Identification and Characterization of Lipopolysaccharide Induced TNFα Factor from Blunt Snout Bream, Megalobrama amblycephala
Abstract
:1. Introduction
2. Results
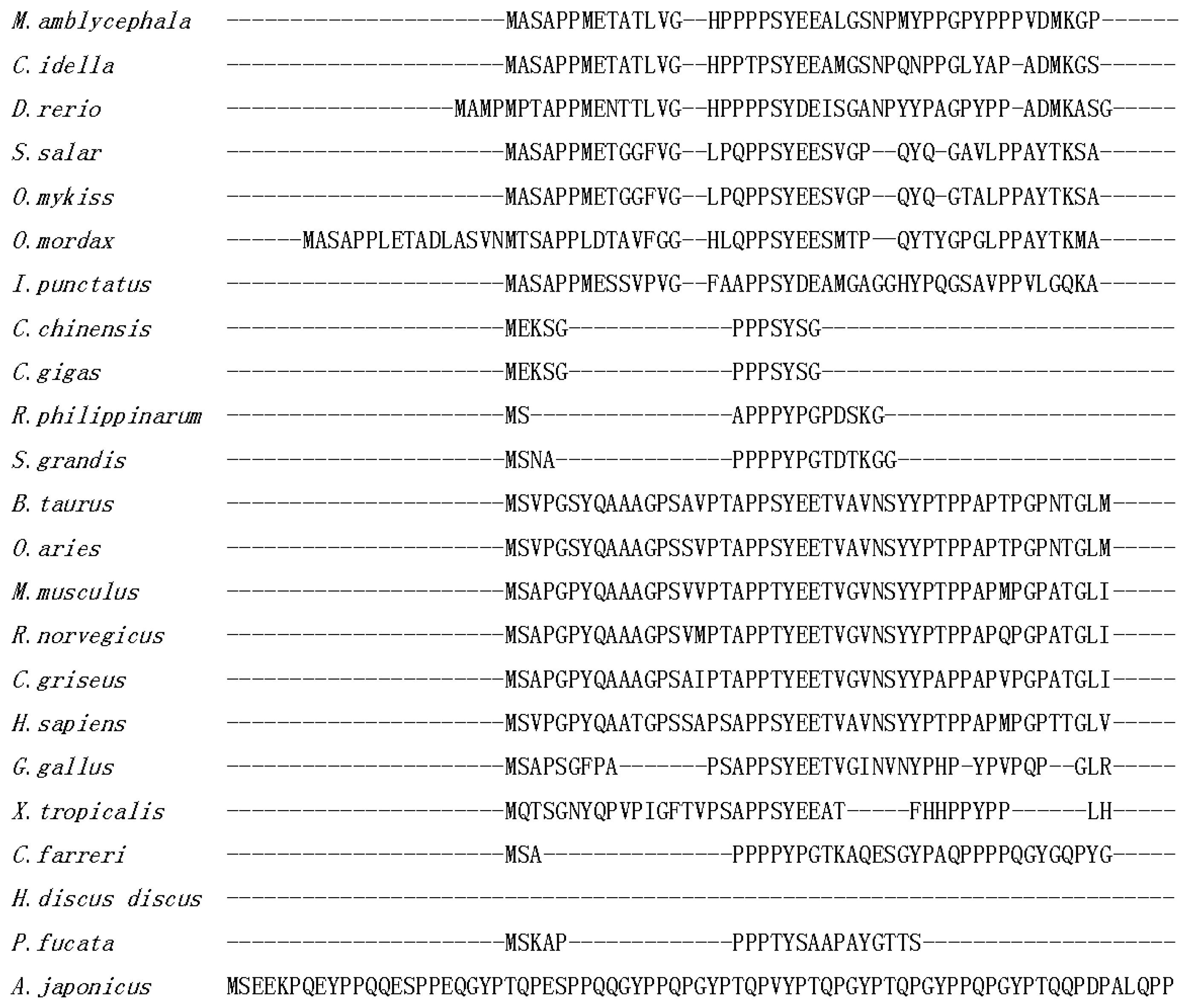
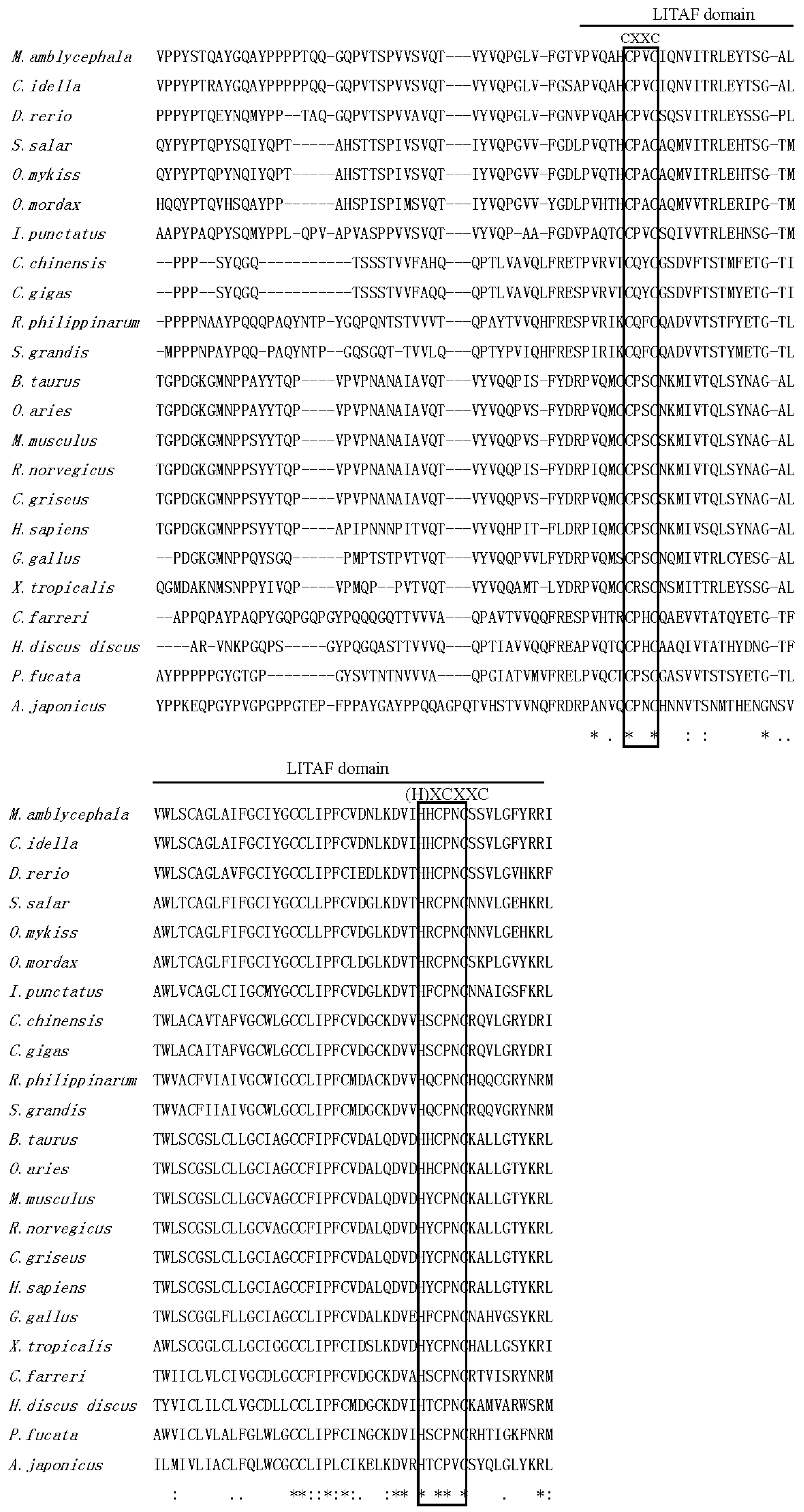
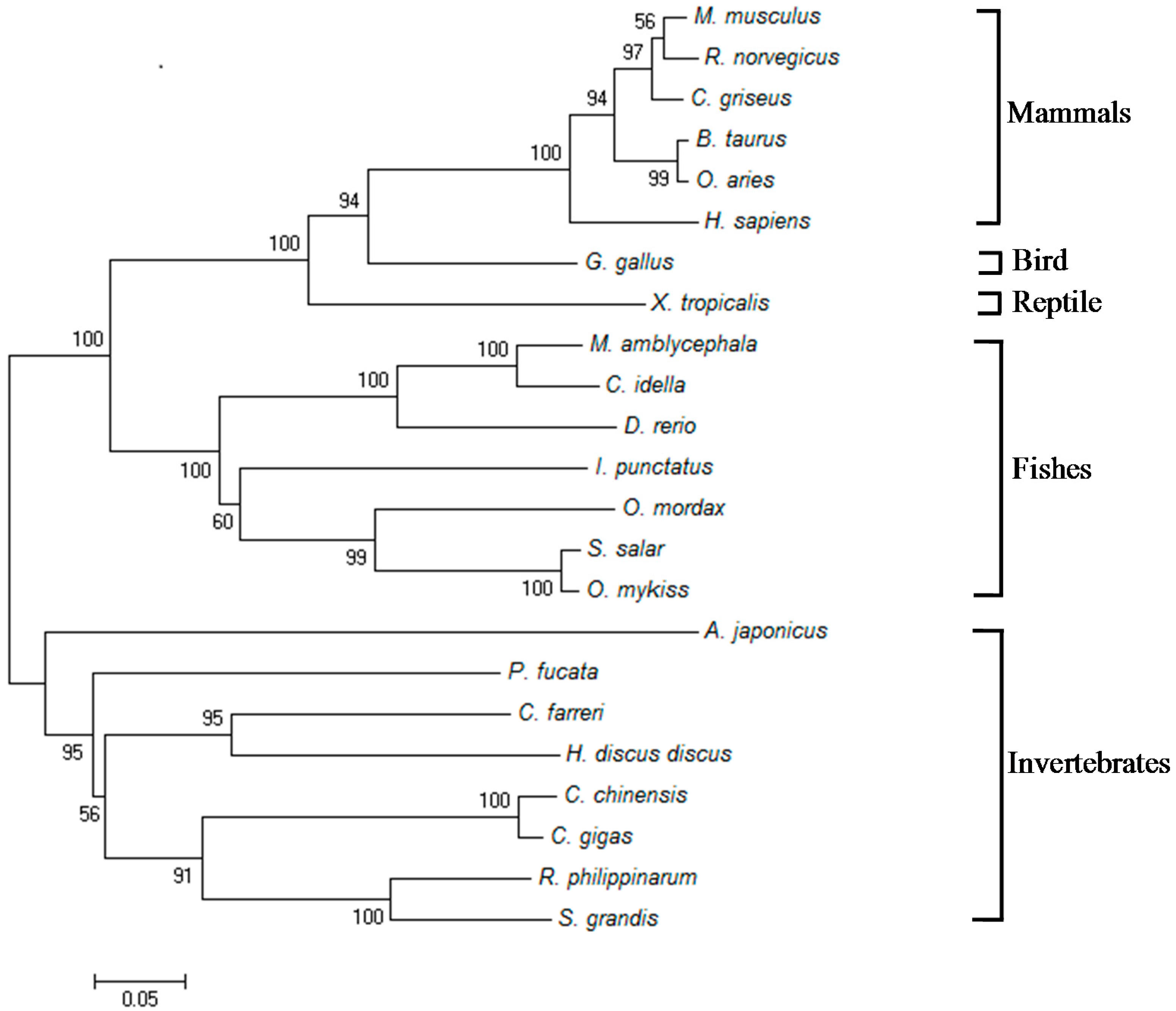
2.1. Isolation and Characterization of Malitaf
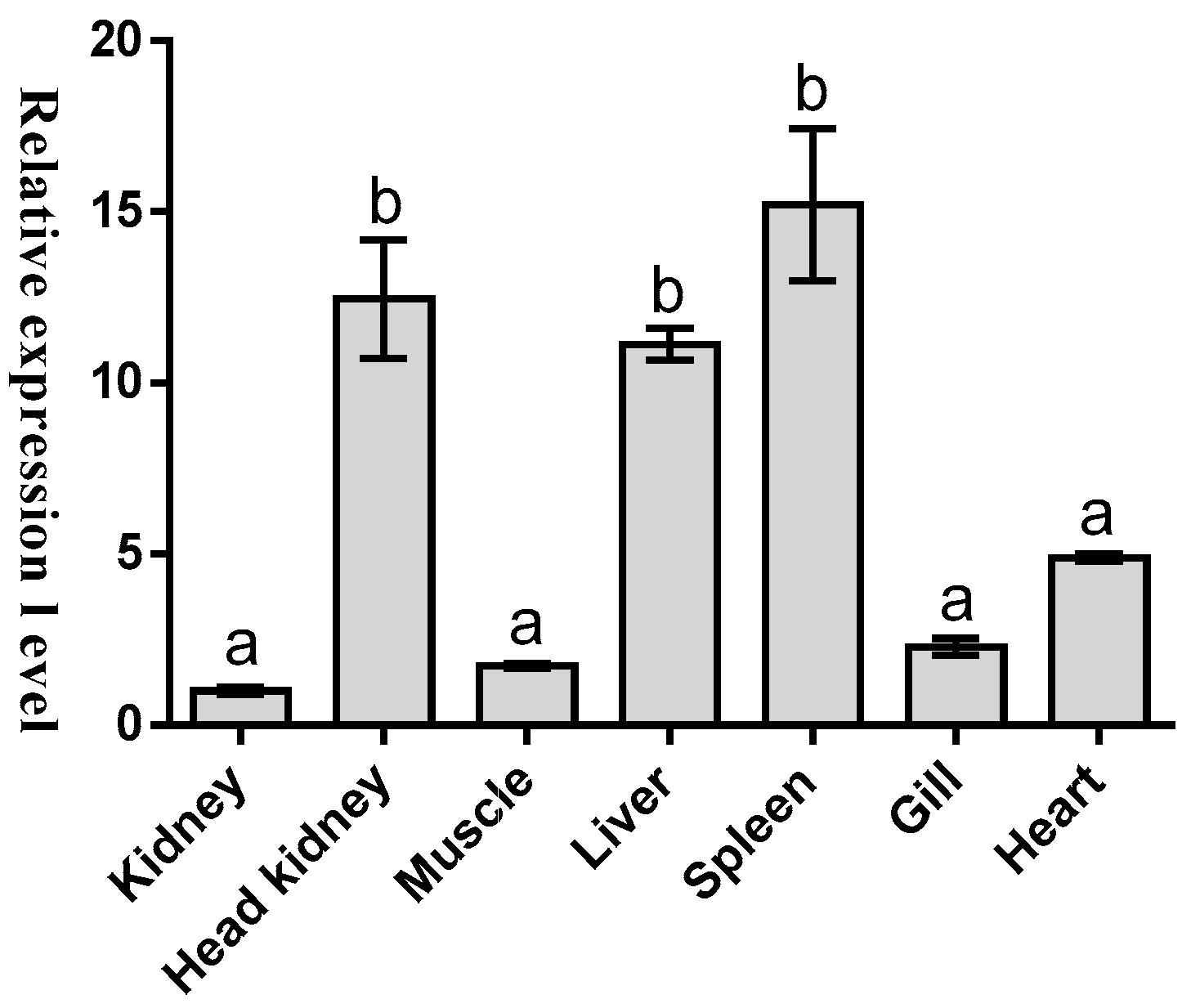
2.2. Tissues Distribution of Malitaf
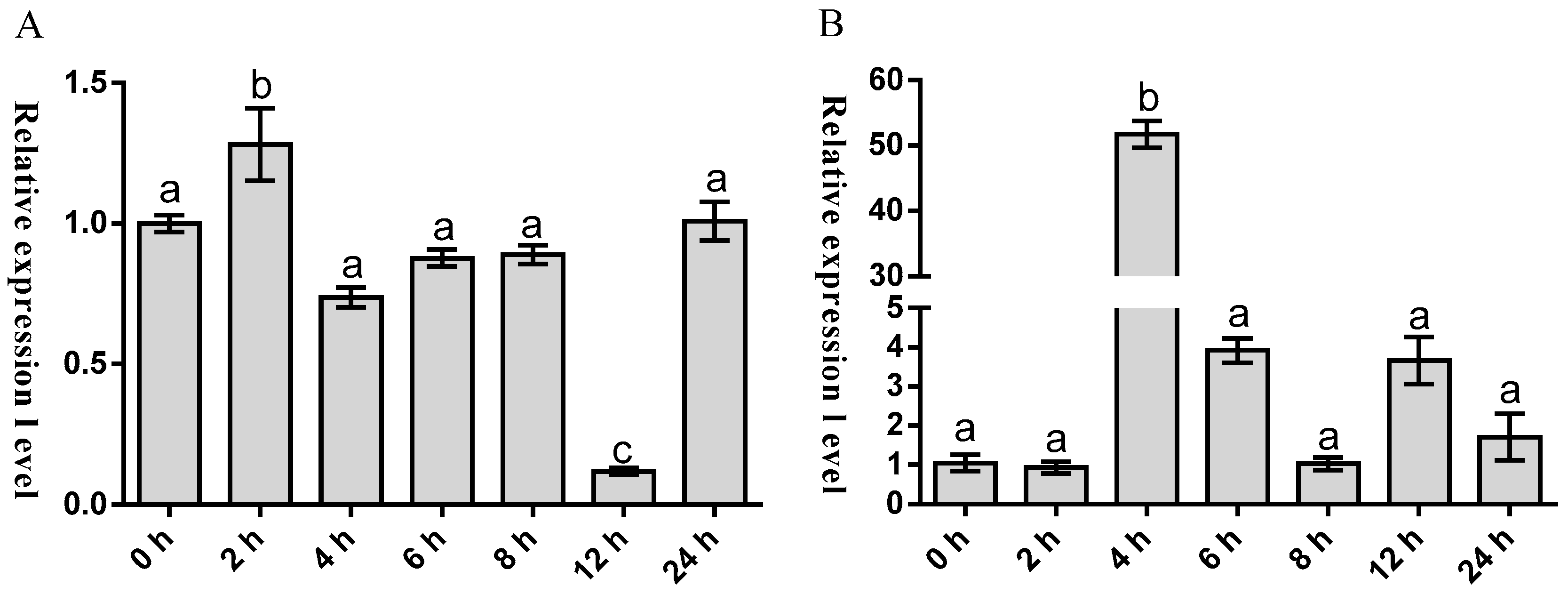
2.3. Temporal Expression of Malitaf and Matnfα after LPS Stimulation
3. Discussion
4. Materials and Methods
4.1. Fish, LPS Stimulation, and RNA Isolation
4.2. Cloning and Characterization of Malitaf
4.3. Spatial and Temporal Expression Analysis of Malitaf and Matnfα
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, S.F.; Cai, W.Q.; Zhou, B.Y. Variation in morphology and biochemical genetic markers among populations of blunt snout bream (Megalobrama amblycephala). Aquaculture 1993, 111, 117–127. [Google Scholar] [CrossRef]
- Tang, L.L.; Liang, Y.H.; Jiang, Y.H.; Liu, S.J.; Zhang, F.Y.; He, X.; Wang, T.Y.; Zhou, Y.; Zhong, H.; Yan, J.P. Identification and expression analysis on bactericidal permeability-increasing protein/lipopolysaccharide-binding protein of blunt snout bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2015, 45, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Tang, L.L.; Zhang, F.Y.; Jiang, H.Y.; Li, X.W.; Yang, L.Y.; Zhang, L.; Mao, J.R.; Yang, J.P. Identification and characterization of immune-related microRNAs in blunt snout bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2016, 49, 470–492. [Google Scholar]
- Bureau, C.F. China Fisheries Yearbook; Chinese Agriculture Press: Beijing, China, 2013. [Google Scholar]
- Nielsen, M.E.; Hoi, L.; Schmidt, A.S.; Qian, D.; Shimada, T.; Shen, J.Y.; Larsen, J.L. Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Organ. 2001, 46, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A.E. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Zhu, L.Y.; Nie, L.; Zhu, G.; Xiang, L.X.; Shao, J.Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.L.; Xiang, X.Y.; Jiang, Y.H.; Lv, Y.N.; Zhou, Y.; Zhong, H.; Xiao, J.; Zhang, F.Y.; Jiang, H.Y.; Yan, J.P. Identification and characterization of a novel Toll-like receptor 4 homologue in blunt snout bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2016, 57, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Walsh, J.G.; Belosevic, M. Characterization and functional analysis of goldfish (Carassius auratus L.) tumor necrosis factor-α. Dev. Comp. Immunol. 2008, 32, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Zhang, J. CsTNF1, a teleost tumor necrosis factor that promotes antibacterial and antiviral immune defense in a manner that depends on the conserved receptor binding site. Dev. Comp. Immunol. 2016, 55, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Baeuerle, P.; Vassalli, P. Regulation of tumor necrosis factor α transcription in macrophages: Involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol. Cell Biol. 1990, 10, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Park, J.; Chung, S.W.; Kang, B.Y.; Kim, S.H.; Kim, T.S. Enhancement of interleukin-4 production in activated CD4+ T cells by diphthalate plasticizers via increased NF-AT binding activity. Int. Arch. Allergy Immunol. 2004, 134, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, M.; D’Isanto, M.; Galdiero, M.; Raieta, K.; Tortora, A.; Rotondo, P.; Peluso, L.; Galdiero, M. Interleukin-8 production by THP-1 cells stimulated by Salmonella enterica serovar Typhimurium porins is mediated by AP-1, NF-κB and MAPK pathways. Cytokine 2004, 27, 15–24. [Google Scholar] [CrossRef]
- Stucchi, A.; Reed, K.; O’Brien, M.; Cerda, S.; Andrews, C.; Gower, A.; Bushell, K.; Amar, S.; Leeman, S.; Becker, J. A new transcription factor that regulates TNF-α gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm. Bowel Dis. 2006, 12, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Metzger, D.; Leeman, S.; Amar, S. LPS-induced TNF-α factor (LITAF)-deficient mice express reduced LPS-induced cytokine: Evidence for LITAF-dependent LPS signaling pathways. Proc. Natl. Acad. Sci. USA 2006, 103, 13777–13782. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Marciano, D.L.; Leeman, S.E.; Amar, S. LPS induces the interaction of a transcription factor, LPS-induced TNF-α factor, and STAT6(B) with effects on multiple cytokines. Proc. Natl. Acad. Sci. USA 2005, 102, 5132–5137. [Google Scholar] [CrossRef] [PubMed]
- Myokai, F.; Takashiba, S.; Lebo, R.; Amar, S. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor α gene expression: Molecular cloning, sequencing, characterization, and chromosomal assignment. Proc. Natl. Acad. Sci. USA 1999, 96, 4518–4523. [Google Scholar] [CrossRef]
- De Zoysa, M.; Nikapitiya, C.; Oh, C.; Whang, I.; Lee, J.S.; Jung, S.J.; Choi, C.Y.; Lee, J. Molecular evidence for the existence of lipopolysaccharide-induced TNF-α factor (LITAF) and Rel/NF-κB pathways in disk abalone (Haliotis discus discus). Fish Shellfish Immunol. 2010, 28, 754–763. [Google Scholar] [CrossRef]
- Li, H.J.; Yang, Q.; Gao, X.G.; Su, H.; Wang, J.; He, C.B. Identification and expression of a putative LPS-induced TNF-α factor from Asiatic hard clam Meretrix meretrix. Mol. Biol. Rep. 2012, 39, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jiang, J.; Jiang, S.; Ma, J.; Su, T.; Qiu, L.; Zhu, C.; Xu, X. Molecular characterization and expression analysis of a putative LPS-induced TNF-α factor (LITAF) from pearl oyster Pinctada fucata. Fish Shellfish Immunol. 2009, 27, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wei, X.; Yang, J.; Yang, J.; Xu, J.; Fang, J.; Wang, S.; Liu, X. Identification of a LPS-induced TNF-α factor (LITAF) from mollusk Solen grandis and its expression pattern towards PAMPs stimulation. Fish Shellfish Immunol. 2013, 35, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, Y.; Yu, Z. Characteristics and expression patterns of the lipopolysaccharide-induced TNF-α factor (LITAF) gene family in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2012, 33, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Qiu, L.; Song, L.; Zhao, J.; Ni, D.; Zhang, Y.; Xu, W. Molecular cloning and characterization of a putative lipopolysaccharide-induced TNF-α factor (LITAF) gene homologue from Zhikong scallop Chlamys farreri. Fish Shellfish Immunol. 2007, 23, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jia, Z.; Li, X.; Geng, X.; Sun, J. Identification and expression analysis of lipopolysaccharide-induced TNF-α factor gene in Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2014, 38, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.H.; Wan, D.H.; Pang, L.R.; Gu, Z.H.; Qiu, W.; Weng, S.P.; Yu, X.Q.; He, J.G. Molecular cloning, characterization and expression analysis of the tumor necrosis factor (TNF) superfamily gene, TNF receptor superfamily gene and lipopolysaccharide-induced TNF-α factor (LITAF) gene from Litopenaeus vannamei. Dev. Comp. Immunol. 2012, 36, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, P.; Li, C.; Li, Y.; Jin, C.; Zhang, W. Characterization of two regulators of the TNF-α signaling pathway in Apostichopus japonicus: LPS-induced TNF-α factor and baculoviral inhibitor of apoptosis repeat-containing 2. Dev. Comp. Immunol. 2015, 48, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Hu, J.; Qian, J.; Chen, L.; Xu, X.; Ma, F. Identification and characterization of a putative lipopolysaccharide-induced TNF-α factor (LITAF) gene from Amphioxus (Branchiostoma belcheri): An insight into the innate immunity of Amphioxus and the evolution of LITAF. Fish Shellfish Immunol. 2012, 32, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Gen, X.; Chen, Y.; Wei, J.; Sun, J. Identification and characterization of lipopolysaccharide-induced TNF-α factor gene from Japanese flounder Paralichthys olivaceus. Vet. Immunol. Immunopathol. 2014, 157, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Huang, Y.; Wei, S.; Ouyang, Z.; Huang, X.; Qin, Q. Characterization of LPS-induced TNF-α factor (LITAF) from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 2013, 35, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.D.; Sang, H.S.; Kwon, M.G.; Chae, Y.S.; Shim, W.J.; Jung, J.H.; Kim, J.W.; Park, C.I. Molecular cloning and expression analysis of two lipopolysaccharide-induced TNF-α factors (LITAFs) from rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol. 2014, 36, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shen, X.; Xu, D.; Lu, L. Lipopolysaccharide-induced TNF-α factor in grass carp (Ctenopharyngodon idella): Evidence for its involvement in antiviral innate immunity. Fish Shellfish Immunol. 2013, 34, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bonecchi, R.; Garlanda, C.; Mantovani, A.; Riva, F. Cytokine decoy and scavenger receptors as key regulators of immunity and inflammation. Cytokine 2016, 87, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Merrill, J.C.; You, J.; Constable, C.; Leeman, S.E.; Amar, S. Whole-body deletion of LPS-induced TNF-α factor (LITAF) markedly improves experimental endotoxic shock and inflammatory arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 21247–21252. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, Y.; Begum, N.A.; Kobayashi, M.; Matsumoto, M.; Toyoshima, K.; Seya, T. Mycobacterium bovis Bacillus Calmette-Guerin and its cell wall complex induce a novel lysosomal membrane protein, SIMPLE, that bridges the missing link between lipopolysaccharide and p53-inducible gene, LITAF(PIG7), and estrogen-inducible gene, EET-1. J. Biol. Chem. 2001, 276, 23065–23076. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Mott, R.; Bork, P.; Copley, R.R. Novel protein domains and repeats in Drosophila melanogaster: Insights into structure, function, and evolution. Genome Res. 2001, 11, 1996–2008. [Google Scholar] [CrossRef] [PubMed]
- Bromage, E.S.; Kaattari, I.M.; Zwollo, P.; Kaattari, S.L. Plasmablast and plasma cell production and distribution in trout immune tissues. J. Immunol. 2004, 173, 7317–7323. [Google Scholar] [CrossRef] [PubMed]
- Bolcatobellemin, A.L.; Mattei, M.G.; Fenton, M.; Amar, S. Molecular cloning and characterization of mouse LITAF cDNA: Role in the regulation of tumor necrosis factor-α (TNF-α) gene expression. J. Endotoxin Res. 2004, 10, 15–23. [Google Scholar]
- Hong, Y.H.; Lillehoj, H.S.; Lee, S.H.; Park, D.W.; Lillehoj, E.P. Molecular cloning and characterization of chicken lipopolysaccharide-induced TNF-α factor (LITAF). Dev. Comp. Immunol. 2006, 30, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Beutler, B.; Cerami, A. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature 1986, 320, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Copley, R.R.; Pils, B.; Pinkert, S.; Schultz, J.; Bork, P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006, 34, D257–D260. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence(5′–3′) | Comment |
|---|---|---|
| LITAF-F | AARCGCTTTGYTTRTCGCTC | Gene cloning |
| LITAF-R | AGGTGYAAMTTGTTTCCCCG | |
| 3′-Adaptor primer | GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18 | 3′RACE |
| 3′-Primer | GCTGTCAACGATACGCTACGTAACG | |
| 3′-Nested primer | CGCTACGTAACGGCATGACAGTG | |
| LITAF-3′-GSP | ACAGCAACACTTGTGGGACACCCGCCT | |
| LITAF-3′-NGSP | AGCCTGGTCTGGTGTTCGGGACTGTT | |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG | 5′RACE |
| AUAP | GGCCACGCGTCGACTAGTAC | |
| LITAF-5′-GSP | CTGTGCCTCCATATTCAACACAAG | |
| LITAF-5′-NGSP | GTTGGGATCAAACCCAATGTACC | |
| LITAF-qF | CACCAGTCCTGTTGTATCGG | Real-time PCR |
| LITAF-qR | CGCACAAGAGAGCCAGACTA | |
| TNFα-qF | CTGCTGTCTGCTTCACGCTC | |
| TNFα-qR | TAAATGGATGGCTGCCTTGG | |
| β-actin-qF | CGGACAGGTCATCACCATTG | |
| β-actin-qR | CGCAAGACTCCATACCCAAGA | |
| 18S rRNA-qF | CAAGACGGACGAGAGCGAAA | |
| 18S rRNA-qR | GCGGGTTGGCATAGTTTACG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, Y.; Xiang, X.; Jiang, Y.; Tang, L.; Zhou, Y.; Zhong, H.; Xiao, J.; Yan, J. Identification and Characterization of Lipopolysaccharide Induced TNFα Factor from Blunt Snout Bream, Megalobrama amblycephala. Int. J. Mol. Sci. 2017, 18, 233. https://doi.org/10.3390/ijms18020233
Lv Y, Xiang X, Jiang Y, Tang L, Zhou Y, Zhong H, Xiao J, Yan J. Identification and Characterization of Lipopolysaccharide Induced TNFα Factor from Blunt Snout Bream, Megalobrama amblycephala. International Journal of Molecular Sciences. 2017; 18(2):233. https://doi.org/10.3390/ijms18020233
Chicago/Turabian StyleLv, Yina, Xinying Xiang, Yuhong Jiang, Leilei Tang, Yi Zhou, Huan Zhong, Jun Xiao, and Jinpeng Yan. 2017. "Identification and Characterization of Lipopolysaccharide Induced TNFα Factor from Blunt Snout Bream, Megalobrama amblycephala" International Journal of Molecular Sciences 18, no. 2: 233. https://doi.org/10.3390/ijms18020233
APA StyleLv, Y., Xiang, X., Jiang, Y., Tang, L., Zhou, Y., Zhong, H., Xiao, J., & Yan, J. (2017). Identification and Characterization of Lipopolysaccharide Induced TNFα Factor from Blunt Snout Bream, Megalobrama amblycephala. International Journal of Molecular Sciences, 18(2), 233. https://doi.org/10.3390/ijms18020233





