Whole-Genome Re-Alignment Facilitates Development of Specific Molecular Markers for Races 1 and 4 of Xanthomonas campestris pv. campestris, the Cause of Black Rot Disease in Brassica oleracea
Abstract
:1. Introduction
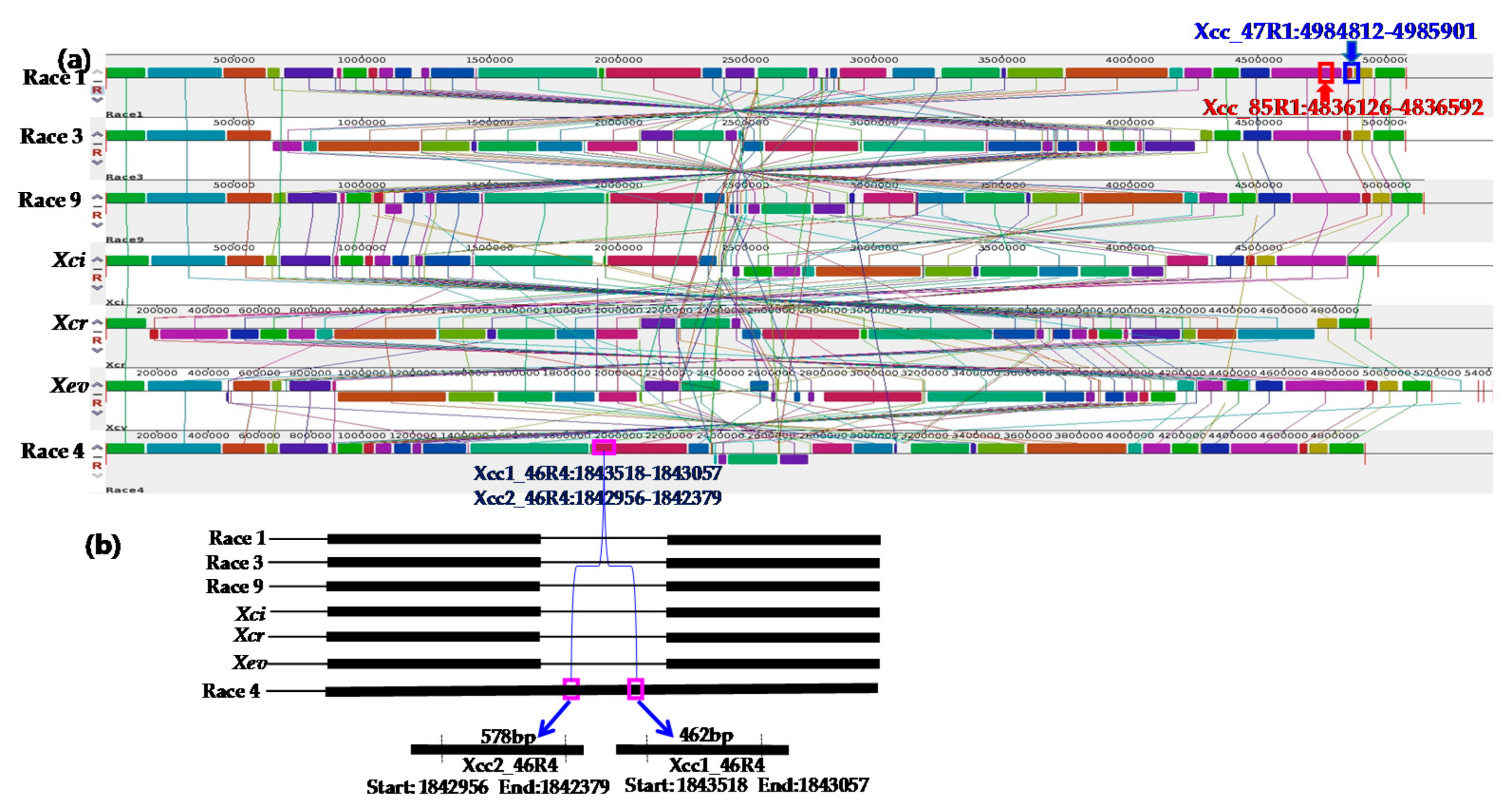
2. Results
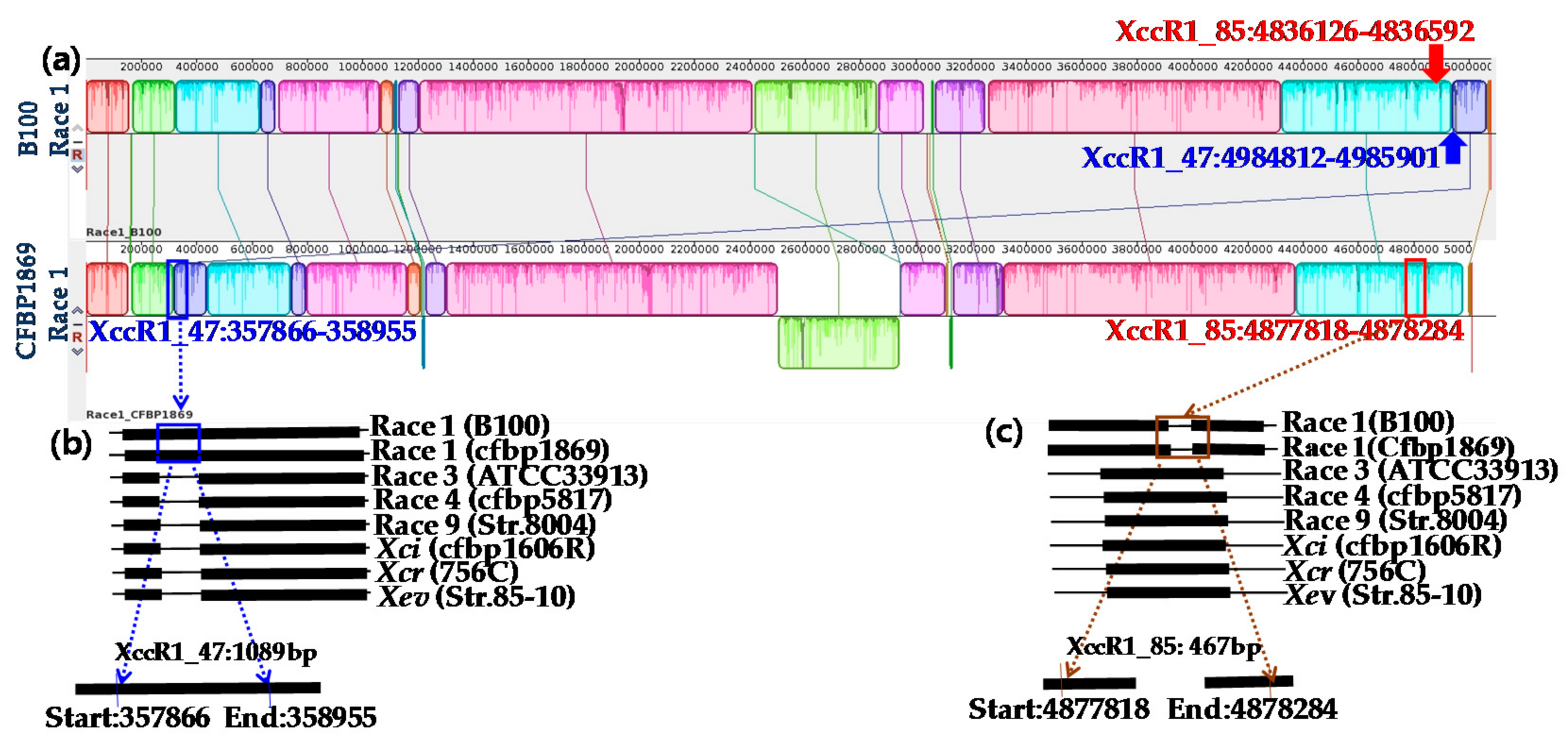
2.1. Specificity of Primers
2.2. Validation of Markers through Bio-PCR in Inoculated Cabbage Leaves
2.3. Race Determination
3. Discussion
3.1. PCR-Based Markers Specifically Detected Xcc Races 1 and 4
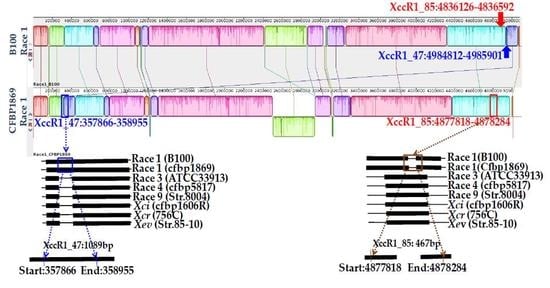
3.2. Whole Genome Sequences of Xcc Enabled Developing Race-Specific Novel Markers
3.3. Direct and Rapid Detection Tool for Xcc Race 4 Developed
3.4. Potential Applications of Race 4 Specific Markers
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Isolation of Total DNA
4.3. Sequence Retrieval and Alignment
4.4. Primer Design and PCR Conditions
4.5. Testing the Specificity of Primers
4.6. Detection of the Race 1 and 4-Specific Pathogen by PCR in Artificially Infected Cabbage Leaves
4.7. Race Identification
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Xcc | Xanthomonas campestris pv. campestris |
| Xci | Xanthomonas campestris pv. incane |
| Xcr | Xanthomonas campestris pv. raphani |
| PCR | Polymerase chain reaction |
| SCAR | Sequence-characterized amplified region |
| InDel | Insertions or deletions |
References
- Vicente, J.G.; Holub, E.B. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 2013, 14, 2–18. [Google Scholar]
- Williams, P.H. Black rot: A continuing threat to world crucifers. Plant Dis. 1980, 64, 736–742. [Google Scholar] [CrossRef]
- Vicente, J.G.; Conway, J.; Roberts, S.J.; Taylor, J.D. Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology 2001, 91, 492–499. [Google Scholar] [PubMed]
- Silva, M.R. Genetic Diversity of Xanthomonas campestris pv. campestris in Brazil. Master’s Thesis, Universidade Federal de Vicosa, Vicosa, Brazil, 2006. [Google Scholar]
- Jensen, B.D.; Vicente, J.G.; Manandhar, H.K.; Roberts, S.J. Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal. Plant Dis. 2010, 94, 298–305. [Google Scholar]
- Popović, T.; Jošić, D.; Starović, M.; Milovanović, P.; Dolovac, N.; Poštić, D.; Stanković, S. Phenotypic and Genotypic Characterization of Xanthomonas campestris pv. campestris Isolated from Cabbage, Kale and Broccoli. Arch. Biol. Sci. 2013, 65, 585–593. [Google Scholar]
- Sewariya, V.K.; Shrivastava, R.; Prasad, G.B.K.S.; Arora, K. In-Vitro Evaluation of Novel Synthetic Compounds against Xanthomonas campestris pv. campestris. Int. J. Pharma Biol. Sci. 2012, 3, 441–453. [Google Scholar]
- Kocks, C.G.; Zadoks, J.C.; Ruissen, M.A. Spatio-temporal development of black rot (X. campestris pv. campestris) in cabbage in relation to initial inoculums levels in field plots in The Netherlands. Plant Pathol. 1999, 48, 176–188. [Google Scholar]
- Kamoun, S.; Kamdar, H.V.; Tola, E.; Kado, C.I. Incompatible interactions between crucifers and Xanthomonas campestris involve a vascular hypersensitive response: Role of the hrpX locus. Mol. Plant-Microbe Interact. 1992, 5, 22–33. [Google Scholar] [CrossRef]
- Fargier, E.; Manceau, C. Pathogenicity assays restrict the species Xanthomonas campestris into three pathovars and reveal nine races within X. campestris pv. campestris. Plant Pathol. 2007, 56, 805–818. [Google Scholar] [CrossRef]
- Cruz, J.; Tenreiro, R.; Cruz, L. Assessment of Diversity Xanthomonas campestris Pathovars Affecting Cruciferous Plants in Portugal and Disclosure of two novel X. campestris pv. campestris races. J. Plant Pathol. 2017, 99. [Google Scholar] [CrossRef]
- Taylor, J.D.; Conway, J.; Roberts, S.J.; Astley, D.; Vicente, J.G. Sources and origin of resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 2002, 92, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Griesbach, E.; Loptien, H.; Miersch, U. Resistance to Xanthomonas campestris pv. campestris (Pammel) Dowson in cabbage Brassica oleracea L. Resistance genes Xanthomonas campestris pv. campestris (Pammel) Dowson im Kohl Brassica oleracea L. J. Plant Dis. Protect. 2003, 5, 461–475. [Google Scholar]
- Dickson, M.D.; Hunter, J.E. Inheritance of resistance in cabbage seedlings to black rot. Hort. Sci. 1987, 22, 108–109. [Google Scholar]
- Ignatov, A.; Hida, K.; Kuginuki, Y. Black rot of crucifers and sources of resistance in Brassica crops. Jpn. Agric. Res. Quart. 1998, 32, 167–172. [Google Scholar]
- Roberts, S.J.; Brough, J.; Everett, B.; Redstone, S. Extraction methods for Xanthomonas campestris pv. campestris from brassica seed. Seed Sci. Technol. 2004, 32, 439–453. [Google Scholar] [CrossRef]
- Koenraadt, H.; van Bilsen, J.G.P.M.; Roberts, S.J. Comparative test of four semi-selective agar media for the detection of Xanthomonas campestris pv. campestris in brassica seeds. Seed Sci. Technol. 2005, 33, 115–125. [Google Scholar] [CrossRef]
- Alvarez, A.M.; Lou, K. Rapid identification of Xanthomonas campestris pv. campestris by ELISA. Plant Dis. 1985, 69, 1082–1086. [Google Scholar] [CrossRef]
- Chitarra, L.G.; Langerak, C.J.; Bergervoet, J.H.W.; Van den Bulk, R.W. Detection of the plant pathogenic bacterium Xanthomonas campestris pv. campestris in seed extracts of Brassica sp. Applying fluorescent antibodies and flow Cytometry. Cytometry 2002, 47, 118–126. [Google Scholar]
- Singh, D.; Raghavendra, B.T.; Singh, R.P.; Singh, H.; Raghuwanshi, R.; Singh, R.P. Detection of black rot disease causing pathogen Xanthomonas campestris pv. campestris by bio-PCR from seeds and plant parts of cole crop. Seed Sci. Technol. 2014, 42, 36–46. [Google Scholar]
- Alvarez, A.M.; Benedict, A.A.; Mizumoto, C.Y.; Hunter, J.E.; Gabriel, D.W. Serological, pathological and genetic diversity among strains of Xanthomonas campestris infecting crucifers. Phytopathology 1994, 84, 1449–1457. [Google Scholar] [CrossRef]
- Berg, T.; Tesoriero, L.A.; Hailstones, D.L. PCR-based detection of Xanthomonas campestris pathovars in Brassica seed. Plant Pathol. 2005, 54, 416–427. [Google Scholar] [CrossRef]
- Leibner-Ciszak, J.; Dobrowolska, A.; Krawczyk, B.; Kaszuba, A.; Staczek, P. Evaluation of a PCR melting profile method for intraspecies differentiation of Trichophyton rubrum and Trichophyton interdigitale. J. Med. Microbiol. 2010, 59, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Kałużna, M.; Puławska, J.; Sobiczewski, P. The use of PCR melting profile for typing of Pseudomonas syringae isolates from stone fruit trees. Eur. J. Plant Pathol. 2010, 126, 437–443. [Google Scholar] [CrossRef]
- Kałużna, M.; Puławska, J.; Waleron, M.; Sobiczewski, P. The genetic characterization of Xanthomonas arboricola pv. juglandis, the causal agent of walnut blight in Poland. Plant Pathol. 2014, 63, 1404–1416. [Google Scholar]
- Tsygankova, S.V.; Ignatov, A.N.; Boulygina, E.S.; Kuznetsov, B.B. Genetic relationships among strains of Xanthomonas campestris pv. campestris revealed by novel rep-PCR primers. Eur. J. Plant Pathol. 2004, 110, 845–853. [Google Scholar]
- Causin, R.; Scopel, C.; Grendene, A.; Montecchio, L. An Improved Method for the Detection of Phytophthora cactorum (L.C.) Schröeter in Infected Plant Tissues Using Scar Markers. J. Plant Pathol. 2005, 87, 25–35. [Google Scholar]
- Väli, U.; Brandström, M.; Johansson, M.; Ellegren, H. Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet. 2008, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Erixon, P.; Oxelman, B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS ONE 2008, 3, e1386. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Muro, T.; Imamura, S.; Yuasa, I. Forensic species identification based on size variation of mitochondrial DNA hypervariable regions. Int. J. Legal Med. 2009, 123, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Carneiro, J.; Matthiesen, R.; van Asch, B.; Pinto, N.; Gusmao, L.; Amorim, A. Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 2010, 38, e203. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Rathaur, P.S.; Vicente, J.G. Characterization, genetic diversity and distribution of Xanthomonas campestris pv. campestris races causing black rot disease in cruciferous crops of India. Plant Pathol. 2016, 65, 1411–1418. [Google Scholar]
- Song, E.S.; Kim, S.Y.; Noh, T.H.; Cho, H.; Chae, S.C.; Lee, B.M. PCR-Based Assay for Rapid and Specific Detection of the New Xanthomonas oryzae pv. oryzae K3a Race Using an AFLP-Derived Marker. Microbiol. Biotechnol. 2014, 24, 732–739. [Google Scholar]
- Wang, B.; Hu, X.; Li, Q.; Hao, B.; Zhang, B.; Li, G.; Kang, Z. Development of race-specific SCAR markers for detection of Chinese races CYR32 and CYR33 of Puccinia striiformis f. sp. tritici. Plant Dis. 2010, 94, 221–228. [Google Scholar] [CrossRef]
- Pasquali, M.; Dematheis, F.; Gullino, M.L.; Garibaldi, A. Identification of race 1 of Fusarium oxysporum f. sp. lactucae on lettuce by inter-retrotransposon sequence-characterized amplified region technique. Phytopathology 2007, 97, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.C.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Jia, Y.; Ren, S.X.; He, Y.Q.; Feng, J.X.; Lu, L.F.; Sun, Q.; Ying, G.; Tang, D.J.; Tang, H.; et al. Comparative and functional genomic analyses of the pathogenicity of Phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005, 15, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Vorholter, F.J.; Schneiker, S.; Goesmann, A.; Krause, L.; Bekel, T.; Kaiser, O.; Linke, B.; Patschkowski, T.; Ruckert, C.; Schmid, J.; et al. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 2008, 134, 33–45. [Google Scholar] [PubMed]
- Bolot, S.; Cerutti, A.; Carrère, S.; Arlat, M.; Saux, M.F.; Portier, P.; Poussier, S.; Jacques, M.; Noëla, L.D. Genome Sequences of the Race 1 and Race 4 Xanthomonas campestris pv. campestris Strains CFBP 1869 and CFBP 5817. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Roux, B.; Bolot, S.; Guy, E.; Denance, N.; Lautier, M.; Jardinaud, M.F.; Fischer-Le Saux, M.; Portier, P.; Jacques, M.A.; Gagnevin, L.; et al. Genomics and transcriptomics of Xanthomonas campestris species challenge the concept of core type III effectome. BMC Genom. 2015, 16, 975. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Koebnik, R.; Lu, H.; Furutani, A.; Angiuoli, S.V.; Patil, P.B.; Van Sluys, M.A.; Ryan, R.P.; Meyer, D.F.; Han, S.W.; et al. Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 2011, 193, 5450–5464. [Google Scholar] [CrossRef] [PubMed]
- Thieme, F.; Koebnik, R.; Bekel, T.; Berger, C.; Boch, J.; Büttner, D.; Caldana, C.; Gaigalat, L.; Goesmann, A.; Kay, S.; et al. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 2005, 187, 54–66. [Google Scholar]
- Cho, M.S.; Kang, M.J.; Kim, C.K.; Seol, Y.J.; Hhan, J.H.; Park, S.C. Sensitive and specific detection of Xanthomonas oryzae pv. oryzae by real-time bio-PCR using pathovars specific primers based on an rhs family gene. Plant Dis. 2011, 95, 589–594. [Google Scholar]
- Laila, R.; Robin, A.H.K.; Yang, K.; Choi, G.J.; Park, J.I.; Nou, I.S. Detection of Ribosomal DNA Sequence Polymorphisms in the Protist Plasmodiophora brassicae for the Identification of Geographical Isolates. Int. J. Mol. Sci. 2017, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- King, E.O.; Ward, M.K.; Raney, D.R. Two simple media for the demonstration of pyrocanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- NCBI. Available online: https://www.ncbi.nlm.nih.gov (accessed on 22 November 2017).
- Primer3Plus. Available online: http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi (accessed on 12 October 2017).
- Xanthomonas campestris pv. campestris (Xcc)—BLAST Tool. Available online: http://210.110.86.160/lab/home.html (accessed on 12 October 2017).
| Strains | Accession | Races | Genome Size (bp) | G + C Content (%) | Reference |
|---|---|---|---|---|---|
| ATCC 33913 (UK) Xanthomonas campestris pv. campestris | AE008922 | 3 | 5,076,188 | 65.1 | [36] |
| Strain 8004 Xanthomonas campestris pv. campestris | NC_007086 | 9 | 5,148,708 | 65.0 | [37] |
| B100 (UK) Xanthomonas campestris pv. campestris | AM920689 | 1 | 5,079,002 | 65.0 | [38] |
| CFBP1869 (France) Xanthomonas campestris pv. campestris | NZ_CM002545 | 1 | 5,008,832 | 65.0 | [39] |
| CFBP 5817 (France) Xanthomonas campestris pv. campestris | NZ_CM002673 | 4 | 4,918,955 | 65.2 | [39] |
| CFBP1606R Xanthomonas campestris pv. incanae | NZ_CM002635 | - | 4,967,288 | 65.0 | [40] |
| 756C Xanthomonas campestris pv. raphani | NC_017271 | - | 4,941,214 | 65.3 | [41] |
| Strain 85–10 Xanthomonas euvesicatoria | NC_007508.1 | - | 5,178,466 | 64.7 | [42] |
| SL. | Bacterial Strains * | Races | Host | Country | Collection Year | Reference |
|---|---|---|---|---|---|---|
| 1 | X. campestris pv. campestris (HRIW-3811) | 1 | B. oleracea | US | 2017 | [3] |
| 2 | X. campestris pv. campestris (HRIW-3849A) | 2 | B. oleracea var. botrytis | US | 2017 | [3] |
| 3 | X. campestris pv. campestris (HRIW-5212) | 3 | B. oleracea var. gemmifera | UK | 2017 | [3] |
| 4 | X. campestris pv. campestris (HRIW-1279A) | 4 | B. oleracea var. capitata | UK | 2017 | [3] |
| 5 | X. campestris pv. campestris (HRIW-3880) | 5 | B. oleracea var. capitata | Australia | 2017 | [3] |
| 6 | X. campestris pv. campestris (HRIW-6181) | 6 | B. rapa | Portugal | 2017 | [3] |
| 7 | X. campestris pv. campestris (HRIW-8450A) | 7 | B. oleracea var. capitata | UK | 2017 | [3] |
| 8 | X. campestris pv. campestris (ICMP8) | - | Brassica oleracea var. capitata | New Zealand | 2016 | This work |
| 9 | X. campestris pv. campestris (KACC19132) | - | B. rapa (Pyeongchang) | South Korea | 2017 | This work |
| 10 | X. campestris pv. campestris (KACC19133) | - | B. rapa (Gangneung) | South Korea | 2017 | This work |
| 11 | X. campestris pv. campestris (KACC19134) | - | - | South Korea | 2017 | This work |
| 12 | X. campestris pv. campestris (KACC19135) | - | - | South Korea | 2017 | This work |
| 13 | X. campestris pv. campestris (KACC19136) | - | - | South Korea | 2017 | This work |
| 14 | X. campestris pv. campestris (KACC10377) | - | Brassica oleracea var. capitata | South Korea | 2017 | This work |
| 15 | X. campestris pv. campestris (KACC17966) | - | - | South Korea | 2017 | This work |
| 16 | X. campestris pv. incane (WHRI-6377) | - | Matthiola incana | UK | 2017 | [3] |
| 17 | X. campestris pv. raphanin (WHRI-8305) | 2 | B. rapa var. perviridis | UK | 2017 | [3] |
| 18 | X. campestris (KACC10490) | - | - | South Korea | 2017 | This work |
| 19 | Pseudomonas syringae pv. maculicola (ICMP13051) | - | Brassica oleracea var. capitata | New Zealand | 2016 | This work |
| 20 | Erwinia carotovora subsp. carotovora (ICMP12464) | - | Brassica oleracea var. capitata | New Zealand | 2016 | This work |
| 21 | Plasmodiophora brassicae (Pathotype1)-Gangneung-1 | - | B. rapa | South Korea | 2016 | [44] |
| 22 | X. euvesicatoria (KACC11153) | - | - | South Korea | 2017 | This work |
| 23 | X. axonopodis pv. dieffenbachiae (KACC17821) | - | Anthurium andraeanum (Yongin) | South Korea | 2017 | This work |
| 24 | X. campestris pv. zinniae (KACC17126) | - | Zinnia elegans (Suwon) | South Korea | 2017 | This work |
| 25 | X. axonopodis pv. glycines (KACC10491) | - | Glycine max | South Korea | 2017 | This work |
| 26 | Didymella bryoniae (NIHHS1326) | - | Cucumis melon | South Korea | 2016 | This work |
| Primer Name | Sequences (5’…3’) | Genomic Position | Gene Name | Description | Base Pair (bp) | Annealing Temperature |
|---|---|---|---|---|---|---|
| Xcc_47R1_F | CCTCCTGAGTCATGGCAATGGC | 498412-4985901 | xcc-b100_4389 | Peptidoglycan binding Protein | 1089 | 65 °C for 40 s |
| Xcc_47R1_R | TAGCAGGGGAGTGCTGCTTGC | |||||
| Xcc_85R1_F | GCGGCTCGGCTTCACGGTCAGC | 4836126-4836592 | xcc-b100_4275 | Membrane protein with arac family transcriptional regulator and peptidase domain | 467 | |
| Xcc_85R1_R | GCCCAGGATGCAGCGCAGCGT | |||||
| Xcc1_46R4_F | GGCATGGGGAATGATCGTTGAC | 1843518-1843057 | Intergenic | - | 462 | 66 °C for 40 s |
| Xcc1_46R4_R | ATGCGGGCGATGGGATGGCCA | |||||
| Xcc2_46R4_F | GCGTAGCGAAAACTGGTAGTTC | 1842956-1842379 | Intergenic | - | 578 | |
| Xcc2_46R4_R | GCACAGGCGCACCAGCATATGGC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubel, M.H.; Robin, A.H.K.; Natarajan, S.; Vicente, J.G.; Kim, H.-T.; Park, J.-I.; Nou, I.-S. Whole-Genome Re-Alignment Facilitates Development of Specific Molecular Markers for Races 1 and 4 of Xanthomonas campestris pv. campestris, the Cause of Black Rot Disease in Brassica oleracea. Int. J. Mol. Sci. 2017, 18, 2523. https://doi.org/10.3390/ijms18122523
Rubel MH, Robin AHK, Natarajan S, Vicente JG, Kim H-T, Park J-I, Nou I-S. Whole-Genome Re-Alignment Facilitates Development of Specific Molecular Markers for Races 1 and 4 of Xanthomonas campestris pv. campestris, the Cause of Black Rot Disease in Brassica oleracea. International Journal of Molecular Sciences. 2017; 18(12):2523. https://doi.org/10.3390/ijms18122523
Chicago/Turabian StyleRubel, Mehede Hassan, Arif Hasan Khan Robin, Sathishkumar Natarajan, Joana G. Vicente, Hoy-Taek Kim, Jong-In Park, and Ill-Sup Nou. 2017. "Whole-Genome Re-Alignment Facilitates Development of Specific Molecular Markers for Races 1 and 4 of Xanthomonas campestris pv. campestris, the Cause of Black Rot Disease in Brassica oleracea" International Journal of Molecular Sciences 18, no. 12: 2523. https://doi.org/10.3390/ijms18122523
APA StyleRubel, M. H., Robin, A. H. K., Natarajan, S., Vicente, J. G., Kim, H.-T., Park, J.-I., & Nou, I.-S. (2017). Whole-Genome Re-Alignment Facilitates Development of Specific Molecular Markers for Races 1 and 4 of Xanthomonas campestris pv. campestris, the Cause of Black Rot Disease in Brassica oleracea. International Journal of Molecular Sciences, 18(12), 2523. https://doi.org/10.3390/ijms18122523