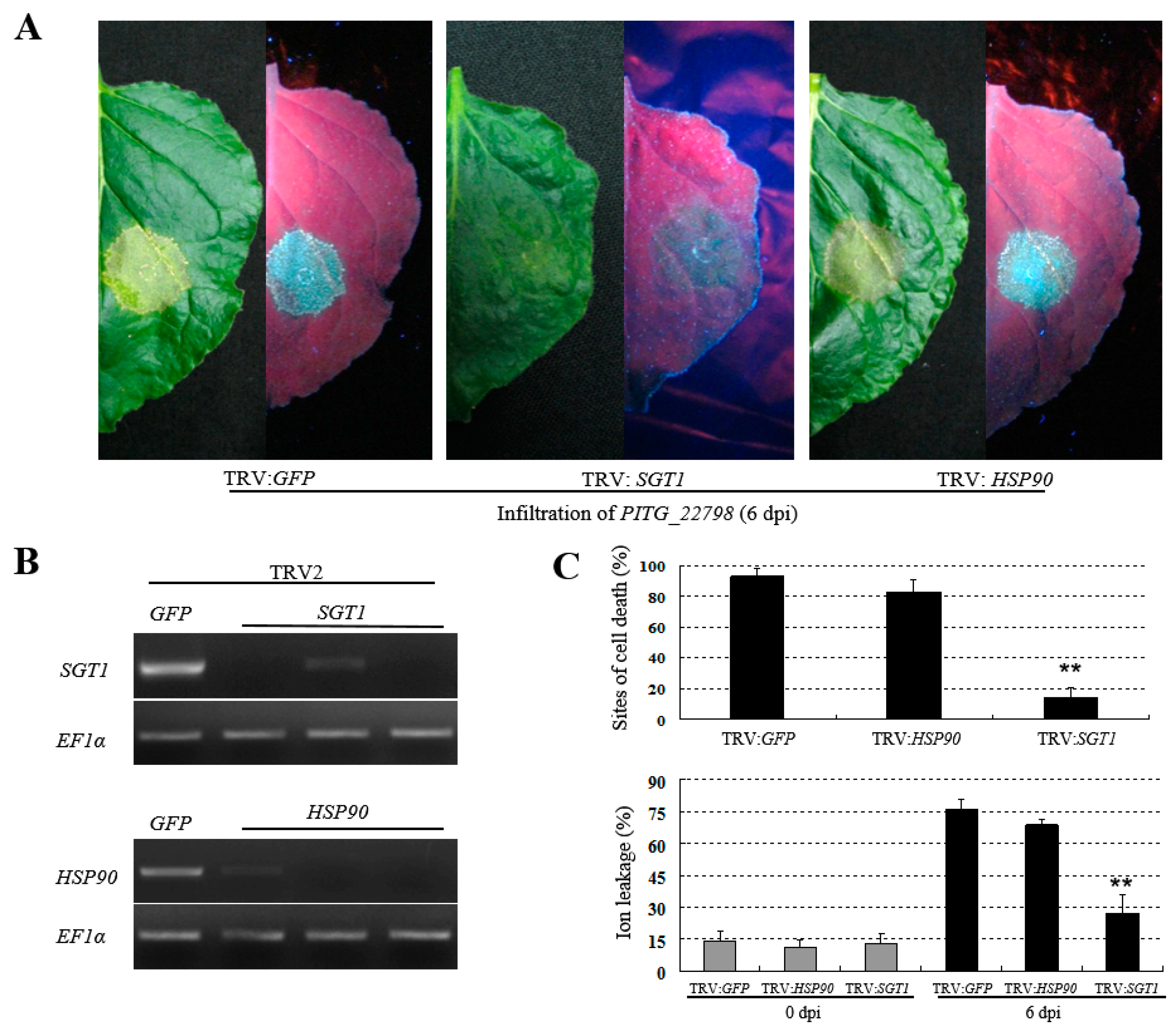
2.5. PITG_22798-Mediated Cell Death Requires SGT1, but Not HSP90
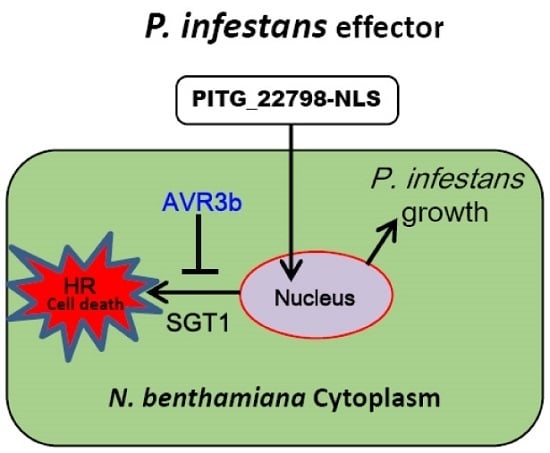
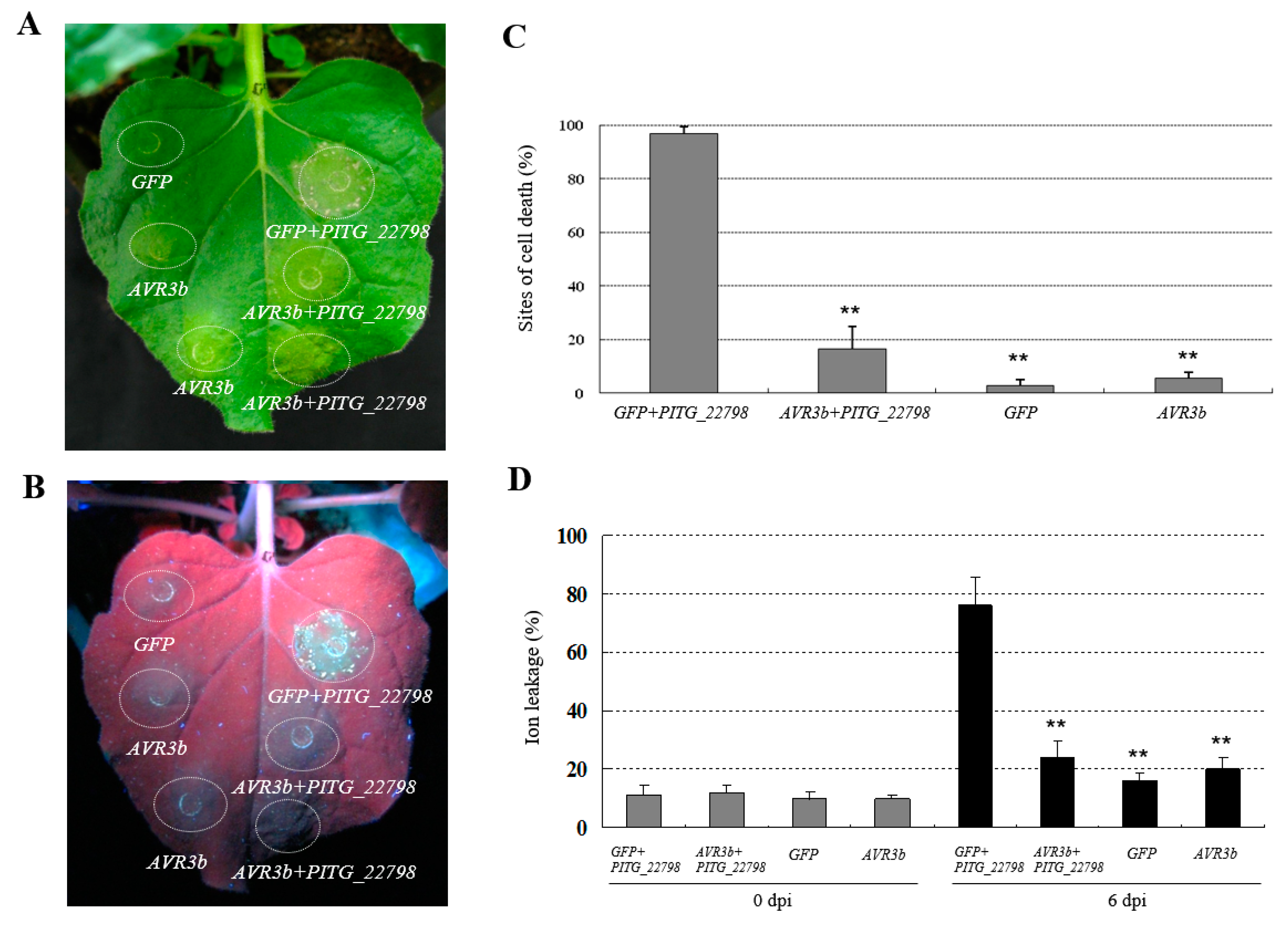
2.6. AVR3b Suppresses PITG_22798-Triggered Cell Death in N. benthamiana
3. Discussion
4. Materials and Methods
4.1. Microbial Strains, Plants, and Culture Conditions
4.2. Construction of Plasmids
4.3. Expression Analysis of PITG_22798 and P. infestans Infection Assay
4.4. Cloning and Sequence Analysis of PITG_22798
4.5. Agroinfiltration and PVX-Agroinfection Assay
4.6. Western Blot and Confocal Microscopic Examination
4.7. Virus-Induced Gene-Silencing (VIGS) Assay
4.8. Measurement of the Ion Leakage
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PTI | Pathogen-associated molecular pattern triggered immunity |
| ETI | Effector-triggered immunity |
| RT-PCR | Reverse transcriptase PCR |
| VIGS | Virus-induced gene silencing |
| NLS | Nuclear localization signal |
| hpi | Hours post inoculation |
| dpi | Days post infiltration |
References
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Panstruga, R.; Dodds, P.N. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 2009, 324, 748–750. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Whisson, S.C.; Boevink, P.C.; Wang, S.; Birch, P.R. The cell biology of late blight disease. Curr. Opin. Microbiol. 2016, 34, 127–135. [Google Scholar] [CrossRef] [PubMed]
- van Damme, M.; Bozkurt, T.O.; Cakir, C.; Schornack, S.; Sklenar, J.; Jones, A.M.; Kamoun, S. The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLoS Pathog. 2012, 8, e1002875. [Google Scholar] [CrossRef] [PubMed]
- Baxter, L.; Tripathy, S.; Ishaque, N.; Boot, N.; Cabral, A.; Kemen, E.; Thines, M.; Ah-Fong, A.; Anderson, R.; Badejoko, W.; et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 2010, 330, 1549–1551. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Meijer, H.J.; Govers, F. At the Frontier; RXLR effectors crossing the Phytophthora-Host interface. Front. Plant Sci. 2011, 2, 75. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Morgan, W.; Bos, J.; Krasileva, K.V.; Cano, L.M.; Chaparro-Garcia, A.; Ammar, R.; Staskawicz, B.J.; Kamoun, S. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 2007, 19, 2349–2369. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.; Raffaele, S.; Vossen, J.H.; Champouret, N.; Oliva, R.; Segretin, M.E.; Rietman, H.; Cano, L.M.; Lokossou, A.; Kessel, G.; et al. Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 2011, 49, 507–531. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.D.; Gu, B.; Capelluto, D.G.; Dou, D.; Feldman, E.; Rumore, A.; Arredondo, F.D.; Hanlon, R.; Fudal, I.; Rouxel, T.; et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 2010, 142, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; Huitema, E.; Cano, L.M.; Bozkurt, T.O.; Oliva, R.; Van Damme, M.; Schwizer, S.; Raffaele, S.; Chaparro-Garcia, A.; Farrer, R.; et al. Ten things to know about oomycete effectors. Mol. Plant Pathol. 2009, 10, 795–803. [Google Scholar] [CrossRef] [PubMed]
- McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Pritchard, L.; Gomez, S.; Morales, J.; Whisson, S.C.; Beynon, J.L.; Birch, P.R. An RxLR effector from Phytophthora infestans prevents relocalisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013, 9, e1003670. [Google Scholar] [CrossRef] [PubMed]
- King, S.R.; McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Bukharova, T.; Sukarta, O.; Win, J.; Kamoun, S.; Birch, P.R.; Banfield, M.J. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK epsilon to suppress plant immune signaling. Plant Cell 2014, 26, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; McLellan, H.; Fraiture, M.; Liu, X.; Boevink, P.C.; Gilroy, E.M.; Chen, Y.; Kandel, K.; Sessa, G.; Birch, P.R.; et al. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 2014, 10, e1004057. [Google Scholar] [CrossRef] [PubMed]
- Boevink, P.C.; Wang, X.; McLellan, H.; He, Q.; Naqvi, S.; Armstrong, M.R.; Zhang, W.; Hein, I.; Gilroy, E.M.; Tian, Z.; et al. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat. Commun. 2016, 7, 10311. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T.D.; Huitema, E.; Cakir, C.; Kamoun, S. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Mol. Plant Microbe Interact. 2006, 19, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Young, C.; Lee, M.; Oliva, R.; Bozkurt, T.O.; Cano, L.M.; Win, J.; Bos, J.I.; Liu, H.Y.; van Damme, M.; et al. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 2009, 21, 2928–2947. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Kawakita, K.; Takemoto, D. SGT1 and HSP90 are essential for age-related non-host resistance of Nicotiana benthamiana against the oomycete pathogen Phytophthora infestans. Physiol. Mol. Plant Pathol. 2011, 75, 120–128. [Google Scholar] [CrossRef]
- Rivas, S. Nuclear dynamics during plant innate immunity. Plant Physiol. 2012, 158, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Rivas, S. The plant cell nucleus. Plant Signal Behav. 2011, 6, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.J.; Muskett, P.; Kahn, K.; Feys, B.J.; Jones, J.D.; Parker, J.E. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 2002, 295, 2077–2080. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.R.; Lu, R.; Sadanandom, A.; Malcuit, I.; Moffett, P.; Brice, D.C.; Schauser, L.; Jaggard, D.A.; Xiao, S.; Coleman, M.J. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 2002, 99, 10865–10869. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Shirasu, K.; Guerois, R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem. Sci. 2010, 35, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Kim, H.; Choi, D. Rpi-blb2-mediated late blight resistance in Nicotiana benthamiana requires SGT1 and salicylic acid-mediated signaling but not RAR1 or HSP90. FEBS Lett. 2014, 588, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Huitema, E.V.; Vleeshouwers, V.G.; Cakir, C.; Kamoun, S.; Govers, F. Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant Microbe Interact. 2005, 18, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Tang, J.; Wang, Q.; Ye, W.; Tao, K.; Duan, S.; Lu, C.; Yang, X.; Dong, S.; Zheng, X.; et al. The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 2012, 196, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Han, C.; Ferreira, A.O.; Yu, X.; Ye, W.; Tripathy, S.; Kale, S.D.; Gu, B.; Sheng, Y.; Sui, Y.; et al. Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 2011, 23, 2064–2086. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; McLellan, H.; Naqvi, S.; He, Q.; Boevink, P.C.; Armstrong, M.; Giuliani, L.M.; Zhang, W.; Tian, Z.; Zhan, J.; et al. Potato NPH3/RPT2-Like Protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 2016, 171, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Kale, S.D.; Wang, X.; Chen, Y.; Wang, Q.; Wang, X.; Jiang, R.H.; Arredondo, F.D.; Anderson, R.G.; Thakur, P.B.; et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 2008, 20, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Huang, S.; Guo, X.; Li, Y.; Yang, Y.; Guo, Z.; Kuang, H.; Rietman, H.; Bergervoet, M.; Vleeshouwers, V.G. Cloning and characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Mol. Plant Microbe Interact. 2011, 24, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, M.R.; Whisson, S.C.; Pritchard, L.; Bos, J.I. B.; Venter, E.; Avrova, A.O.; Rehmany, A.P.; Bohme, U.; Brooks, K.; Cherevach, I.; et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 2005, 102, 7766–7771. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, E.M.; Breen, S.; Whisson, S.C.; Squires, J.; Hein, I.; Kaczmarek, M.; Turnbull, D.; Boevink, P.C.; Lokossou, A.; Cano, L.M.; et al. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011, 191, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Halterman, D.A.; Chen, Y.; Sopee, J.; Berduo-Sandoval, J.; Sanchez-Perez, A. Competition between Phytophthora infestans effectors leads to increased aggressiveness on plants containing broad-spectrum late blight resistance. PLoS ONE 2010, 5, e10536. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.; Dooijeweert, W.V.; Keizer, L.C. P.; Sijpkes, L.; Govers, F.; Colon, L.T. A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur. J. Plant Pathol. 1999, 105, 241–250. [Google Scholar] [CrossRef]
- Wang, H.; Sun, C.; Jiang, R.; He, Q.; Yang, Y.; Tian, Z.; Tian, Z.; Xie, C. The dihydrolipoyl acyltransferase gene BCE2 participates in basal resistance against Phytophthora infestans in potato and Nicotiana benthamiana. J. Plant Physiol. 2014, 171, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Rietman, H.; Vleeshouwers, V.G. Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J. Vis. Exp. 2014, e50971. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple hidden markov model for nuclear localization signal prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Tian, Z.; Liu, J.; Vleeshouwers, V.G.; Shi, X.; Xie, C. Functional analysis of potato genes involved in quantitative resistance to Phytophthora infestans. Mol. Biol. Rep. 2013, 40, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.S.; Quartin, V.; Ramalho, J.C.; Nunes, M.A. Electrolyte leakage and lipid degradation account for cold sensitivity in leaves of Coffea sp. plants. J. Plant Physiol. 2003, 160, 283–292. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Ren, Y.; Zhou, J.; Du, J.; Hou, J.; Jiang, R.; Wang, H.; Tian, Z.; Xie, C. The Cell Death Triggered by the Nuclear Localized RxLR Effector PITG_22798 from Phytophthora infestans Is Suppressed by the Effector AVR3b. Int. J. Mol. Sci. 2017, 18, 409. https://doi.org/10.3390/ijms18020409
Wang H, Ren Y, Zhou J, Du J, Hou J, Jiang R, Wang H, Tian Z, Xie C. The Cell Death Triggered by the Nuclear Localized RxLR Effector PITG_22798 from Phytophthora infestans Is Suppressed by the Effector AVR3b. International Journal of Molecular Sciences. 2017; 18(2):409. https://doi.org/10.3390/ijms18020409
Chicago/Turabian StyleWang, Hongyang, Yajuan Ren, Jing Zhou, Juan Du, Juan Hou, Rui Jiang, Haixia Wang, Zhendong Tian, and Conghua Xie. 2017. "The Cell Death Triggered by the Nuclear Localized RxLR Effector PITG_22798 from Phytophthora infestans Is Suppressed by the Effector AVR3b" International Journal of Molecular Sciences 18, no. 2: 409. https://doi.org/10.3390/ijms18020409
APA StyleWang, H., Ren, Y., Zhou, J., Du, J., Hou, J., Jiang, R., Wang, H., Tian, Z., & Xie, C. (2017). The Cell Death Triggered by the Nuclear Localized RxLR Effector PITG_22798 from Phytophthora infestans Is Suppressed by the Effector AVR3b. International Journal of Molecular Sciences, 18(2), 409. https://doi.org/10.3390/ijms18020409