Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging in the Study of Gastric Cancer: A Mini Review
Abstract
1. Introduction
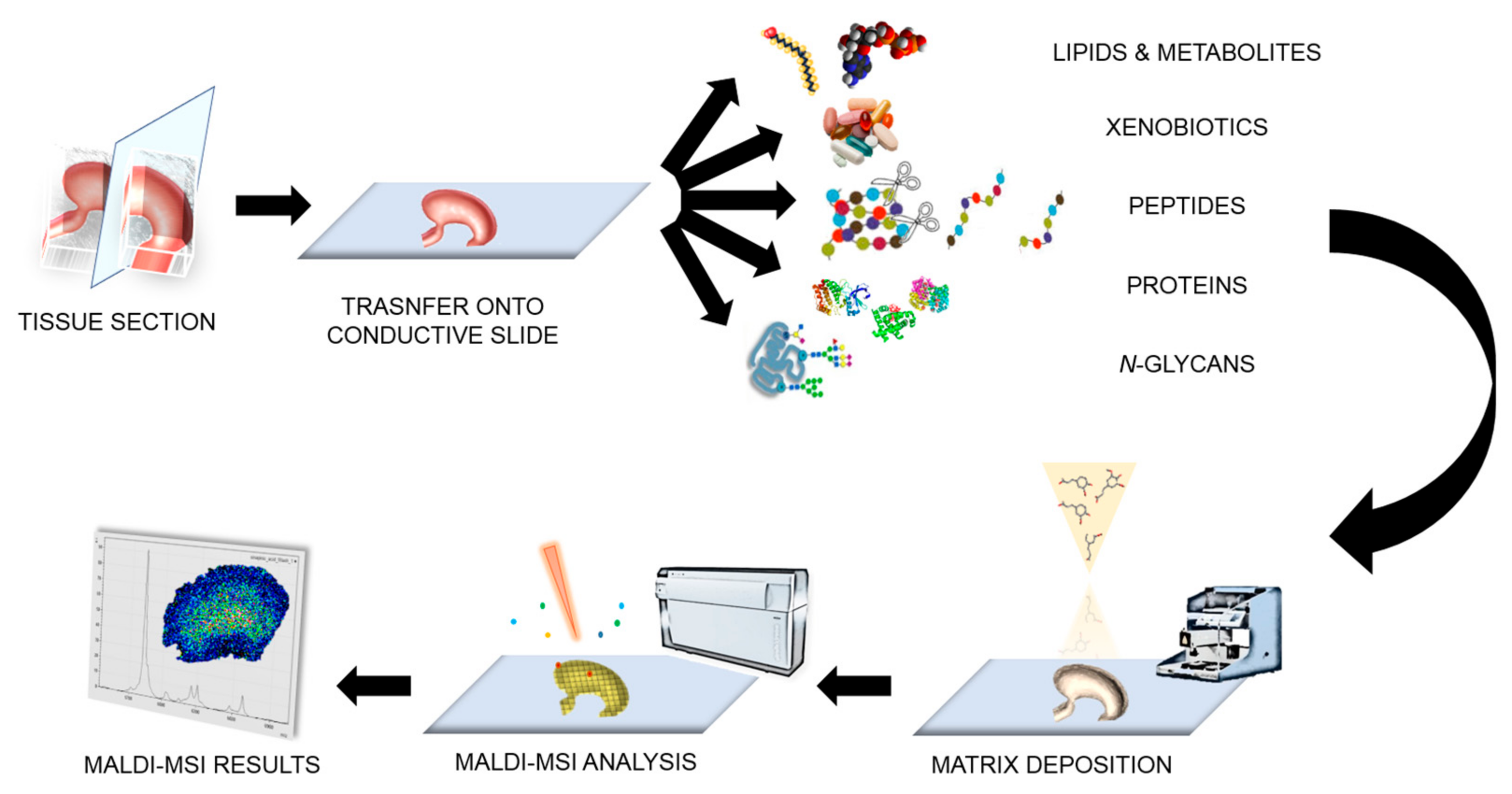
2. Matrix-Assisted Laser Desorption/Ionisation-Mass Spectrometry Imaging (MALDI-MSI) in a Nutshell
2.1. Sample Preparation
2.2. Instrumental Advancements
2.3. Statistical Analysis and Data Elaboration
3. Applications in Gastric Cancer
4. Concluding Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| FF | Fresh-frozen |
| FFPE | Formalin-fixed paraffin-embedded |
| FTICR | Fourier transform ion cyclotron resonance |
| GC | Gastric cancer |
| HCA | Hierarchical clustering analysis |
| HER2/neu | Human epidermal growth factor receptor 2 |
| HNP | Human neutrophil peptide |
| MALDI | Matrix-assisted laser desorption/ionisation |
| MSI | Mass spectrometry imaging |
| PCA | Principal component analysis |
| ROCK 1/2 | RHO-associated protein kinase 1 and 2 |
| ToF | Time of flight |
| t-SNE | t-Distributed stochastic neighbour embedding |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012: Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J. Gastroenterol. 2014, 20, 13648–13657. [Google Scholar] [CrossRef] [PubMed]
- Chugtai, K.; Heeren, R. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 2010, 110, 3237–3277. [Google Scholar] [CrossRef] [PubMed]
- Casadonte, R.; Longuespée, R.; Kriegsmann, J.; Kriegsmann, M. MALDI IMS and Cancer Tissue Microarrays. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 134, pp. 173–200. ISBN 978-0-12-805249-5. [Google Scholar]
- Kriegsmann, J.; Kriegsmann, M.; Casadonte, R. MALDI TOF imaging mass spectrometry in clinical pathology: A valuable tool for cancer diagnostics. Int. J. Oncol. 2015, 46, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Lee, Y.; Lee, J.E. Recent advances in mass spectrometry-based proteomics of gastric cancer. World J. Gastroenterol. 2016, 22, 8283–8293. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M.; Walch, A. MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Investig. 2015, 95, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Caprioli, R.M.; Farmer, T.B.; Gile, J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997, 69, 4751–4760. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.C.; Han, J.; Borchers, C.H. Recent advancements in matrix-assisted laser desorption/ionization mass spectrometry imaging. Curr. Opin. Biotechnol. 2017, 43, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.J.A.; Nilsson, A.; Borg, D.; Langridge-Smith, P.R.R.; Harrison, D.J.; Mackay, C.L.; Iverson, S.L.; Andrén, P.E. Conductive carbon tape used for support and mounting of both whole animal and fragile heat-treated tissue sections for MALDI MS imaging and quantitation. J. Proteom. 2012, 75, 4912–4920. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Goodwin, R.J.A.; Shariatgorji, M.; Vallianatou, T.; Webborn, P.J.H.; Andrén, P.E. Mass Spectrometry Imaging in Drug Development. Anal. Chem. 2015, 87, 1437–1455. [Google Scholar] [CrossRef] [PubMed]
- Greer, T.; Sturm, R.; Li, L. Mass spectrometry imaging for drugs and metabolites. J. Proteom. 2011, 74, 2617–2631. [Google Scholar] [CrossRef] [PubMed]
- Giordano, S.; Morosi, L.; Veglianese, P.; Licandro, S.A.; Frapolli, R.; Zucchetti, M.; Cappelletti, G.; Falciola, L.; Pifferi, V.; Visentin, S.; et al. 3D Mass Spectrometry Imaging Reveals a very Heterogeneous Drug Distribution in Tumors. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.G.; Bokhart, M.T.; Sykes, C.; Adamson, L.; Fedoriw, Y.; Luciw, P.A.; Muddiman, D.C.; Kashuba, A.D.M.; Rosen, E.P. Mass Spectrometry Imaging Reveals Heterogeneous Efavirenz Distribution within Putative HIV Reservoirs. Antimicrob. Agents Chemother. 2015, 59, 2944–2948. [Google Scholar] [CrossRef] [PubMed]
- Chumbley, C.W.; Reyzer, M.L.; Allen, J.L.; Marriner, G.A.; Via, L.E.; Barry, C.E.; Caprioli, R.M. Absolute Quantitative MALDI Imaging Mass Spectrometry: A Case of Rifampicin in Liver Tissues. Anal. Chem. 2016, 88, 2392–2398. [Google Scholar] [CrossRef] [PubMed]
- Groseclose, M.R.; Castellino, S. A Mimetic Tissue Model for the Quantification of Drug Distributions by MALDI Imaging Mass Spectrometry. Anal. Chem. 2013, 85, 10099–10106. [Google Scholar] [CrossRef] [PubMed]
- Longuespée, R.; Casadonte, R.; Kriegsmann, M.; Pottier, C.; Picard de Muller, G.; Delvenne, P.; Kriegsmann, J.; De Pauw, E. MALDI mass spectrometry imaging: A cutting-edge tool for fundamental and clinical histopathology. Proteom. Clin. Appl. 2016, 10, 701–719. [Google Scholar] [CrossRef] [PubMed]
- Stauber, J.; MacAleese, L.; Franck, J.; Claude, E.; Snel, M.; Kaletas, B.K.; Wiel, I.M.V.D.; Wisztorski, M.; Fournier, I.; Heeren, R.M.A. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 338–347. [Google Scholar] [CrossRef] [PubMed]
- De Sio, G.; Smith, A.J.; Galli, M.; Garancini, M.; Chinello, C.; Bono, F.; Pagni, F.; Magni, F. A MALDI-Mass Spectrometry Imaging method applicable to different formalin-fixed paraffin-embedded human tissues. Mol. Biosyst. 2015, 11, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Gorzolka, K.; Walch, A. MALDI mass spectrometry imaging of formalin-fixed paraffin-embedded tissues in clinical research. Histol. Histopathol. 2014, 29, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Buck, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.-P.; Kuppen, P.J.K.; van de Velde, C.J.H.; Weirich, G.; et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat. Protoc. 2016, 11, 1428–1443. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.T.; Ho, Y.Y.; Kaur, G.; Oehler, M.K.; Everest-Dass, A.V.; Packer, N.H.; Hoffmann, P. N-Glycan matrix-assisted laser desorption/ionization mass spectrometry imaging protocol for formalin-fixed paraffin-embedded tissues. Rapid Commun. Mass Spectrom. 2017, 31, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Heijs, B.; Holst, S.; Briaire-de Bruijn, I.H.; van Pelt, G.W.; de Ru, A.H.; van Veelen, P.A.; Drake, R.R.; Mehta, A.S.; Mesker, W.E.; Tollenaar, R.A.; et al. Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Anal. Chem. 2016, 88, 7745–7753. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.A.; Reyzer, M.L.; Caprioli, R.M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: Practical aspects of sample preparation. J. Mass Spectrom. 2003, 38, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.J.A.; Pennington, S.R.; Pitt, A.R. Protein and peptides in pictures: Imaging with MALDI mass spectrometry. Proteomics 2008, 8, 3785–3800. [Google Scholar] [CrossRef] [PubMed]
- Patel, E. Fresh Frozen Versus Formalin-Fixed Paraffin Embedded for Mass Spectrometry Imaging. Methods Mol. Biol. 2017, 1618, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, R.; Wisztorski, M.; Desmons, A.; Tabet, J.C.; Day, R.; Salzet, M.; Fournier, I. MALDI-MS direct tissue analysis of proteins: Improving signal sensitivity using organic treatments. Anal. Chem. 2006, 78, 7145–7153. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.J.; Liu, C.B.; Wu, H.-W. A simple desalting method for direct MALDI mass spectrometry profiling of tissue lipids. J. Lipid Res. 2011, 52, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Chaurand, P. Advances in tissue section preparation for MALDI imaging MS. Bioanalysis 2014, 6, 967–982. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.J.A. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. J. Proteom. 2012, 75, 4893–4911. [Google Scholar] [CrossRef] [PubMed]
- Boskamp, T.; Lachmund, D.; Oetjen, J.; Cordero Hernandez, Y.; Trede, D.; Maass, P.; Casadonte, R.; Kriegsmann, J.; Warth, A.; Dienemann, H.; et al. A new classification method for MALDI imaging mass spectrometry data acquired on formalin-fixed paraffin-embedded tissue samples. Biochim. Biophys. Acta 2017, 1865, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Groseclose, M.R.; Massion, P.P.; Chaurand, P.; Caprioli, R.M. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 2008, 8, 3715–3724. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, J.O.R.; Oehler, M.K.; McColl, S.R.; Hoffmann, P. Citric acid antigen retrieval (CAAR) for tryptic peptide imaging directly on archived formalin-fixed paraffin-embedded tissue. J. Proteome Res. 2010, 9, 4315–4328. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Z.; Nichols, A.; Liu, D. Direct Identification and Quantification of Aspartyl Succinimide in an IgG2 mAb by RapiGest Assisted Digestion. Anal. Chem. 2009, 81, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Patel, E.; Clench, M.R.; West, A.; Marshall, P.S.; Marshall, N.; Francese, S. Alternative surfactants for improved efficiency of in situ tryptic proteolysis of fingermarks. J. Am. Soc. Mass Spectrom. 2015, 26, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Franck, J.; Arafah, K.; Barnes, A.; Wisztorski, M.; Salzet, M.; Fournier, I. Improving Tissue Preparation for Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging. Part 1: Using Microspotting. Anal. Chem. 2009, 81, 8193–8202. [Google Scholar] [CrossRef] [PubMed]
- Beine, B.; Diehl, H.C.; Meyer, H.E.; Henkel, C. Tissue MALDI Mass Spectrometry Imaging (MALDI MSI) of Peptides. In Proteomis in Systems Biology; Reinders, J., Ed.; Springer: New York, NY, USA, 2016; Volume 1394, pp. 129–150. ISBN 978-1-4939-3339-6. [Google Scholar]
- Hankin, J.A.; Barkley, R.M.; Murphy, R.C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007, 18, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Caprioli, R.M. Matrix pre-coated targets for high throughput MALDI imaging of proteins: Matrix pre-coated targets for MALDI imaging MS. J. Mass Spectrom. 2014, 49, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Grove, K.J.; Frappier, S.L.; Caprioli, R.M. Matrix Pre-Coated MALDI MS Targets for Small Molecule Imaging in Tissues. J. Am. Soc. Mass Spectrom. 2011, 22, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Kaletaş, B.K.; van der Wiel, I.M.; Stauber, J.; Dekker, L.J.; Güzel, C.; Kros, J.M.; Luider, T.M.; Heeren, R.M.A. Sample preparation issues for tissue imaging by imaging MS. Proteomics 2009, 9, 2622–2633. [Google Scholar] [CrossRef] [PubMed]
- Franck, J.; Longuespée, R.; Wisztorski, M.; Van Remoortere, A.; Van Zeijl, R.; Deelder, A.; Salzet, M.; McDonnell, L.; Fournier, I. MALDI mass spectrometry imaging of proteins exceeding 30,000 daltons. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2010, 16, BR293–BR299. [Google Scholar]
- Van Remoortere, A.; van Zeijl, R.J.M.; van den Oever, N.; Franck, J.; Longuespée, R.; Wisztorski, M.; Salzet, M.; Deelder, A.M.; Fournier, I.; McDonnell, L.A. MALDI imaging and profiling MS of higher mass proteins from tissue. J. Am. Soc. Mass Spectrom. 2010, 21, 1922–1929. [Google Scholar] [CrossRef] [PubMed]
- Mainini, V.; Bovo, G.; Chinello, C.; Gianazza, E.; Grasso, M.; Cattoretti, G.; Magni, F. Detection of high molecular weight proteins by MALDI imaging mass spectrometry. Mol. Biosyst. 2013, 9, 1101. [Google Scholar] [CrossRef] [PubMed]
- Calvano, C.D.; Carulli, S.; Palmisano, F. Aniline/α-cyano-4-hydroxycinnamic acid is a highly versatile ionic liquid for matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Fagerer, S.; Nielsen, S.; Ibáñez, A.; Zenobi, R. Matrix-assisted laser desorption/ionization matrices for negative-mode metabolomics. Eur. J. Mass Spectrom. 2013, 19, 39. [Google Scholar] [CrossRef]
- Garate, J.; Fernández, R.; Lage, S.; Bestard-Escalas, J.; Lopez, D.H.; Reigada, R.; Khorrami, S.; Ginard, D.; Reyes, J.; Amengual, I.; et al. Imaging mass spectrometry increased resolution using 2-mercaptobenzothiazole and 2,5-diaminonaphtalene matrices: Application to lipid distribution in human colon. Anal. Bioanal. Chem. 2015, 407, 4697–4708. [Google Scholar] [CrossRef] [PubMed]
- Spraggins, J.M.; Rizzo, D.G.; Moore, J.L.; Noto, M.J.; Skaar, E.P.; Caprioli, R.M. Next-generation technologies for spatial proteomics: Integrating ultra-high speed MALDI-TOF and high mass resolution MALDI FTICR imaging mass spectrometry for protein analysis. Proteomics 2016, 16, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Cornett, D.S.; Frappier, S.L.; Caprioli, R.M. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 2008, 80, 5648–5653. [Google Scholar] [CrossRef] [PubMed]
- Römpp, A.; Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013, 139, 759–783. [Google Scholar] [CrossRef] [PubMed]
- Kettling, H.; Vens-Cappell, S.; Soltwisch, J.; Pirkl, A.; Haier, J.; Müthing, J.; Dreisewerd, K. MALDI Mass Spectrometry Imaging of Bioactive Lipids in Mouse Brain with a Synapt G2-S Mass Spectrometer Operated at Elevated Pressure: Improving the Analytical Sensitivity and the Lateral Resolution to Ten Micrometers. Anal. Chem. 2014, 86, 7798–7805. [Google Scholar] [CrossRef] [PubMed]
- Ogrinc Potočnik, N.; Porta, T.; Becker, M.; Heeren, R.M.A.; Ellis, S.R. Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun. Mass Spectrom. 2015, 29, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Oetjen, J.; Veselkov, K.; Watrous, J.; McKenzie, J.S.; Becker, M.; Hauberg-Lotte, L.; Kobarg, J.H.; Strittmatter, N.; Mróz, A.K.; Hoffmann, F.; et al. Benchmark datasets for 3D MALDI- and DESI-imaging mass spectrometry. GigaScience 2015, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.H.; Doonan, R.J.; Daskalopoulou, S.S.; Dufresne, M.; Lenglet, S.; Montecucco, F.; Thomas, A.; Chaurand, P. Three-dimensional imaging MS of lipids in atherosclerotic plaques: Open-source methods for reconstruction and analysis. Proteomics 2016, 16, 1642–1651. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.G.; Van de Plas, R.; Rose, K.L.; Hill, S.; Schey, K.L.; Solga, A.C.; Gutmann, D.H.; Caprioli, R.M. 3-D imaging mass spectrometry of protein distributions in mouse Neurofibromatosis 1 (NF1)-associated optic glioma. J. Proteom. 2016, 149, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Trede, D.; Kobarg, J.H.; Oetjen, J.; Thiele, H.; Maass, P.; Alexandrov, T. On the Importance of Mathematical Methods for Analysis of MALDI-Imaging Mass Spectrometry Data. J. Integr. Bioinform. 2012, 9, 1–11. [Google Scholar]
- Ràfols, P.; Vilalta, D.; Brezmes, J.; Cañellas, N.; del Castillo, E.; Yanes, O.; Ramírez, N.; Correig, X. Signal preprocessing, multivariate analysis and software tools for MA(LDI)-TOF mass spectrometry imaging for biological applications. Mass Spectrom. Rev. 2016, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.L.; Cornett, D.S.; Mobley, J.A.; Andersson, M.; Seeley, E.H.; Chaurand, P.; Caprioli, R.M. Processing MALDI Mass Spectra to Improve Mass Spectral Direct Tissue Analysis. Int. J. Mass Spectrom. 2007, 260, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Zoppis, I.; Smith, A.; Magni, F.; Mauri, G. Machine learning approaches in MALDI-MSI: Clinical applications. Expert Rev. Proteom. 2016, 13, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Deininger, S.-O.; Cornett, D.S.; Paape, R.; Becker, M.; Pineau, C.; Rauser, S.; Walch, A.; Wolski, E. Normalization in MALDI-TOF imaging datasets of proteins: Practical considerations. Anal. Bioanal. Chem. 2011, 401, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.A.; Deininger, S.-O.; Hogendoorn, P.C.W.; Deelder, A.M.; McDonnell, L.A. Imaging mass spectrometry statistical analysis. J. Proteom. 2012, 75, 4962–4989. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, F.; Contreras, P. Algorithms for hierarchical clustering: An overview. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 86–97. [Google Scholar] [CrossRef]
- Alexandrov, T.; Becker, M.; Deininger, S.-O.; Ernst, G.; Wehder, L.; Grasmair, M.; von Eggeling, F.; Thiele, H.; Maass, P. Spatial Segmentation of Imaging Mass Spectrometry Data with Edge-Preserving Image Denoising and Clustering. J. Proteome Res. 2010, 9, 6535–6546. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Abdelmoula, W.M.; Balluff, B.; Englert, S.; Dijkstra, J.; Reinders, M.J.T.; Walch, A.; McDonnell, L.A.; Lelieveldt, B.P.F. Data-driven identification of prognostic tumor subpopulations using spatially mapped t-SNE of mass spectrometry imaging data. Proc. Natl. Acad. Sci. USA 2016, 113, 12244–12249. [Google Scholar] [CrossRef] [PubMed]
- Kotsiantis, S.B. Supervised Machine Learning: A Review of Classification Techniques. Informatica 2007, 31, 249–268. [Google Scholar]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000; ISBN 978-0-511-80138-9. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Takahashi, T.; Saikawa, Y.; Kitagawa, Y. Gastric Cancer: Current Status of Diagnosis and Treatment. Cancers 2013, 5, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, M.; Nishizawa, T.; Ueda, H.; Gotoh, K.; Tanaka, A.; Hayashi, A.; Yamamoto, S.; Tatsuno, K.; Katoh, H.; Watanabe, Y.; et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 2014, 46, 583–587. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Wei, L.; Surma, M.; Shi, S.; Lambert-Cheatham, N.; Shi, J. Novel Insights into the Roles of Rho Kinase in Cancer. Arch. Immunol. Ther. Exp. 2016, 64, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Hinsenkamp, I.; Schulz, S.; Roscher, M.; Suhr, A.-M.; Meyer, B.; Munteanu, B.; Fuchser, J.; Schoenberg, S.O.; Ebert, M.P.A.; Wängler, B.; et al. Inhibition of Rho-Associated Kinase 1/2 Attenuates Tumor Growth in Murine Gastric Cancer. Neoplasia 2016, 18, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Kikuchi, H.; Miyazaki, S.; Iino, I.; Setoguchi, T.; Hiramatsu, Y.; Ohta, M.; Kamiya, K.; Morita, Y.; Tanaka, H.; et al. Overexpression of Lysophosphatidylcholine Acyltransferase 1 and Concomitant Lipid Alterations in Gastric Cancer. Ann. Surg. Oncol. 2016, 23, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2015, 4, 5959. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Heiden, M.G.V. Altered metabolite levels in cancer: Implications for tumour biology and cancer therapy. Nat. Rev. Cancer 2016, 16, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Electric Field-Assisted Matrix Coating Method Enhances the Detection of Small Molecule Metabolites for Mass Spectrometry Imaging. Anal. Chem. 2015, 87, 5860–5865. [Google Scholar] [CrossRef] [PubMed]
- Deininger, S.-O.; Ebert, M.P.; Fütterer, A.; Gerhard, M.; Röcken, C. MALDI Imaging Combined with Hierarchical Clustering as a New Tool for the Interpretation of Complex Human Cancers. J. Proteome Res. 2008, 7, 5230–5236. [Google Scholar] [CrossRef] [PubMed]
- Balluff, B.; Rauser, S.; Meding, S.; Elsner, M.; Schöne, C.; Feuchtinger, A.; Schuhmacher, C.; Novotny, A.; Jütting, U.; Maccarrone, G.; et al. MALDI Imaging Identifies Prognostic Seven-Protein Signature of Novel Tissue Markers in Intestinal-Type Gastric Cancer. Am. J. Pathol. 2011, 179, 2720–2729. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Freire, J.M.; Pacheco, T.R.; Barata, J.T.; Castanho, M.A.R.B. Apoptotic human neutrophil peptide-1 anti-tumor activity revealed by cellular biomechanics. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Kemik, O.; Kemik, A.; Sumer, A.; Begenik, H.; Purisa, S.; Tuzun, S. Human neutrophil peptides 1, 2 and 3 (HNP 1–3): Elevated serum levels in colorectal cancer and novel marker of lymphatic and hepatic metastasis. Hum. Exp. Toxicol. 2013, 32, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Lundy, F.T.; Orr, D.F.; Gallagher, J.R.; Maxwell, P.; Shaw, C.; Napier, S.S.; Gerald Cowan, C.; Lamey, P.-J.; Marley, J.J. Identification and overexpression of human neutrophil α-defensins (human neutrophil peptides 1, 2 and 3) in squamous cell carcinomas of the human tongue. Oral Oncol. 2004, 40, 139–144. [Google Scholar] [CrossRef]
- Balluff, B.; Frese, C.K.; Maier, S.K.; Schöne, C.; Kuster, B.; Schmitt, M.; Aubele, M.; Höfler, H.; Deelder, A.M.; Heck, A.J.; et al. De novo discovery of phenotypic intratumour heterogeneity using imaging mass spectrometry. J. Pathol. 2015, 235, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-C.; Chang, J.; Chen, L.-Y.; Ho, A.-S.; Ker-Jer, H.; Lee, S.-C.; Mai, F.-D.; Chang, C.-C. Human neutrophil peptides 1-3 as gastric cancer tissue markers measured by MALDI-imaging mass spectrometry: Implications for infiltrated neutrophils as a tumor target. Dis. Markers 2012, 32, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Reyzer, M.L.; Choi, I.J.; Kim, C.G.; Kim, H.S.; Oshima, A.; Chertov, O.; Colantonio, S.; Fisher, R.J.; Allen, J.L.; et al. Gastric Cancer-Specific Protein Profile Identified Using Endoscopic Biopsy Samples via MALDI Mass Spectrometry. J. Proteome Res. 2010, 9, 4123–4130. [Google Scholar] [CrossRef] [PubMed]
- Elsner, M.; Rauser, S.; Maier, S.; Schöne, C.; Balluff, B.; Meding, S.; Jung, G.; Nipp, M.; Sarioglu, H.; Maccarrone, G.; et al. MALDI imaging mass spectrometry reveals COX7A2, TAGLN2 and S100-A10 as novel prognostic markers in Barrett’s adenocarcinoma. J. Proteom. 2012, 75, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M.; Elsner, M.; Ludyga, N.; Feuchtinger, A.; Zangen, V.; Maier, S.K.; Balluff, B.; Schöne, C.; Hierber, L.; Braselmann, H.; et al. Clinical response to chemotherapy in oesophageal adenocarcinoma patients is linked to defects in mitochondria: Mitochondrial defects predict chemotherapy response. J. Pathol. 2013, 230, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Ikegami, K.; Goto-Inoue, N.; Hayasaka, T.; Zaima, N.; Tanaka, H.; Uehara, T.; Setoguchi, T.; Sakaguchi, T.; Igarashi, H.; et al. Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray. Cancer Sci. 2010, 101, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, B.; Meyer, B.; von Reitzenstein, C.; Burgermeister, E.; Bog, S.; Pahl, A.; Ebert, M.P.; Hopf, C. Label-Free in Situ Monitoring of Histone Deacetylase Drug Target Engagement by Matrix-Assisted Laser Desorption Ionization-Mass Spectrometry Biotyping and Imaging. Anal. Chem. 2014, 86, 4642–4647. [Google Scholar] [CrossRef] [PubMed]
- Meding, S.; Nitsche, U.; Balluff, B.; Elsner, M.; Rauser, S.; Schöne, C.; Nipp, M.; Maak, M.; Feith, M.; Ebert, M.P.; et al. Tumor Classification of Six Common Cancer Types Based on Proteomic Profiling by MALDI Imaging. J. Proteome Res. 2012, 11, 1996–2003. [Google Scholar] [CrossRef] [PubMed]
- Balluff, B.; Elsner, M.; Kowarsch, A.; Rauser, S.; Meding, S.; Schuhmacher, C.; Feith, M.; Herrmann, K.; Röcken, C.; Schmid, R.M.; et al. Classification of HER2/neu Status in Gastric Cancer Using a Breast-Cancer Derived Proteome Classifier. J. Proteome Res. 2010, 9, 6317–6322. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Duarte, H.O.; Balmaña, M.; Mereiter, S.; Osório, H.; Gomes, J.; Reis, C.A. Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor. Int. J. Mol. Sci. 2017, 18, 2262. [Google Scholar] [CrossRef] [PubMed]
- Kunzke, T.; Balluff, B.; Feuchtinger, A.; Buck, A.; Langer, R.; Luber, B.; Lordick, F.; Zitzelsberger, H.; Aichler, M.; Walch, A. Native glycan fragments detected by MALDI-FT-ICR mass spectrometry imaging impact gastric cancer biology and patient outcome. Oncotarget 2017, 8, 68012–68025. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, A.; Piga, I.; Galli, M.; Stella, M.; Denti, V.; Del Puppo, M.; Magni, F. Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging in the Study of Gastric Cancer: A Mini Review. Int. J. Mol. Sci. 2017, 18, 2588. https://doi.org/10.3390/ijms18122588
Smith A, Piga I, Galli M, Stella M, Denti V, Del Puppo M, Magni F. Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging in the Study of Gastric Cancer: A Mini Review. International Journal of Molecular Sciences. 2017; 18(12):2588. https://doi.org/10.3390/ijms18122588
Chicago/Turabian StyleSmith, Andrew, Isabella Piga, Manuel Galli, Martina Stella, Vanna Denti, Marina Del Puppo, and Fulvio Magni. 2017. "Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging in the Study of Gastric Cancer: A Mini Review" International Journal of Molecular Sciences 18, no. 12: 2588. https://doi.org/10.3390/ijms18122588
APA StyleSmith, A., Piga, I., Galli, M., Stella, M., Denti, V., Del Puppo, M., & Magni, F. (2017). Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry Imaging in the Study of Gastric Cancer: A Mini Review. International Journal of Molecular Sciences, 18(12), 2588. https://doi.org/10.3390/ijms18122588







