Environmental Burkholderia cenocepacia Strain Enhances Fitness by Serial Passages during Long-Term Chronic Airways Infection in Mice
Abstract
1. Introduction
2. Results
2.1. Characteristics of B. cenocepacia Mex1 Strain
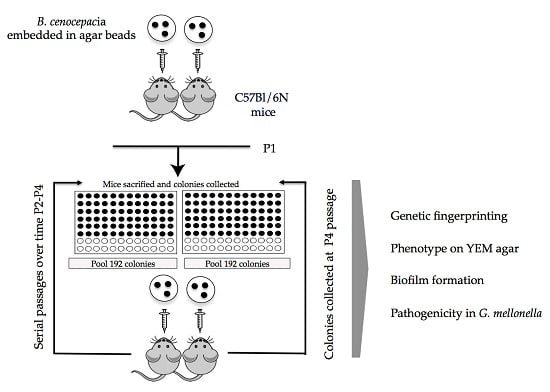
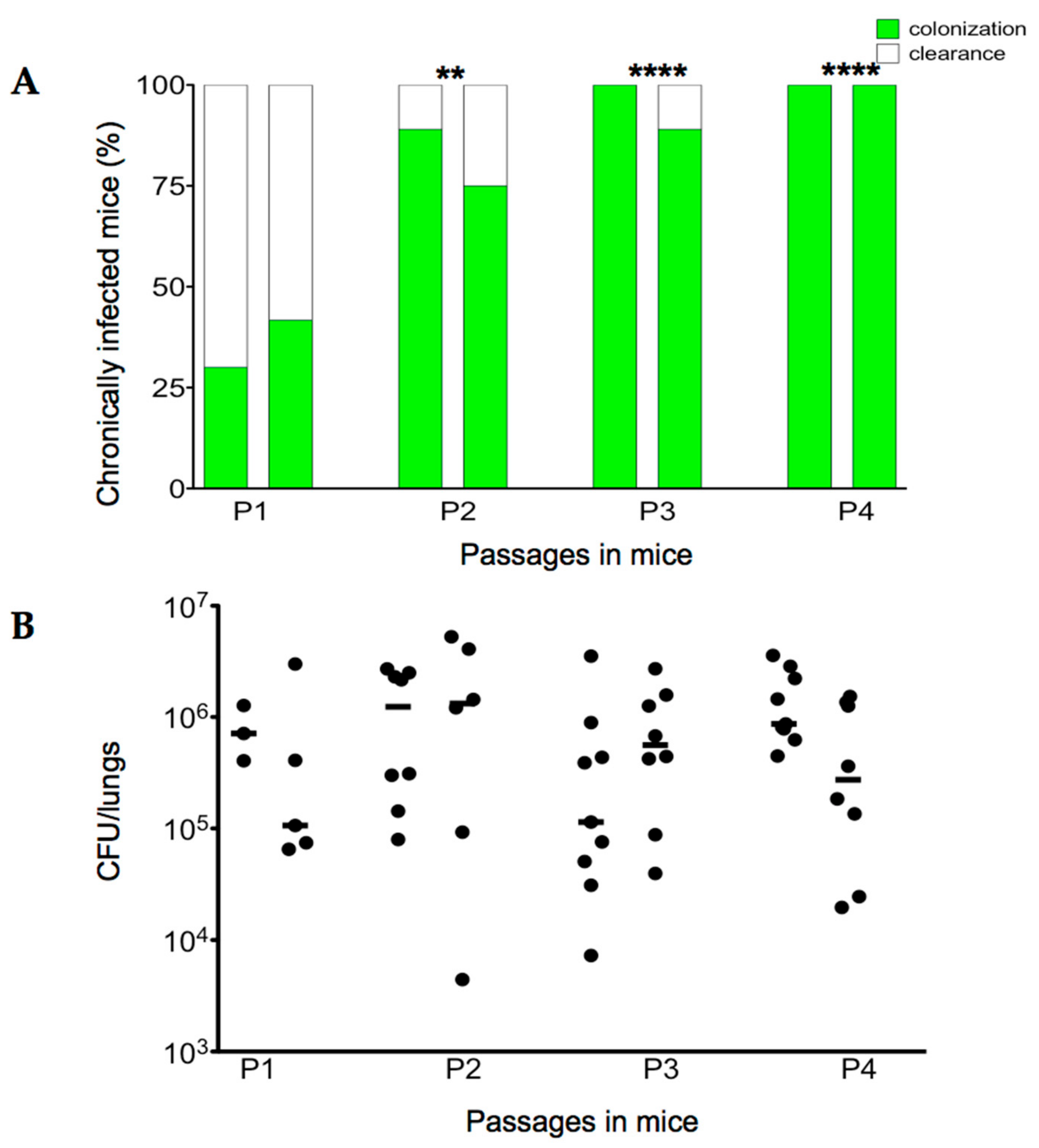
2.2. Serial Passages of the Environmental B. cenocepacia Mex1 in Mice
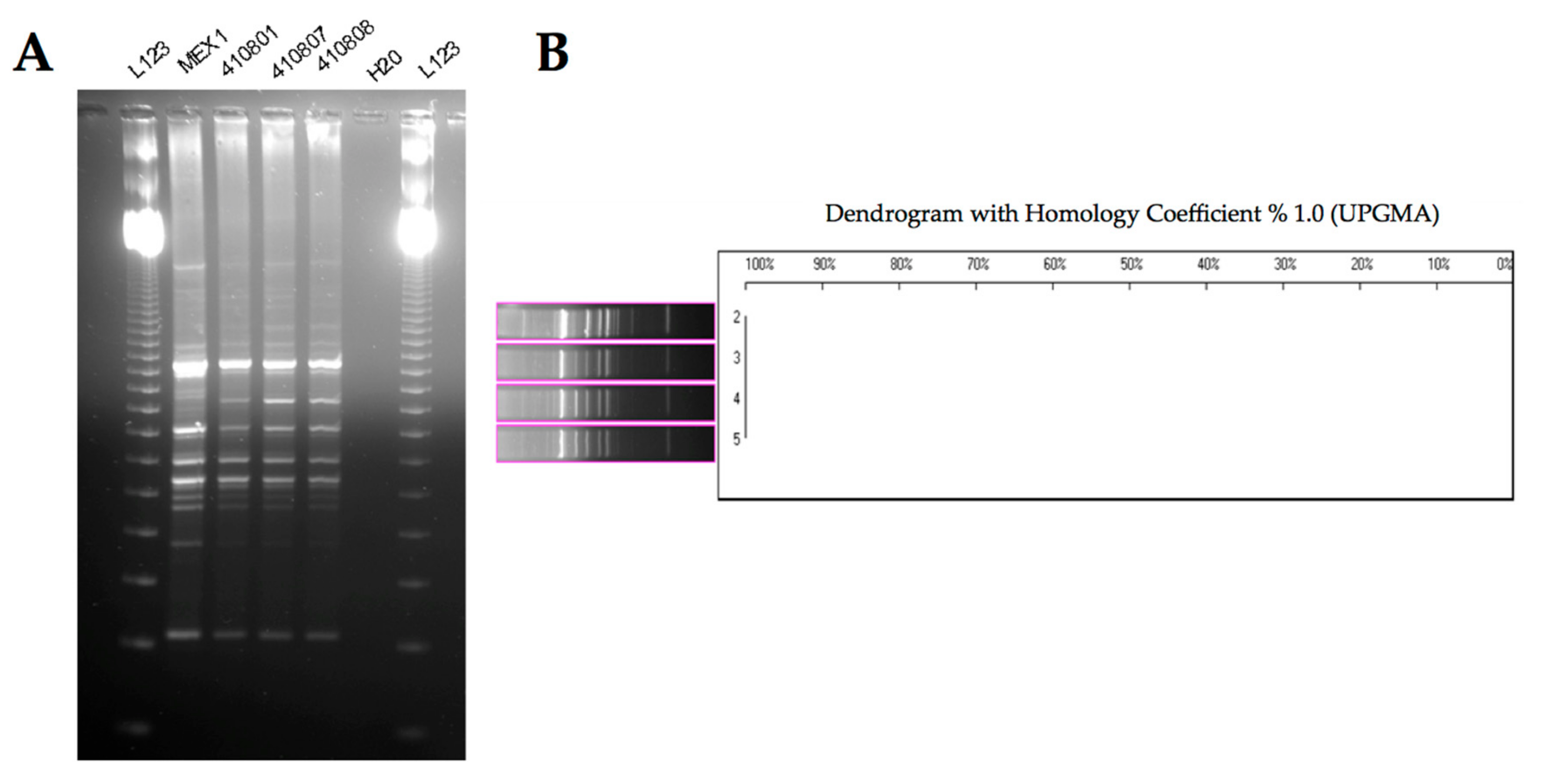
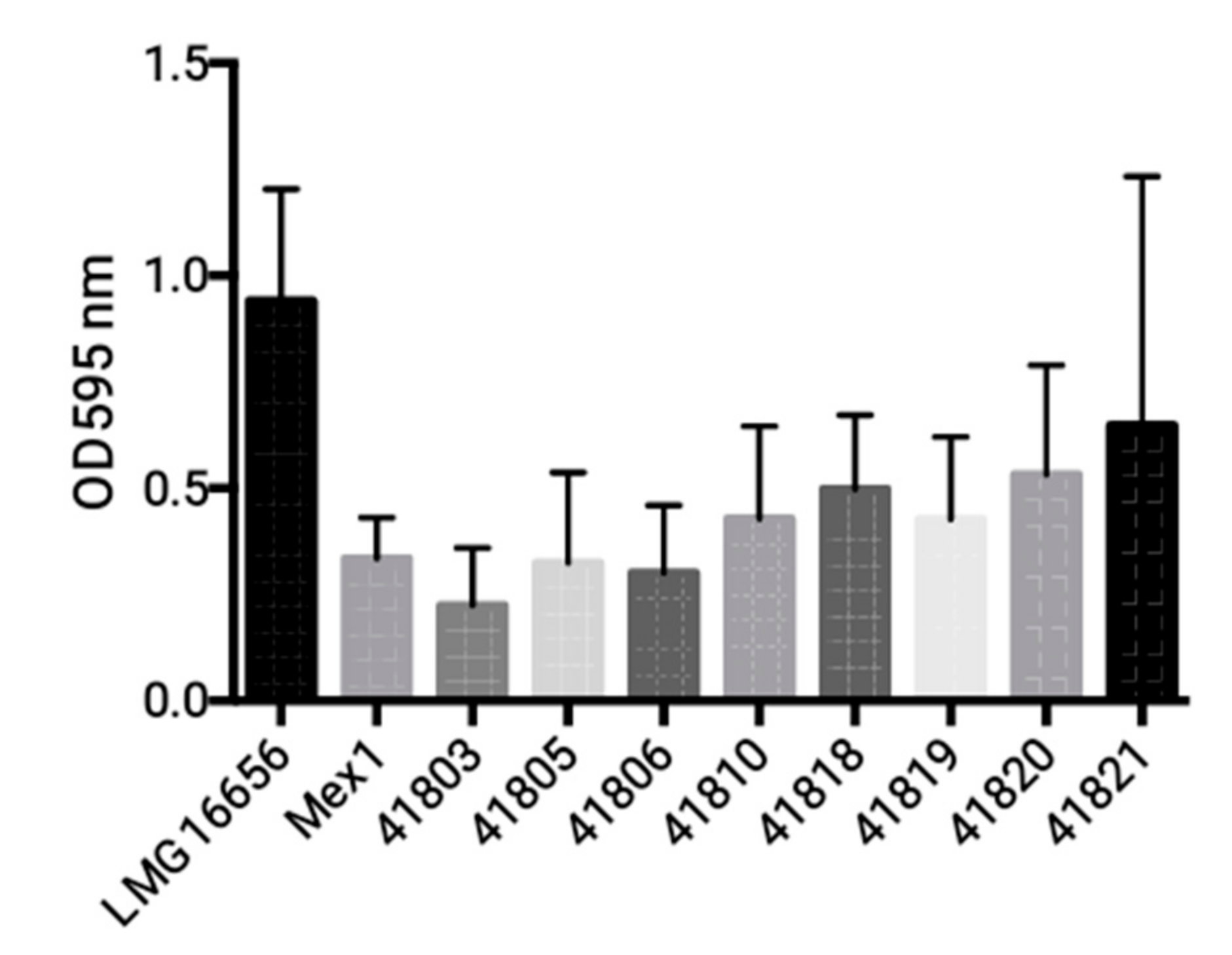
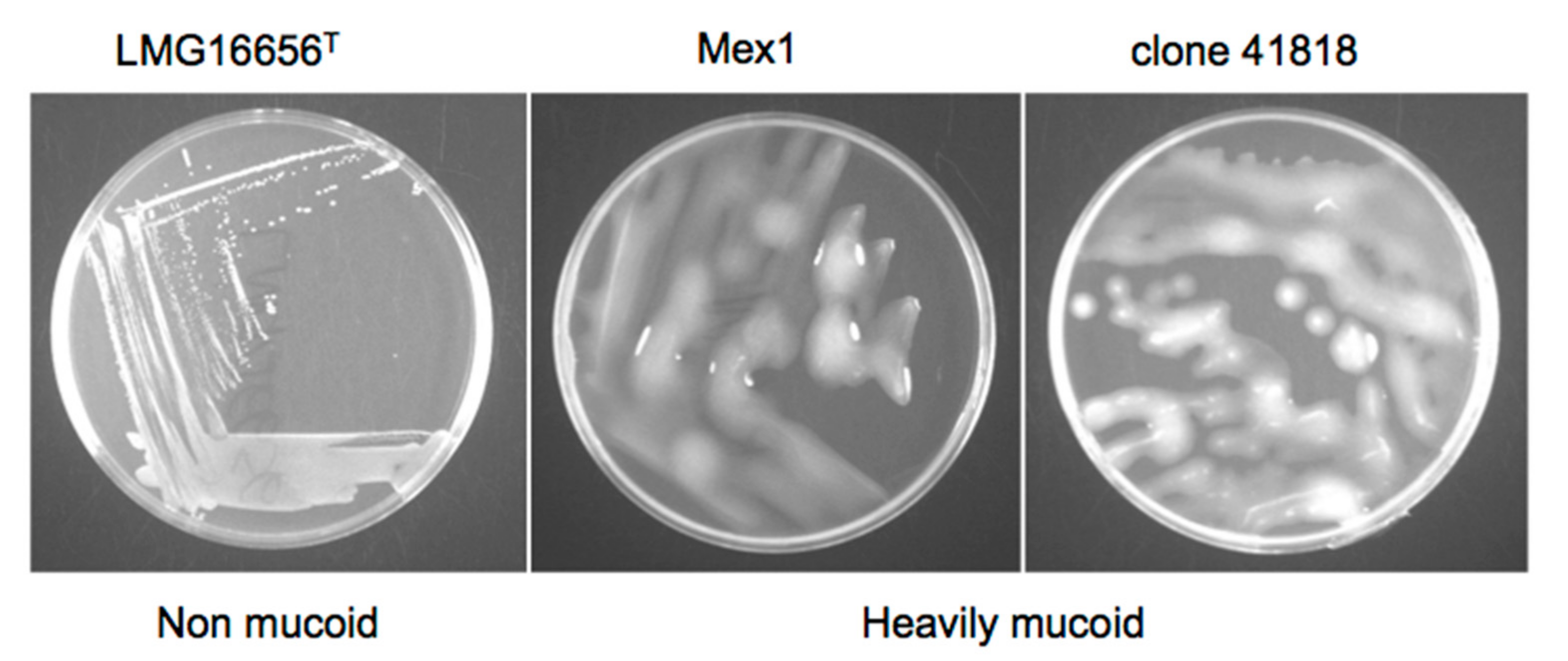
2.3. Phenotypic and Molecular Characterization of Mex1 Adapted to Murine Lungs
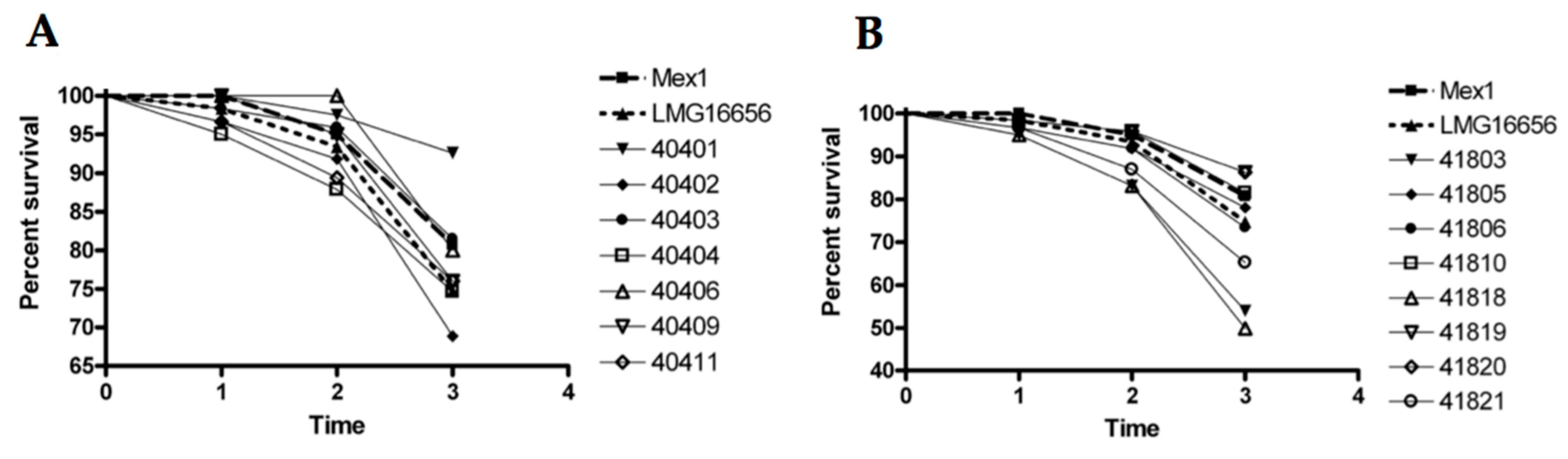
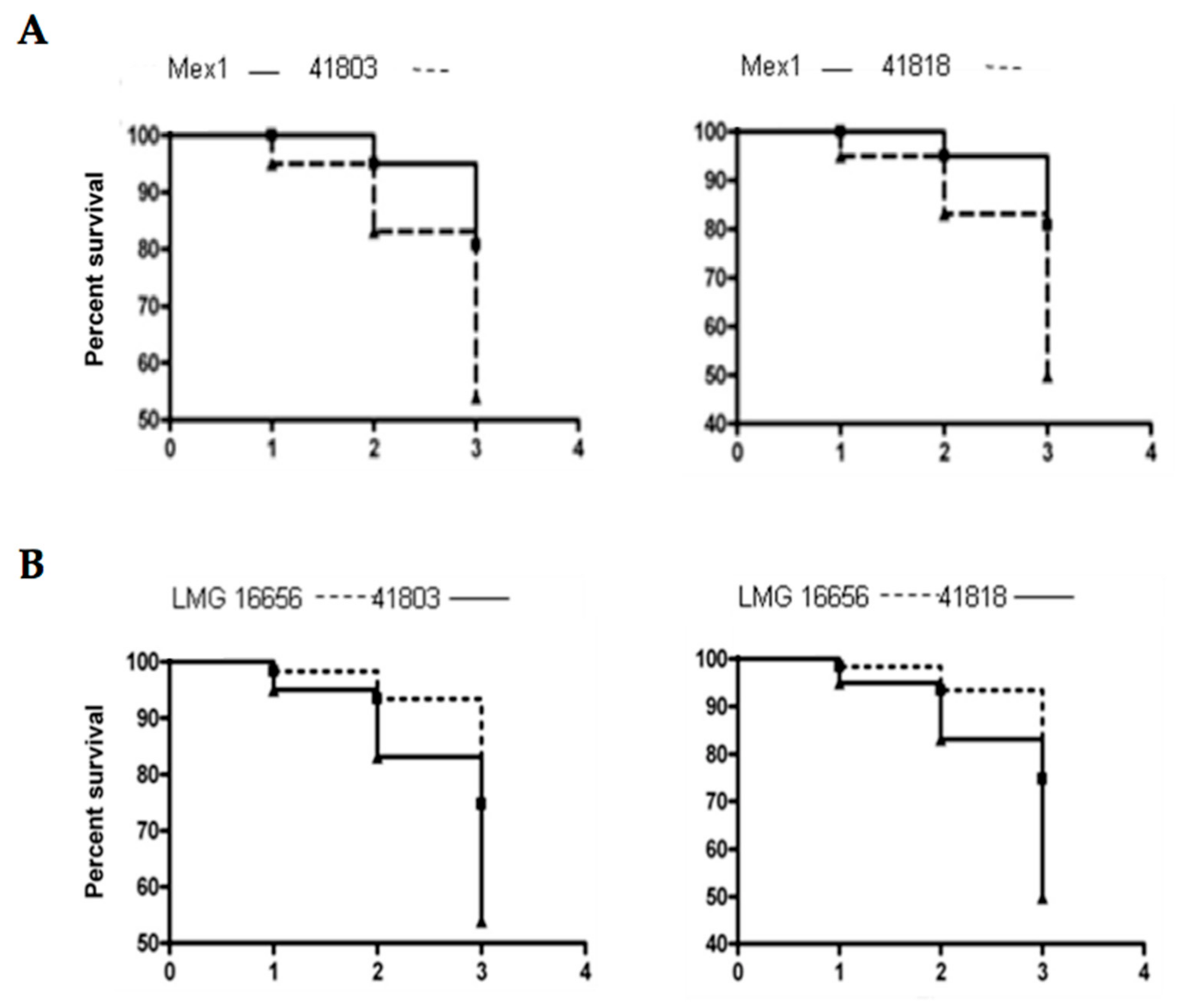
2.4. Survival and Relative Bacterial Loads of Mex1 and Its Derivative Clones in Infected G. mellonella Larvae
3. Discussion
4. Materials and Methods
4.1. Bacteria and Culture Conditions
4.2. Ethical Statement
4.3. Sequential Infection in Mice
4.4. Molecular Characterization of Mex1-Derived Clones
4.4.1. Random Amplified Polymorphic DNA (RAPD) Fingerprinting
4.4.2. PCR Amplification of cblA Gene
4.5. Biofilm Formation
4.6. Growth on YEM Yeast Extract-Mannitol (YEM) Agar
4.7. Screening of Mex1-Derived Clones in the G. mellonella Model
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ratjen, F.; Döring, G. Cystic fibrosis. Lancet 2003, 361, 681–689. [Google Scholar] [CrossRef]
- Worlitzsch, D.; Tarran, R.; Ulrich, M.; Schwab, U.; Cekici, A.; Meyer, K.C.; Birrer, P.; Bellon, G.; Berger, J.; Weiss, T.; et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 2002, 109, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Döring, G.; Gulbins, E. Cystic fibrosis and innate immunity: How chloride channel mutations provoke lung disease. Cell. Microbiol. 2009, 11, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Cystic Fibrosis Foundation Patient Registry. Annual Data Report. 2015. Available online: https://www.cff.org/Our-Research/CF-Patient-Registry/2015-Patient-Registry-Annual-Data-Report.pdf (accessed on 1 August 2017).
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Conrad, D.; Haynes, M.; Salamon, P.; Rainey, P.B.; Youle, M.; Rohwer, F. Cystic fibrosis therapy: A community ecology perspective. Am. J. Respir. Cell Mol. Biol. 2013, 48, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Lorè, N.I.; Cigana, C.; De Fino, I.; Riva, C.; Juhas, M.; Schwager, S.; Eberl, L.; Bragonzi, A. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS ONE 2012, 7, e35648. [Google Scholar] [CrossRef] [PubMed]
- Döring, G.; Conway, S.P.; Heijerman, H.G.; Hodson, M.E.; Hoiby, N.; Smyth, A.; Touw, D.J. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: A European consensus. Eur. Respir. J. 2000, 16, 749–767. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.E.; Buckley, D.G.; Wu, Z.; Saenphimmachak, C.; Hoffman, L.R.; D’Argenio, D.A.; Miller, S.I.; Ramsey, B.W.; Speert, D.P.; Moskowitz, S.M.; et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2006, 103, 8487–8492. [Google Scholar] [CrossRef] [PubMed]
- Chicurel, M. Can organisms speed their own evolution? Science 2001, 292, 1824–1827. [Google Scholar] [CrossRef] [PubMed]
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016, 24, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Fothergill, J.L.; Walshaw, M.J.; Winstanley, C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur. Respir. J. 2012, 40, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Parkins, M.D.; Glezerson, B.A.; Sibley, C.D.; Sibley, K.A.; Duong, J.; Purighalla, S.; Mody, C.H.; Workentine, M.L.; Storey, D.G.; Surette, M.G.; et al. Twenty-five-year outbreak of Pseudomonas aeruginosa infecting individuals with cystic fibrosis: Identification of the prairie epidemic strain. J. Clin. Microbiol. 2014, 52, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Drevinek, P.; Mahenthiralingam, E. Burkholderia cenocepacia in cystic fibrosis: Epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 2010, 16, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Courtney, J.M.; Dunbar, K.E.; McDowell, A.; Moore, J.E.; Warke, T.J.; Stevenson, M.; Elborn, J.S. Clinical outcome of Burkholderia cepacia complex infection in cystic fibrosis adults. J. Cyst. Fibros. 2004, 3, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Tabacchioni, S.; Bevivino, A.; Dalmastri, C.; Chiarini, L. Burkholderia cepacia complex in the rhizosphere: A minireview. Ann. Microbiol. 2002, 52, 103–117. [Google Scholar]
- Pirone, L.; Chiarini, L.; Dalmastri, C.; Bevivino, A.; Tabacchioni, S. Detection of cultured and uncultured Burkholderia cepacia complex bacteria naturally occurring in the maize rhizosphere. Environ. Microbiol. 2005, 7, 1734–1742. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Visca, P. Burkholderia cepacia complex species: Health hazards and biotechnological potential. Trends Microbiol. 2006, 14, 277–286. [Google Scholar] [CrossRef] [PubMed]
- LiPuma, J.J.; Spilker, T.; Coenye, T.; Gonzalez, C.F. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 2002, 359, 2002–2003. [Google Scholar] [CrossRef]
- Baldwin, A.; Mahenthiralingam, E.; Drevinek, P.; Vandamme, P.; Govan, J.R.; Waine, D.J.; LiPuma, J.J.; Chiarini, L.; Dalmastri, C.; Henry, D.A.; et al. Environmental Burkholderia cepacia complex isolates in human infections. Emerg. Infect. Dis. 2007, 13, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Alavi, M.; Schmid, M.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. In Molecular Microbial Ecology of the Rhizosphere, 1st ed.; de Bruijn Frans, J., Ed.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2013; Volume 2, pp. 1209–1216. ISBN 9781118296172. [Google Scholar]
- Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Chiarini, L.; Belli, M.L.; Piana, S.; Materazzo, A.; Vandamme, P.; Manno, G. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: Genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 2002, 40, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Pirone, L.; Bragonzi, A.; Farcomeni, A.; Paroni, M.; Auriche, C.; Conese, M.; Chiarini, L.; Dalmastri, C.; Bevivino, A.; Ascenzioni, F. Burkholderia cenocepacia strains isolated from cystic fibrosis patients are apparently more invasive and more virulent than rhizosphere strains. Environ. Microbiol. 2008, 10, 2773–2784. [Google Scholar] [CrossRef] [PubMed]
- Bevivino, A.; Pirone, L.; Pilkington, R.; Cifani, N.; Dalmastri, C.; Callaghan, M.; Ascenzioni, F.; McClean, S. Interaction of environmental Burkholderia cenocepacia strains with cystic fibrosis and non-cystic fibrosis bronchial epithelial cells in vitro. Microbiology 2012, 158, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.W.; Altman, E.; Beveridge, T.J.; Speert, D.P. Colonial morphology of Burkholderia cepacia complex genomovar III: Implications in exopolysaccharide production, pilus expression, and persistence in the mouse. Infect. Immun. 2003, 71, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Bragonzi, A.; Farulla, I.; Paroni, M.; Twomey, K.B.; Pirone, L.; Lorè, N.I.; Bianconi, I.; Dalmastri, C.; Ryan, R.P.; Bevivino, A. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE 2012, 7, e52330. [Google Scholar] [CrossRef] [PubMed]
- Pirone, L. Pathogenicity of Clinical and Environmental Burkholderia cenocepacia Strains: Influence of Pseudomonas aeruginosa and Cystic Fibrosis Host. Ph.D. Thesis, University of Rome, Rome, Italy, 2009. [Google Scholar]
- Bragonzi, A.; Worlitzsch, D.; Pier, G.B.; Timpert, P.; Ulrich, M.; Hentzer, M.; Andersen, J.B.; Givskov, M.; Conese, M.; Döring, G. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 2005, 192, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Seed, K.D.; Dennis, J.J. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 2008, 76, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Roger, F.; Bourdier, A.; Lotthé, A.; Lamy, B.; Marchandin, H.; Jumas-Bilak, E. From environment to man: Genome evolution and adaptation of human opportunistic bacterial pathogens. Genes 2012, 3, 191–232. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Laevens, S.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Vandamme, P.; Chiarini, L. Burkholderia cepacia complex: Distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 2001, 3, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Dalmastri, C.; Baldwin, A.; Tabacchioni, S.; Bevivino, A.; Mahenthiralingam, E.; Chiarini, L.; Dowson, C. Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ. Microbiol. 2007, 9, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Manno, G.; Dalmastri, C.; Tabacchioni, S.; Vandamme, P.; Lorini, R.; Minicucci, L.; Romano, L.; Giannattasio, A.; Chiarini, L.; Bevivino, A. Epidemiology and clinical course of Burkholderia cepacia complex infections, particularly those caused by different Burkholderia cenocepacia strains, among patients attending an Italian cystic fibrosis center. J. Clin. Microbiol. 2004, 42, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.V.; Leitão, J.H.; Mahenthiralingam, E.; Vandamme, P.; Lito, L.; Barreto, C.; Salgado, M.J.; Sá-Correia, I. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: A 7-year study. J. Clin. Microbiol. 2003, 41, 4113–4120. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, E.P.; Sokol, P.A. Burkholderia cenocepacia differential gene expression during host–pathogen interactions and adaptation to the host environment. Front. Cell. Infect. Microbiol. 2011, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.; Feliciano, J.; Pita, T.; Guerreiro, S.; Leitão, J.H. Burkholderia cepacia complex regulation of virulence gene expression: A review. Genes 2017, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.N.; Cooper, V.S. Experimental adaptation of Burkholderia cenocepacia to onion medium reduces host range. Appl. Environ. Microbiol. 2010, 76, 2387–2396. [Google Scholar] [CrossRef] [PubMed]
- Sokol, P.A. Burkholderia: Molecular Microbiology and Genomics. In Burkholderia: Molecular Microbiology and Genomics; Coenye, T., Vandamme, P., Eds.; Horizon Bioscience; Taylor and Francis, Inc.: Wymondham, UK, 2007; pp. 229–250. ISBN 1904933289. [Google Scholar]
- Zlosnik, J.E.A.; Hird, T.J.; Fraenkel, M.C.; Moreira, L.M.; Henry, D.A.; Speert, D.P. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J. Clin. Microbiol. 2008, 46, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.; Chewapreecha, C.; Bentley, S.D. Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol. 2012, 7, 1283–1296. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Van den Driessche, F.; Vanhoutte, B.; Brackman, G.; Crabbé, A.; Rigole, P.; Vercruysse, J.; Verstraete, G.; Cappoen, D.; Vervaet, C.; Cos, P.; et al. Evaluation of combination therapy for Burkholderia cenocepacia lung infection in different in vitro and in vivo models. PLoS ONE 2017, 12, e0172723. [Google Scholar] [CrossRef] [PubMed]
- Koskiniemi, S.; Gibbons, H.S.; Sandegren, L.; Anwar, N.; Ouellette, G.; Broomall, S.; Karavis, M.; McGregor, P.; Liem, A.; Fochler, E.; et al. Pathoadaptive mutations in Salmonella enterica isolated after serial passage in mice. PLoS ONE 2013, 8, e70147. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Simpson, D.A.; Speert, D.P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 1997, 35, 808–816. [Google Scholar] [PubMed]
- Clode, F.E.; Kaufmann, M.E.; Malnick, H.; Pitt, T.L. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 2000, 38, 1763–1766. [Google Scholar] [PubMed]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Uehlinger, S.; Schwager, S.; Bernier, S.P.; Riedel, K.; Nguyen, D.T.; Sokol, P.A.; Eberl, L. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 2009, 77, 4102–4110. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bragonzi, A.; Paroni, M.; Pirone, L.; Coladarci, I.; Ascenzioni, F.; Bevivino, A. Environmental Burkholderia cenocepacia Strain Enhances Fitness by Serial Passages during Long-Term Chronic Airways Infection in Mice. Int. J. Mol. Sci. 2017, 18, 2417. https://doi.org/10.3390/ijms18112417
Bragonzi A, Paroni M, Pirone L, Coladarci I, Ascenzioni F, Bevivino A. Environmental Burkholderia cenocepacia Strain Enhances Fitness by Serial Passages during Long-Term Chronic Airways Infection in Mice. International Journal of Molecular Sciences. 2017; 18(11):2417. https://doi.org/10.3390/ijms18112417
Chicago/Turabian StyleBragonzi, Alessandra, Moira Paroni, Luisa Pirone, Ivan Coladarci, Fiorentina Ascenzioni, and Annamaria Bevivino. 2017. "Environmental Burkholderia cenocepacia Strain Enhances Fitness by Serial Passages during Long-Term Chronic Airways Infection in Mice" International Journal of Molecular Sciences 18, no. 11: 2417. https://doi.org/10.3390/ijms18112417
APA StyleBragonzi, A., Paroni, M., Pirone, L., Coladarci, I., Ascenzioni, F., & Bevivino, A. (2017). Environmental Burkholderia cenocepacia Strain Enhances Fitness by Serial Passages during Long-Term Chronic Airways Infection in Mice. International Journal of Molecular Sciences, 18(11), 2417. https://doi.org/10.3390/ijms18112417