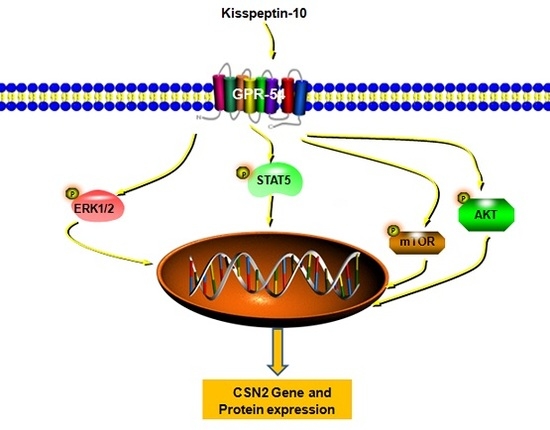
Kisspeptin-10 Induces β-Casein Synthesis via GPR54 and Its Downstream Signaling Pathways in Bovine Mammary Epithelial Cells
Abstract
1. Introduction
2. Results
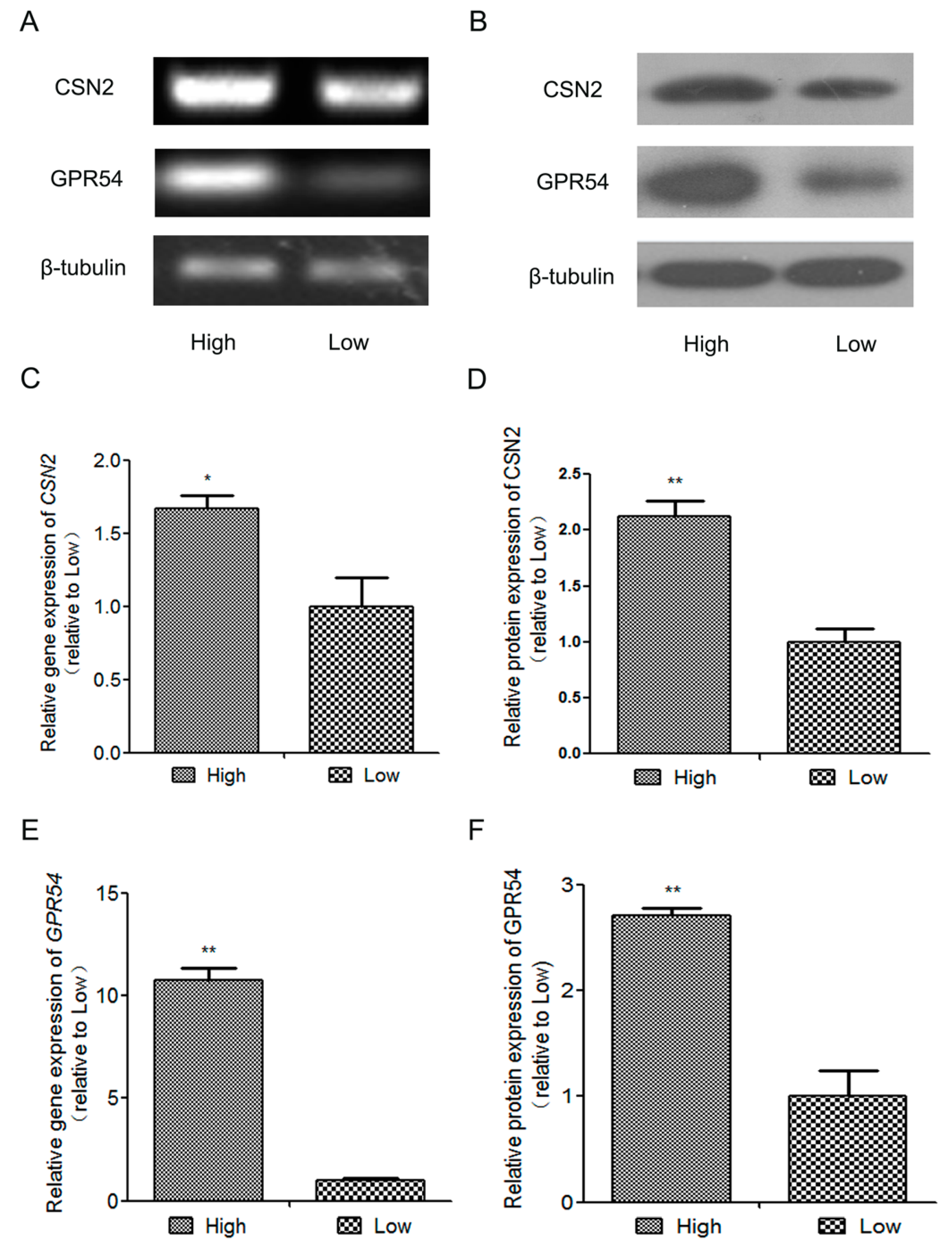
2.1. The Expression of β-Casein(CSN2) and GPR54 in High-Quality Milk and Low-Quality Milk Mammary Gland Tissues from Lactating Holstein Dairy Cows
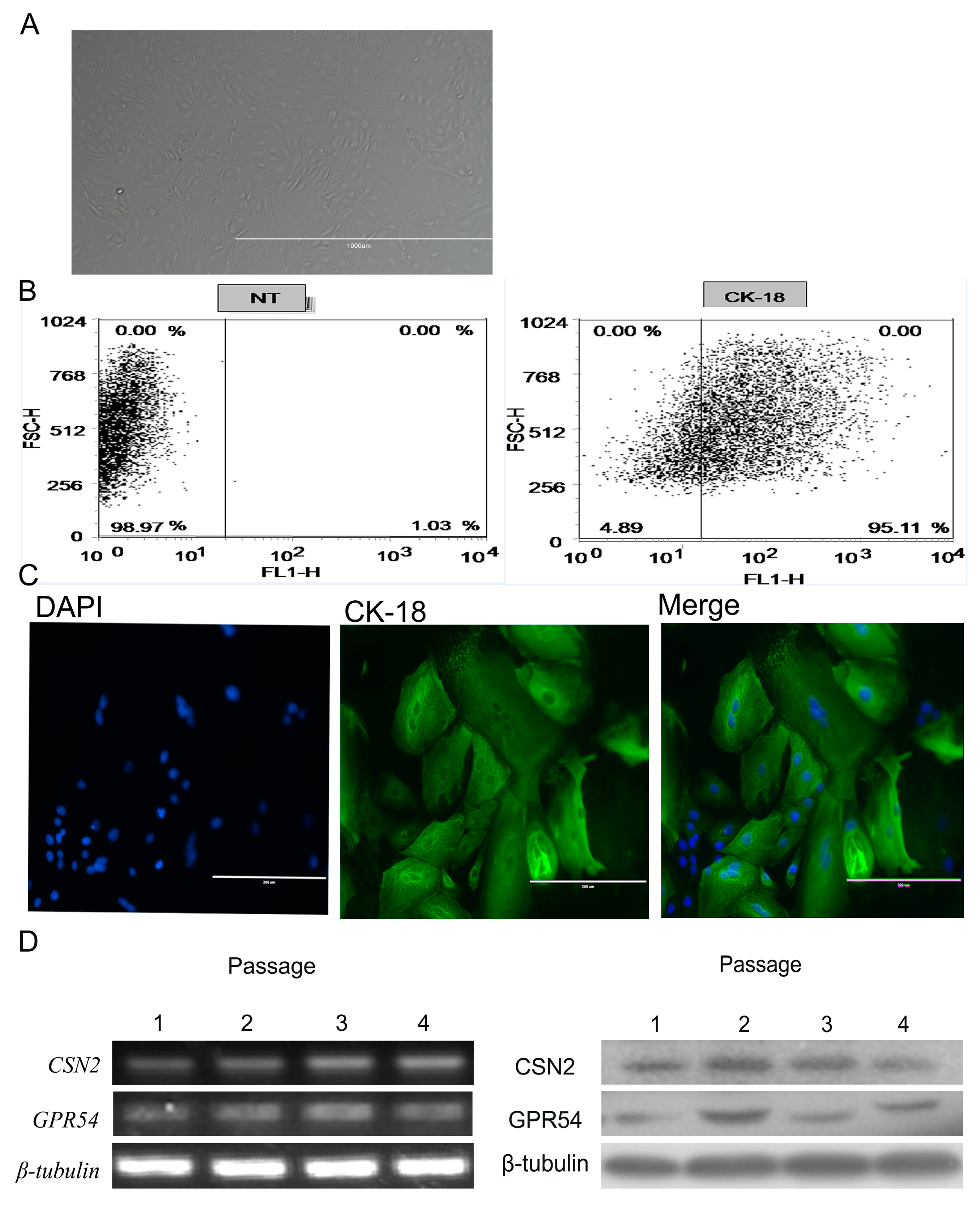
2.2. BMECs Culture and Identification
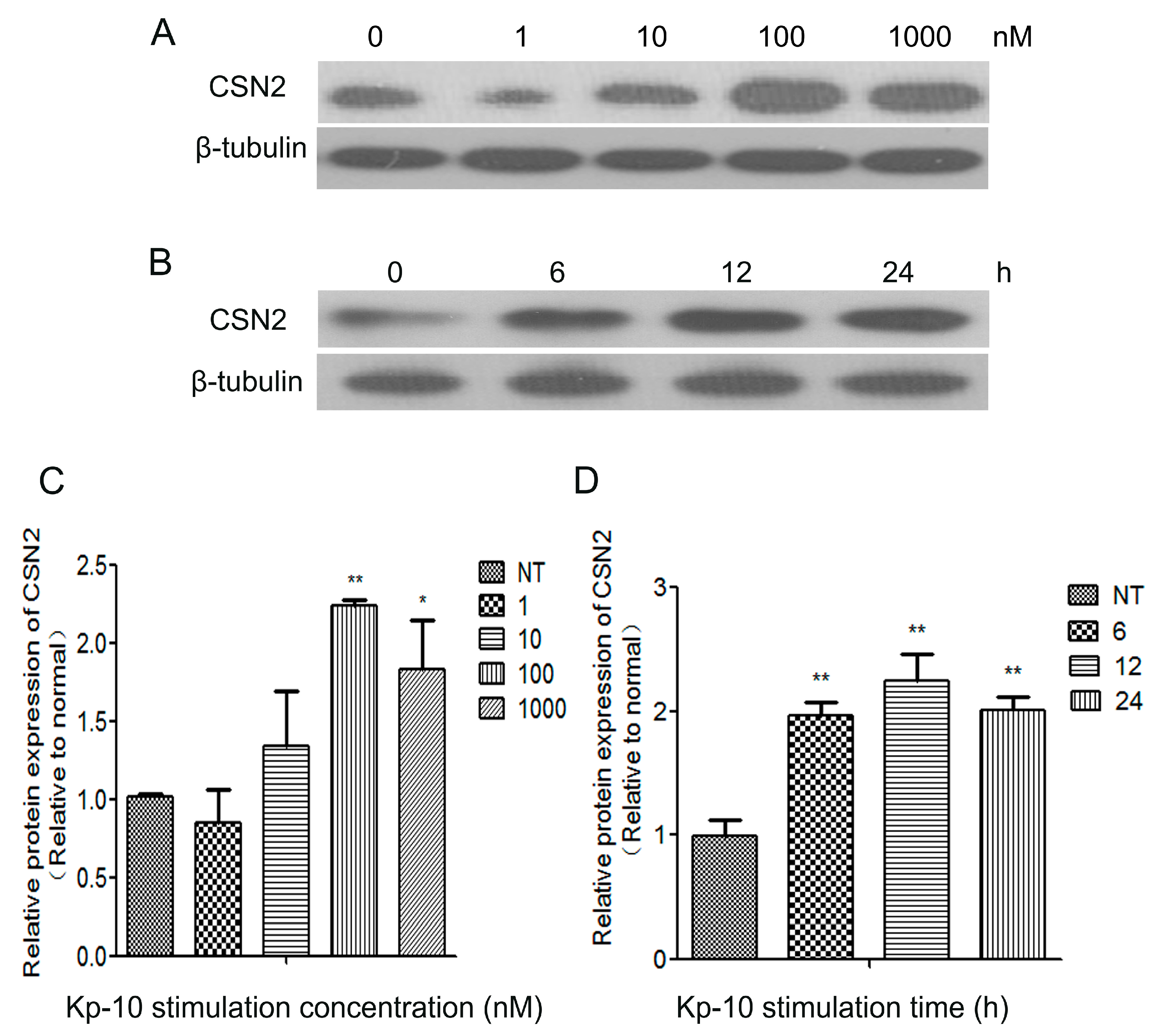
2.3. Kp-10 Promoted CSN2 mRNA and Protein Expression in Bovine Mammary Epithelial Cells (bMECs)
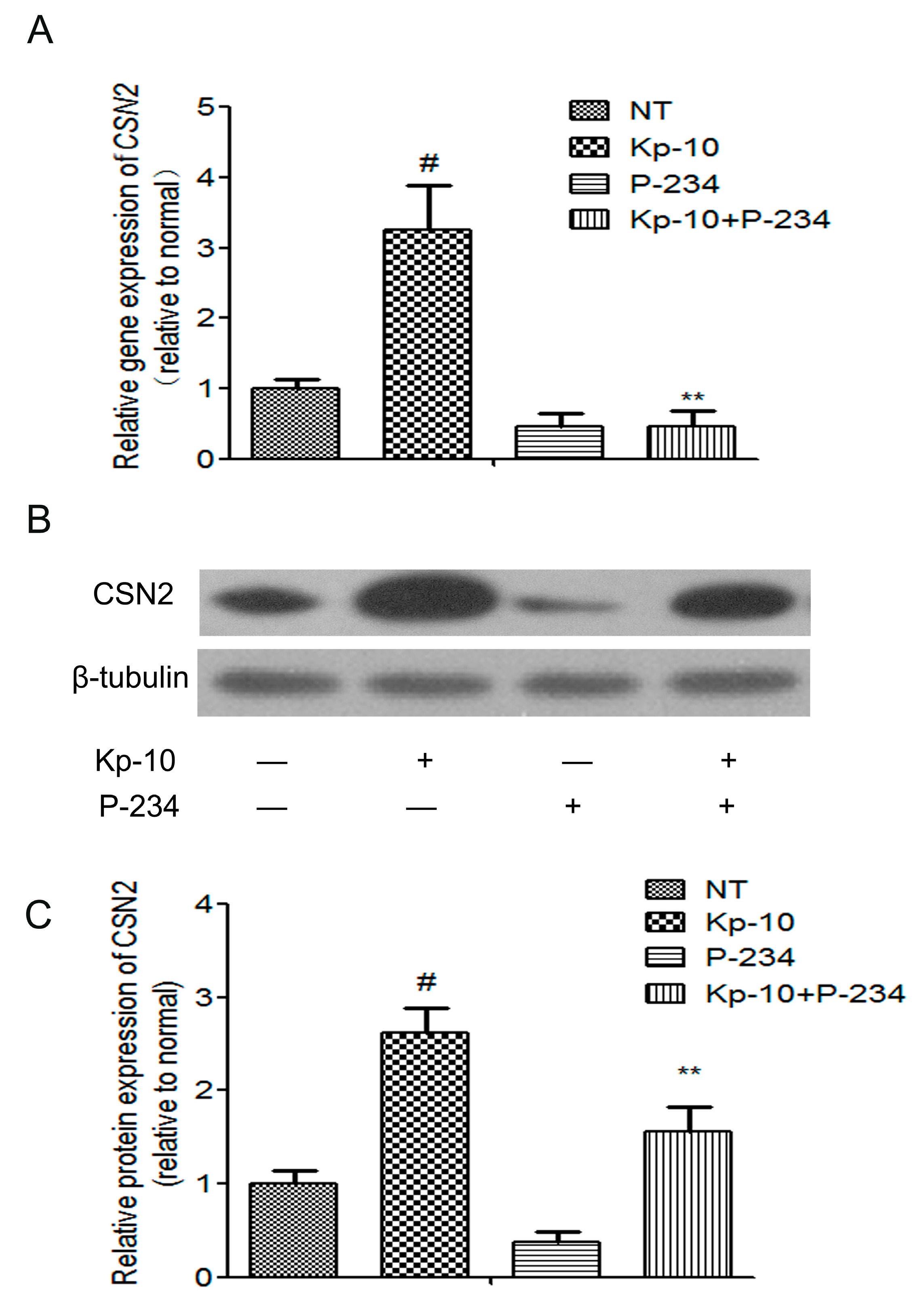
2.4. Peptide-234 Inhibited the Kp-10-Induced mRNA and Protein Expression of CSN2 in BMECs
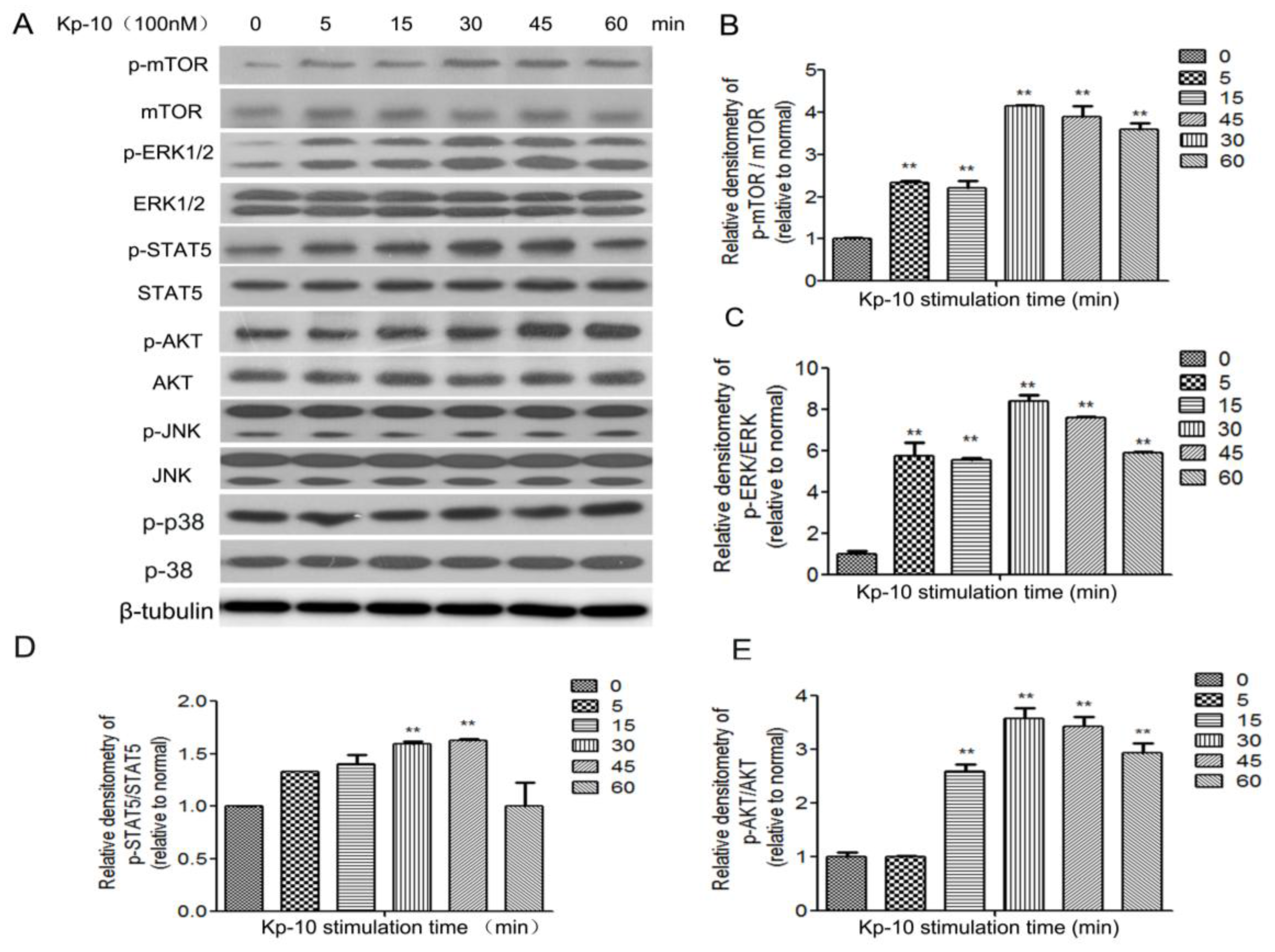
2.5. Kp-10 Phosphorylates mTOR, ERK1/2, STAT5 and AKT in BMECs
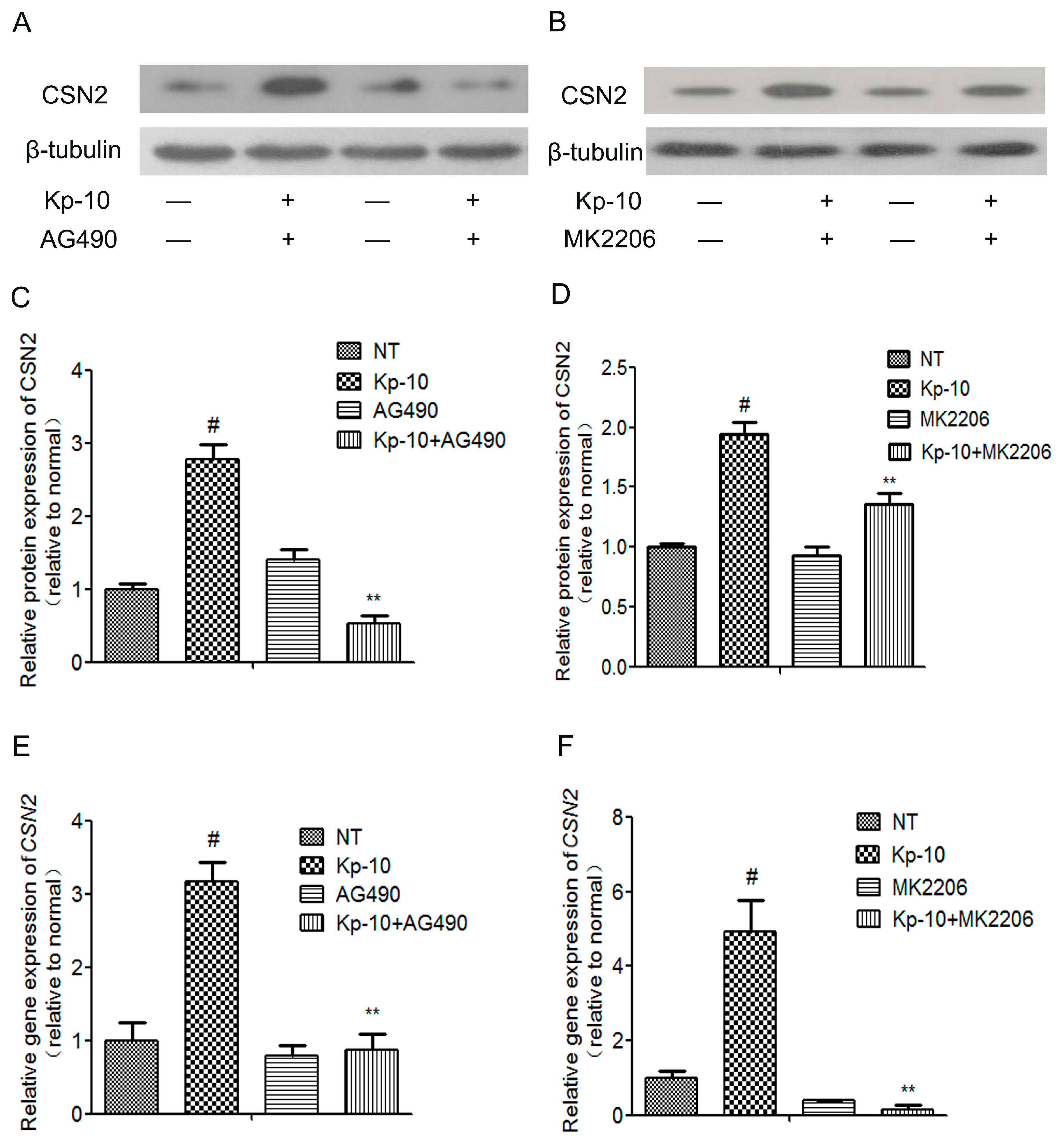
2.6. The mTOR, ERK1/2, STAT5 and AKT Inhibitors Attenuated the Kp-10-Induced Gene and Protein Expression of CSN2 in BMECs
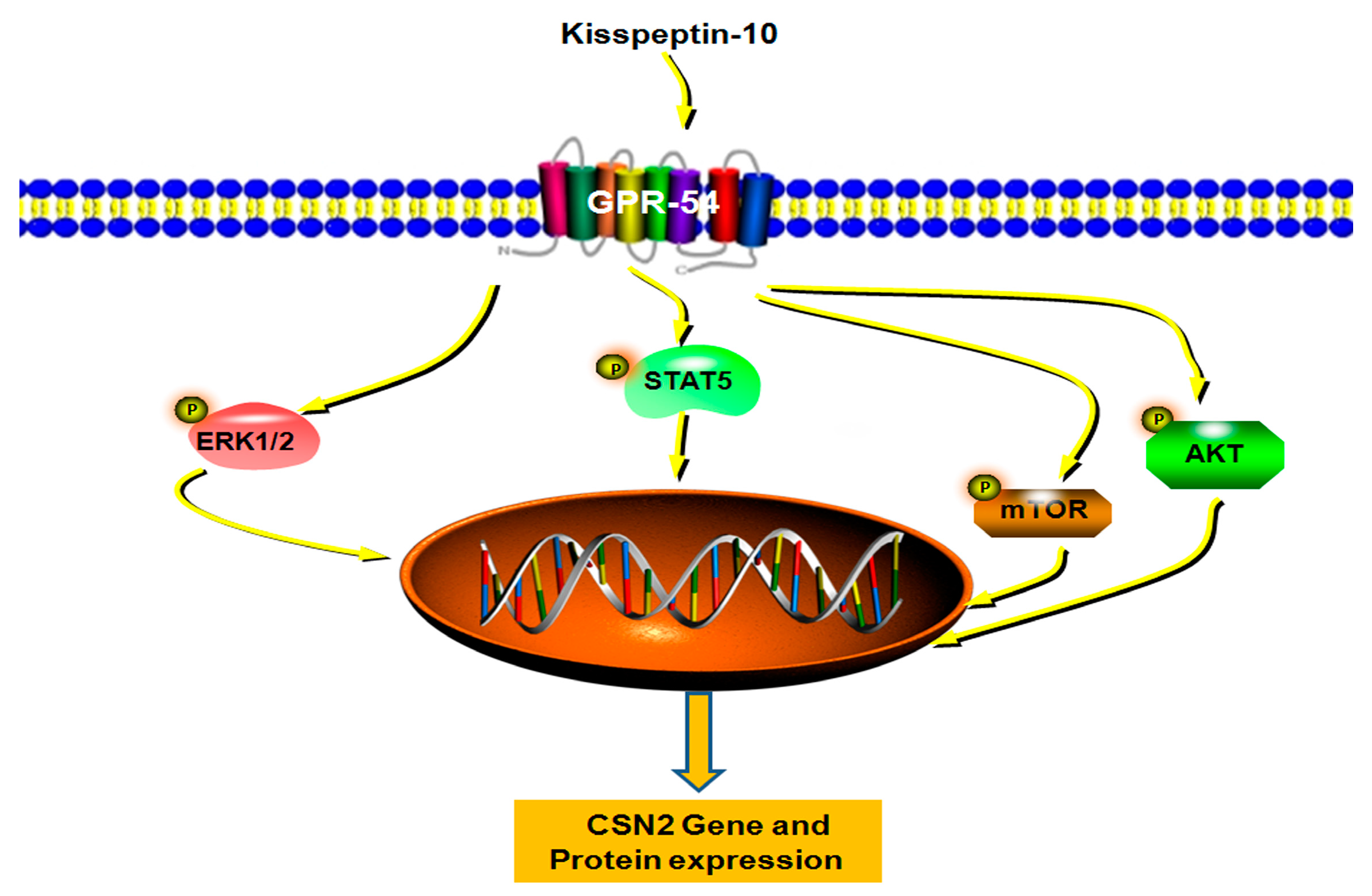
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Antibodies
4.3. Ethics Statement
4.4. Tissue Acquisition, bMECs Culture and Treatments
4.5. RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR
4.6. Western Blot Analysis
4.7. Immunofluorescence Assay
4.8. Flow Cytometry Assay
4.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Kp-10 | Kisspeptin-10 |
| CSN2 | β-casein |
| BMECs | Bovine mammary epithelial cells |
| GPR54 | Guanosine-binding protein coupled receptor 54 |
References
- Han, X.; Fang, Z.; Wang, H.; Jiao, R.; Zhou, J.; Fang, N. CUL4A functions as an oncogene in ovarian cancer and is directly regulated by miR-494. Biochem. Biophys. Res. Commun. 2016, 480, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform. Biol. Insights 2011, 5, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Selvaggi, M.; D’Alessandro, A.G.; Dario, C. Environmental and genetic factors affecting milk yield and quality in three Italian sheep breeds. J. Dairy Res. 2017, 84, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Huderson, B.P.; Velayudhan, B.T.; Pearson, R.E.; Ellis, S.E.; Akers, R.M. Effect of exogenous somatotropin in Holstein calves on mammary gland composition and proliferation. J. Dairy Sci. 2011, 94, 5005–5016. [Google Scholar] [CrossRef] [PubMed]
- McCoard, S.A.; Hayashi, A.A.; Sciascia, Q.; Rounce, J.; Sinclair, B.; McNabb, W.C.; Roy, N.C. Mammary transcriptome analysis of lactating dairy cows following administration of bovine growth hormone. Animal 2016, 10, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, J.; Lv, Q.; Zhang, C.; Xu, S.; Yang, D.; Huang, B.; Zeng, Y.; Gao, Y.; Wang, W. AG and UAG induce β-casein expression via activation of ERK1/2 and AKT pathways. J. Mol. Endocrinol. 2016, 56, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Shahed, A.; Young, K.A. Differential Ovarian Expression of KiSS-1 and GPR-54 During the Estrous Cycle and Photoperiod Induced Recrudescence in Siberian Hamsters (Phodopus sungorus). Mol. Reprod. Dev. 2009, 76, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Goertzen, C.G.; Dragan, M.; Turley, E.; Babwah, A.V.; Bhattacharya, M. KISS1R signaling promotes invadopodia formation in human breast cancer cell via β-arrestin2/ERK. Cell. Signal. 2016, 28, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Martino, N.A.; Rizzo, A.; Pizzi, F.; Dell’Aquila, M.E.; Sciorsci, R.L. Effects of kisspeptin-10 on in vitro proliferation and kisspeptin receptor expression in primary epithelial cell cultures isolated from bovine placental cotyledons of fetuses at the first trimester of pregnancy. Theriogenology 2015, 83, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Radwanska, P.; Kosior-Korzecka, U. Relationships between leptin, KiSS-1/GPR54 expression and TSH secretion from pituitary cells of pubertal ewes in vitro. Res. Vet. Sci. 2016, 105, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, A.A.; Saito, H.; Sawada, T.; Yaegashi, T.; Goto, Y.; Nakajima, Y.; Jin, J.; Yamashita, T.; Sawai, K.; Hashizume, T. The role of sexual steroid hormones in the direct stimulation by Kisspeptin-10 of the secretion of luteinizing hormone, follicle-stimulating hormone and prolactin from bovine anterior pituitary cells. Anim. Reprod. Sci. 2010, 121, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Kadokawa, H.; Suzuki, S.; Hashizume, T. Kisspeptin-10 stimulates the secretion of growth hormone and prolactin directly from cultured bovine anterior pituitary cells. Anim. Reprod. Sci. 2008, 105, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Scott, V.; Brown, C.H. Beyond the GnRH axis: Kisspeptin regulation of the oxytocin system in pregnancy and lactation. Adv. Exp. Med. Biol. 2013, 784, 201–218. [Google Scholar] [PubMed]
- Luque, R.M.; Cordoba-Chacon, J.; Gahete, M.D.; Navarro, V.M.; Tena-Sempere, M.; Kineman, R.D.; Castano, J.P. Kisspeptin Regulates Gonadotroph and Somatotroph Function in Nonhuman Primate Pituitary via Common and Distinct Signaling Mechanisms. Endocrinology 2011, 152, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Kanasaki, H.; Oride, A.; Hara, T.; Mijiddorj, T.; Sukhbaatar, U.; Kyo, S. Interactions between Two Different G Protein-Coupled Receptors in Reproductive Hormone-Producing Cells: The Role of PACAP and Its Receptor PAC1R. Int. J. Mol. Sci. 2016, 17, 1635. [Google Scholar] [CrossRef] [PubMed]
- Kotani, M.; Katagiri, F.; Hirai, T.; Kagawa, J.; Tanaka, I. Plasma kisspeptin levels in lactational amenorrhea. Gynecol. Endocrinol. 2017, 33. [Google Scholar] [CrossRef] [PubMed]
- Papaoiconomou, E.; Lymperi, M.; Petraki, C.; Philippou, A.; Msaouel, P.; Michalopoulou, F.; Kafiri, G.; Vassilakos, G.; Zografos, G.; Koutsilieris, M. Kiss-1/GPR54 protein expression in breast cancer. Anticancer Res. 2014, 34, 1401–1407. [Google Scholar] [PubMed]
- Manjarin, R.; Steibel, J.P.; Kirkwood, R.N.; Taylor, N.P.; Trottier, N.L. Transcript abundance of hormone receptors, mammalian target of rapamycin pathway-related kinases, insulin-like growth factor I, and milk proteins in porcine mammary tissue. J. Anim. Sci. 2012, 90, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Khudhair, N.; Luo, C.; Khalid, A.; Zhang, L.; Zhang, S.; Ao, J.; Li, Q.; Gao, X. 14-3-3γ affects mTOR pathway and regulates lactogenesis in dairy cow mammary epithelial cells. In Vitro Cell. Dev. Biol. Anim. 2015, 51, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shao, W.; Luo, C.; Wu, K.; Yu, X. Co-culture with umbilical cord mesenchymal stem cells promotes the synthesis and mechnism of milk protein in bovine mammary epithelial cells. Chin. J. Cell. Mol. Immunol. 2017, 33, 185–189. [Google Scholar]
- Huang, Y.L.; Zhao, F.; Luo, C.C.; Zhang, X.; Si, Y.; Sun, Z.; Zhang, L.; Li, Q.Z.; Gao, X.J. SOCS3-mediated blockade reveals major contribution of JAK2/STAT5 signaling pathway to lactation and proliferation of dairy cow mammary epithelial cells in vitro. Molecules 2013, 18, 12987–13002. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Chung, H.K.; Lee, H.G.; Lee, H.C.; Woo, J.S.; Lee, S.; Jo, S.J.; Chang, W.K.; Lee, H.T.; Kwon, M.; et al. Cloning and characterization of 5′-untranslated region of porcine β casein gene (CSN2). Domest. Anim. Endocrinol. 2008, 35, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Robinson, G.W.; Wagner, K.U.; Garrett, L.; Wynshaw-Boris, A.; Hennighausen, L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997, 11, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Riedlinger, G.; Miyoshi, K.; Tang, W.; Li, C.; Deng, C.X.; Robinson, G.W.; Hennighausen, L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 2004, 24, 8037–8047. [Google Scholar] [CrossRef] [PubMed]
- Oliver, C.H.; Watson, C.J. Making milk: A new link between STAT5 and Akt1. JAK—STAT 2013, 2, e23228. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.M.; Rudolph, M.C.; McManaman, J.L.; Neville, M.C. Key stages in mammary gland development. Secretory activation in the mammary gland: It’s not just about milk protein synthesis! Breast Cancer Res. 2007, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Murney, R.; Stelwagen, K.; Wheeler, T.T.; Margerison, J.K.; Singh, K. The effects of milking frequency on insulin-like growth factor I signaling within the mammary gland of dairy cows. J. Dairy Sci. 2015, 98, 5422–5428. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.E.; Cooke, J.H.; Baxter, J.E.; Parkinson, J.R.; Bataveljic, A.; Ghatei, M.A.; Bloom, S.R.; Murphy, K.G. A kisspeptin-10 analog with greater in vivo bioactivity than kisspeptin-10. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E296–E303. [Google Scholar] [CrossRef] [PubMed]
- Ahow, M.; Min, L.; Pampillo, M.; Nash, C.; Wen, J.; Soltis, K.; Carroll, R.S.; Glidewell-Kenney, C.A.; Mellon, P.L.; Bhattacharya, M.; et al. KISS1R signals independently of Gαq/11 and triggers LH secretion via the beta-arrestin pathway in the male mouse. Endocrinology 2014, 155, 4433–4446. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.K.; Hiney, J.K.; Dees, W.L. Manganese-Stimulated Kisspeptin Is Mediated by the IGF-1/Akt/Mammalian Target of Rapamycin Pathway in the Prepubertal Female Rat. Endocrinology 2016, 157, 3233–3241. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.A.; Furigo, I.C.; Zampieri, T.T.; Bohlen, T.M.; de Paula, D.G.; Franci, C.R.; Donato, J.; Frazao, R. STAT5 signaling in kisspeptin cells regulates the timing of puberty. Mol. Cell. Endocrinol. 2017, 448, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kadokawa, H.; Hashizume, T. Direct kisspeptin-10 stimulation on luteinizing hormone secretion from bovine and porcine anterior pituitary cells. Anim. Reprod. Sci. 2008, 103, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Gottsch, M.L.; Clifton, D.K.; Steiner, R.A. Kisspepeptin-GPR54 signaling in the neuroendocrine reproductive axis. Mol. Cell. Endocrinol. 2006, 254–255, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Pompolo, S.; Pereira, A.; Estrada, K.M.; Clarke, I.J. Colocalization of kisspeptin and gonadotropin-releasing hormone in the ovine brain. Endocrinology 2006, 147, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Haug, A.; Hostmark, A.T.; Harstad, O.M. Bovine milk in human nutrition—A review. Lipids Health Dis. 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Wlodek, M.E.; Westcott, K.T.; Serruto, A.; O’Dowd, R.; Wassef, L.; Ho, P.W.; Moseley, J.M. Impaired mammary function and parathyroid hormone-related protein during lactation in growth-restricted spontaneously hypertensive rats. J. Endocrinol. 2003, 178, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Barash, I.; Rhoads, R.E. Insulin and prolactin synergistically stimulate β-casein messenger ribonucleic acid translation by cytoplasmic polyadenylation. Mol. Endocrinol. 2004, 18, 1670–1686. [Google Scholar] [CrossRef] [PubMed]
- Elovic, A.; Wong, D.T.; Weller, P.F.; Matossian, K.; Galli, S.J. Expression of transforming growth factors-α and β1 messenger RNA and product by eosinophils in nasal polyps. J. Allergy Clin. Immunol. 1994, 93, 864–869. [Google Scholar] [CrossRef]
- Robinson, S.D.; Roberts, A.B.; Daniel, C.W. TGF β suppresses casein synthesis in mouse mammary explants and may play a role in controlling milk levels during pregnancy. J. Cell Biol. 1993, 120, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wang, Y.; Yu, Z.; Hu, L.; Liu, C.; Gao, X.; Zheng, S. WISP3 (CCN6) Regulates Milk Protein Synthesis and Cell Growth Through mTOR Signaling in Dairy Cow Mammary Epithelial Cells. DNA Cell Biol. 2015, 34, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.X.; Wei, C.J.; Si, Y.; Luo, C.C.; Lv, W.; Lin, Y.; Cui, Y.J.; Gao, X.J. Tudor-SN Regulates Milk Synthesis and Proliferation of Bovine Mammary Epithelial Cells. Int. J. Mol. Sci. 2015, 16, 29936–29947. [Google Scholar] [CrossRef] [PubMed]
- Hanchate, N.K.; Parkash, J.; Bellefontaine, N.; Mazur, D.; Colledge, W.H.; de Tassigny, X.D.; Prevot, V. Kisspeptin-GPR54 Signaling in Mouse NO-Synthesizing Neurons Participates in the Hypothalamic Control of Ovulation. J. Neurosci. 2012, 32, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Roa, J.; Garcia-Galiano, D.; Varela, L.; Sanchez-Garrido, M.A.; Pineda, R.; Castellano, J.M.; Ruiz-Pino, F.; Romero, M.; Aguilar, E.; Lopez, M.; et al. The Mammalian Target of Rapamycin as Novel Central Regulator of Puberty Onset via Modulation of Hypothalamic Kiss1 System. Endocrinology 2009, 150, 5016–5026. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S.E.; Herbison, A.E.; Grattan, D.R. Prolactin Regulation of Kisspeptin Neurones in the Mouse Brain and its Role in the Lactation-Induced Suppression of Kisspeptin Expression. J. Neuroendocrinol. 2014, 26, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Liu, H.Y.; Zhou, M.M.; Liu, J.X. Establishment and characterization of a lactating bovine mammary epithelial cell model for the study of milk synthesis. Cell Biol. Int. 2010, 34, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Baumgard, L.H.; Matitashvili, E.; Corl, B.A.; Dwyer, D.A.; Bauman, D.E. trans-10, cis-12 conjugated linoleic acid decreases lipogenic rates and expression of genes involved in milk lipid synthesis in dairy cows. J. Dairy Sci. 2002, 85, 2155–2163. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, G.; Ao, C.; Zhang, S.; Qiu, M.; Wang, X.; Wu, Y.; Erdene, K.; Jin, L.; Lei, C.; et al. Feeding a high-concentrate corn straw diet increased the release of endotoxin in the rumen and pro-inflammatory cytokines in the mammary gland of dairy cows. BMC Vet. Res. 2014, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Qiu, M.; Ao, C.; Zhou, J.; Khas, E.; Wang, X.; Zhang, Z.; Yang, Y. Feeding a high-concentrate corn straw diet induced epigenetic alterations in the mammary tissue of dairy cows. PLoS ONE 2014, 9, e107659. [Google Scholar] [CrossRef] [PubMed]

| Gene | Sequences | Length (bp) |
|---|---|---|
| GPR54 | (F) 5′-TGGCAATGCAGCCCTTGT-3′ | 149 |
| (R) 5′-GAAAAGCGGCACCAACCAG-3′ | ||
| CSN2 | (F) 5′-AACAGCCTCCCACAAAC-3′ | 158 |
| (R) 5′-AGCCATAGCCTCCTTCAC-3′ | ||
| β-tubulin | (F) 5′-TCACCAACTGGGACGACA-3′ | 206 |
| (R) 5′-GCATACAGGGACAGCACA-3′ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Liu, J.; Huang, B.; Kan, X.; Chen, G.; Wang, W.; Fu, S. Kisspeptin-10 Induces β-Casein Synthesis via GPR54 and Its Downstream Signaling Pathways in Bovine Mammary Epithelial Cells. Int. J. Mol. Sci. 2017, 18, 2621. https://doi.org/10.3390/ijms18122621
Sun J, Liu J, Huang B, Kan X, Chen G, Wang W, Fu S. Kisspeptin-10 Induces β-Casein Synthesis via GPR54 and Its Downstream Signaling Pathways in Bovine Mammary Epithelial Cells. International Journal of Molecular Sciences. 2017; 18(12):2621. https://doi.org/10.3390/ijms18122621
Chicago/Turabian StyleSun, Jianhua, Juxiong Liu, Bingxu Huang, Xingchi Kan, Guangxin Chen, Wei Wang, and Shoupeng Fu. 2017. "Kisspeptin-10 Induces β-Casein Synthesis via GPR54 and Its Downstream Signaling Pathways in Bovine Mammary Epithelial Cells" International Journal of Molecular Sciences 18, no. 12: 2621. https://doi.org/10.3390/ijms18122621
APA StyleSun, J., Liu, J., Huang, B., Kan, X., Chen, G., Wang, W., & Fu, S. (2017). Kisspeptin-10 Induces β-Casein Synthesis via GPR54 and Its Downstream Signaling Pathways in Bovine Mammary Epithelial Cells. International Journal of Molecular Sciences, 18(12), 2621. https://doi.org/10.3390/ijms18122621