iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai
Abstract
1. Introduction
2. Results
2.1. Protein Profiling
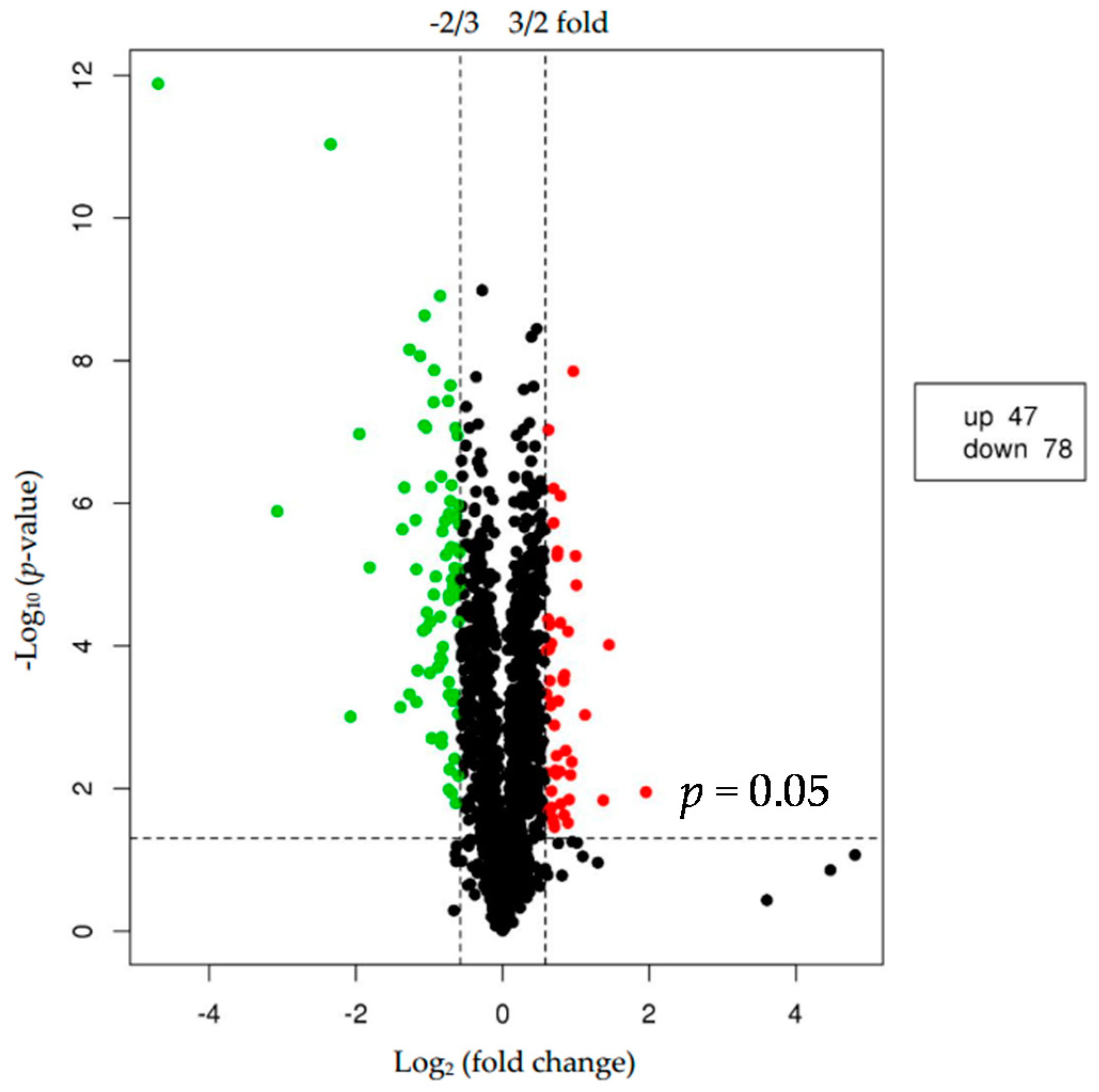
2.2. Differentially Expressed Proteins (DEPs)
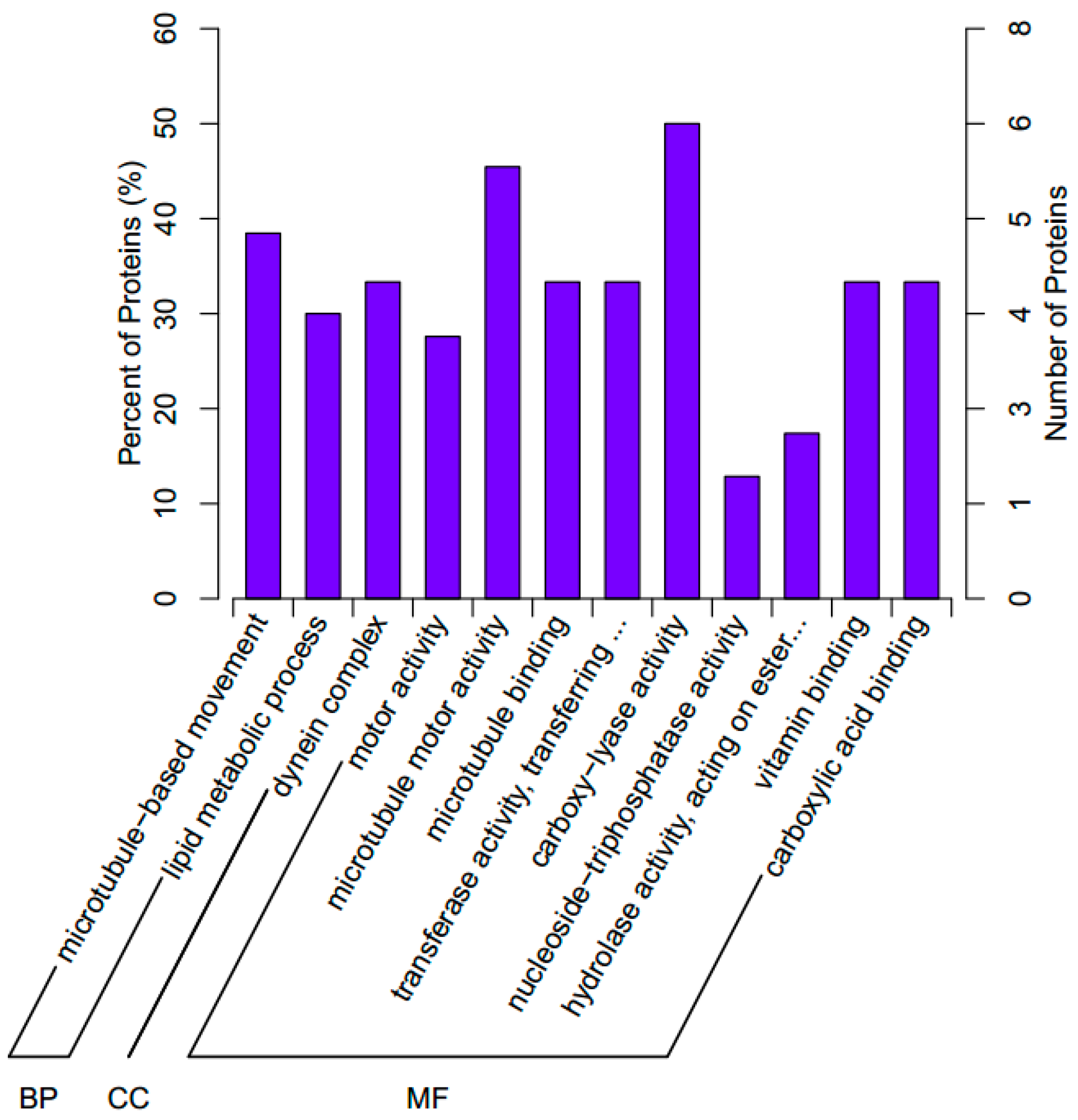
2.3. GO Functional Classification
2.4. KEGG Pathway Analysis
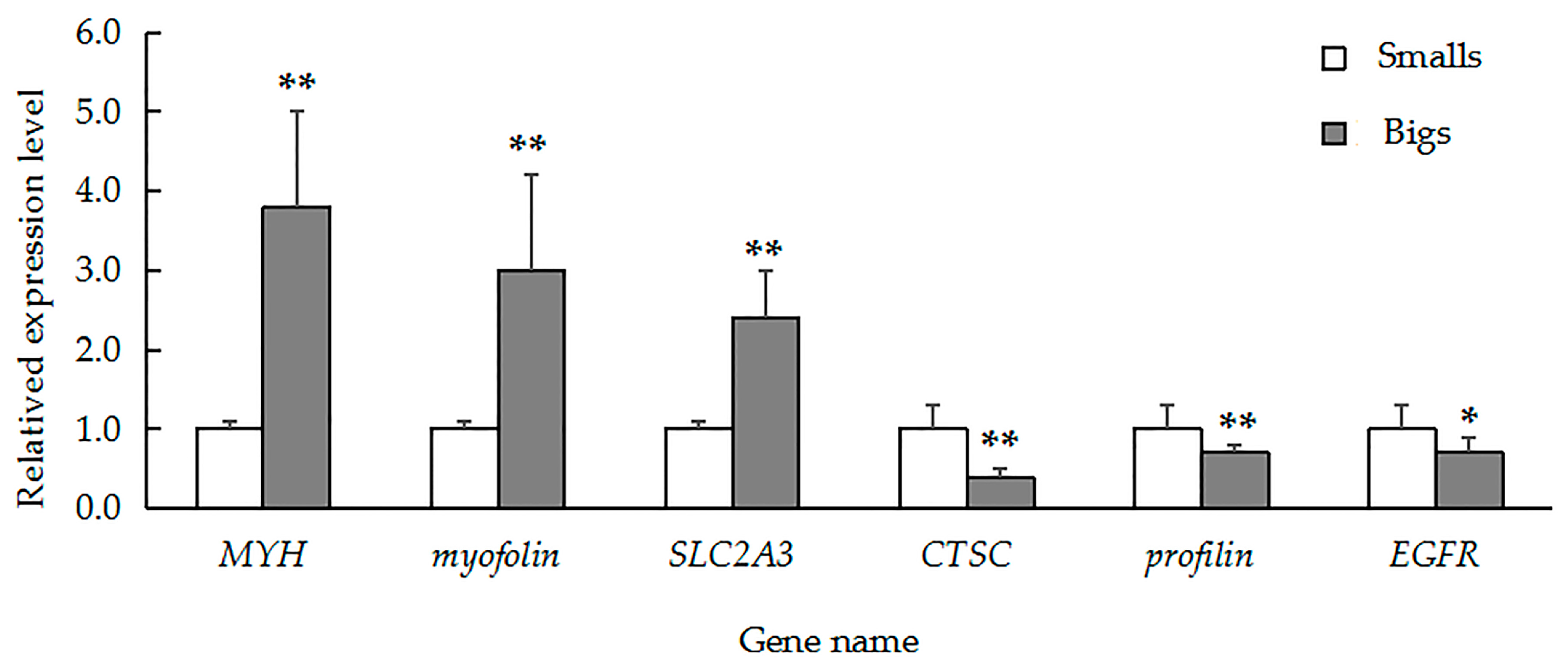
2.5. qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Experimental Tissue
4.2. Protein Preparation
4.3. iTRAQ Labeling of Peptides
4.4. HPLC Fractionation
4.5. LC-MS/MS Analysis
4.6. Data Analysis
4.7. qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guo, X.M.; Ford, S.E.; Zhang, F.S. Molluscan aquaculture in China. J. Shellfish Res. 1999, 18, 19–31. [Google Scholar]
- Luo, X.; Ke, C.H.; You, W.W.; Wang, D.X.; Chen, F. Molecular identification of interspecific hybrids between Haliotis discus hannai Ino and Haliotis gigantea Gmelin usingamplified fragment-length polymorphism and microsatellite markers. Aquacult. Res. 2010, 41, 1827–1834. [Google Scholar] [CrossRef]
- Chen, N.; Luo, X.; Gu, Y.T.; Han, G.D.; Dong, Y.W.; You, W.W.; Ke, C.H. Assessment of the thermal tolerance of abalone based on cardiac performance in Haliotis discus hannai, H. gigantea and their interspecific hybrid. Aquaculture 2016, 465, 258–264. [Google Scholar] [CrossRef]
- Guo, Y.F.; Zhao, W.W.; Gao, H.Q.; Wang, S.; Yu, P.M.; Yu, H.S.; Wang, D.; Wang, Q.; Wang, J.X.; Wang, Z.F.; et al. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2017. [Google Scholar]
- Naipil, C.C.; Muñoz, V.V.; Valdés, J.A.; Molina, A.; Escárate, C.G. RNA interference in Haliotis rufescens myostatin evidences upregulation of insulin signaling pathway. Agri Gene 2016, 1, 93–99. [Google Scholar] [CrossRef]
- Elliott, N.G. Genetic improvement programmes in abalone: What is the future? Aquac. Res. 2000, 31, 51–59. [Google Scholar] [CrossRef]
- Di, G.; Luo, X.; You, W.; Zhao, J.; Kong, X.; Ke, C. Proteomic analysis of muscle between hybrid abalone andparental lines Haliotis gigantea Reeve and Haliotis discus hannai Ino. Heredity 2015, 114, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Meng, B.; Chen, W.H.; Ge, X.M.; Liu, S.Q.; Yu, J. A proteomic study on postdiapaused embryonic development of brine shrimp (Artemia franciscana). Proteomics 2007, 7, 3580–3591. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Fernández, M.; Rodríguez-Piñeiro, A.M.; Oliveira, E.; Páez de la Cadena, M.; Rolán-Alvarez, E. Proteomic Comparison between Two Marine Snail Ecotypes Reveals Details about the Biochemistry of Adaptation. J. Proteome Res. 2008, 7, 4926–4934. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Y.; Thiyagarajan, V.; Qian, P.Y.; Qiu, J.W. Protein expression during the embryonic development of a gastropod. Proteomics 2010, 10, 2701–2711. [Google Scholar] [CrossRef] [PubMed]
- Karp, N.A.; Huber, W.; Sadowski, P.G.; Charles, P.D.; Hester, S.V.; Lilley, K.S. Addressing Accuracy and Precision Issues in iTRAQ Quantitation. Mol. Cell. Proteom. 2010, 9, 1885–1897. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Ba, H.X.; Zhang, W.; Coates, D.; Li, C.Y. iTRAQ-Based Quantitative Proteomic Analysis of the Potentiated and Dormant Antler Stem Cells. Int. J. Mol. Sci. 2016, 17, 1778. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.; Unwin, R.D.; Evans, C.A.; Griffiths, S.; Carney, L.; Zhang, L.; Jaworska, E.; Lee, C.F.; Blinco, D.; Okoniewski, M.J.; et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteom. 2008, 7, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.S.; Liu, X.H.; Liu, L.X.; Lou, W.H.; Jin, D.Y.; Yang, P.Y.; Wang, X.L. iTRAQ-based quantitative proteomics reveals myoferlin as a novel prognostic predictor in pancreatic adenocarcinoma. J. Proteom. 2013, 91, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Malécot, M.; Marie, A.; Dao, S.P.; Edery, M. iTRAQ-based proteomic study of the effectsof microcystin-LR on medaka fish liver. Proteomics 2011, 11, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Marancik, D.P.; Fast, M.D.; Camus, A.C. Proteomic characterization of the acute-phase response of yellow stingrays Urobatis jamaicensis after injection with a Vibrio anguillarum-ordalii bacterin. Fish Shellfish Immunol. 2013, 34, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Lü, A.; Hu, X.C.; Wang, Y.; Shen, X.J.; Li, X.; Zhu, A.H.; Tian, J.; Ming, Q.L.; Feng, Z.J. iTRAQ analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 2014, 36, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, J.; Mu, H.W.; Li, J.; Zhang, Y.H.; Xu, F.J.; Xiang, Z.M.; Qin, P.Y.; Qiu, J.W.; Yu, Z.N. Proteomic Basis of Stress Responses in the Gills of the Pacific Oyster Crassostrea gigas. J. Proteom. Res. 2015, 14, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.Y.; Yu, X.; Gao, B.Q.; Li, J.; Liu, P. iTRAQ-based identification of differentially expressed proteins related to growth in the swimming crab, Portunus trituberculatus. Aquac. Res. 2016, 1–11. [Google Scholar] [CrossRef]
- Xu, D.X.; Sun, L.N.; Liu, S.L.; Zhang, L.B.; Yang, H.S. Understanding the Heat Shock Response in the Sea Cucumber Apostichopus japonicus, Using iTRAQ-Based Proteomics. Int. J. Mol. Sci. 2016, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Miranda, D.V.; Portilla, M.D.; Escárate, C.G. Characterization of the growth-related transcriptome in California red abalone (Haliotis rufescens) through RNA-Seq analysis. Mar. Genom. 2015, 24, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Kim, G.D.; Kim, J.M.; Lim, H.K. Differentially-Expressed Genes Associated with Faster Growth of the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2015, 16, 27520–27534. [Google Scholar] [CrossRef] [PubMed]
- Hevrøy, E.M.; Jordal, A.E.O.; Hordvik, I.; Espe, M.; Hemre, G.I.; Olsvik, P.A. Myosin heavy chain mRNA expression correlates higher with muscle protein accretion than growth in Atlantic salmon, Salmo salar. Aquaculture 2006, 252, 453–461. [Google Scholar] [CrossRef]
- Gauvry, L.; Fauconneau, B. Cloning of a trout fast skeletal myosin heavy chain expressed both in embryo and adult muscles and in myotubes neoformed in vitro. Comp. Biochem. Physiol. 1996, 115, 183–190. [Google Scholar] [CrossRef]
- Young, R.B.; Hsieh, M.Y.; Hudson, J.R., Jr.; Richter, H.E.; Scott, M. Expression pattern and partial sequence analysis of a fetal bovine myosin heavy chain gene. J. Anim. Sci. 1994, 72, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Dong, J.J.; Sun, C.F.; Tian, Y.Y.; Ye, X. cDNA Cloning and Analyses of Two Myosin Heavy Chain Isoforms of Mandarin Fish (Siniperca chuatsi) Based on Transcriptome Sequencing. Prog. Fish. Sci. 2017, 38, 51–61. [Google Scholar] [CrossRef]
- Chen, T.L.; Kowalczyk, P.A.; Ho, G.; Chisholm, R.L. Targeted disruption of the Dictyostelium myosin essential light chain gene produces cells defective in cytokinesis and morphogenesis. J. Cell Sci. 1995, 108, 3207–3218. [Google Scholar] [PubMed]
- Greenfield, N.J.; Montelione, G.T.; Farid, R.S.; Hitchcock-DeGregori, S.E. The structure of the N-terminus of striated muscle α-tropomyosin in a chimeric peptide: Nuclear magnetic resonance structure and circular dichroism studies. Biochemistry 1998, 37, 7834–7843. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Vandekerckhove, J. Stucture and function of actin. Ann. Rev. Biophys. 1992, 21, 49–76. [Google Scholar]
- Patwary, M.U.; Reith, M.; Kenchington, E.L. Isolation and charcterization of a cDNA encoding an actin gene from sea scallop (Placopecten magellanicus). J. Shellfish Res. 1996, 15, 265–270. [Google Scholar]
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- DesGroseillers, L.; Auclair, D.; Wickham, L. Nucleotide sequence of an actin cDNA gene from Aplysia californica. Nucleic Acids Res. 1990, 18, 3654. [Google Scholar] [CrossRef] [PubMed]
- DesGroseillers, L.; Auclair, D.; Wickham, L.; Maalouf, M. A novel actin cDNA is expressed in the neurons of Aplysia californica. Biochim. Biophys. Acta 1994, 1217, 322–324. [Google Scholar] [CrossRef]
- Miyamoto, H.; Hamaguchi, M.; Okoshi, K. Analysis of genes expressed in the mantle of oyster Crassostreagigas. Fish Sci. 2002, 68, 651–658. [Google Scholar] [CrossRef]
- VanLoon, A.E.; Goedemans, H.J.; Daemen, A.J.J.M.; van de Kamp, A.J.; van den Biggelaar, J.A.M. Actin genes expressed during early development of Patella vulgata. Rouxs Arch. Dev. Biol. 1993, 202, 77–84. [Google Scholar] [CrossRef]
- Bryant, M.J.; Flint, H.J.; Sin, F.Y.T. Isolation, Characterization, and Expression Analysis of Three Actin Genes in the New Zealand Black-Footed Abalone, Haliotis iris. Mar. Biotechnol. 2006, 8, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.T.; Zhang, Y.Z. Progress in Profilin. Chin. J. Cell Biol. 2007, 29, 325–330. [Google Scholar]
- Pernier, J.; Shekhar, S.; Jegou, A.; Guichard, B.; Carlier, M.F. Profilin Interaction with Actin Filament Barbed End Controls Dynamic Instability, Capping, Branching, and Motility. Dev. Cell 2016, 36, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Carroll, R.T.; Burke, T.A.; Christensen, J.R.; Bestul, A.J.; Sees, J.A.; James, M.L.; Sirotkin, V.; Kovar, D.R. Profilin Regulates F-Actin Network Homeostasis by Favoring Formin over Arp2/3 Complex. Dev. Cell 2015, 32, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rotty, J.D.; Wu, C.; Haynes, E.M.; Suarez, C.; Winkelman, J.D.; Johnson, H.E.; Haugh, J.M.; Kovar, D.R.; Bear, J.E. Profilin-1 Serves as a Gatekeeper for Actin Assembly by Arp2/3-Dependent and Independent Pathways. Dev. Cell 2015, 32, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Sohn, R.H.; Clermont, P.J. Profilin: At the crossroads of signal transduction and the actin cytoskeleton. BioEssays 1994, 16, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Gasser, R.B.; Jones, M.K.; Lightowlers, M.W. Identification and characterization of myophilin, a muscle-specific antigen of Echinococcus granulosus. Mol. Biochem. Parasitol. 1995, 70, 139–148. [Google Scholar] [CrossRef]
- Martin, R.M.; Colebrook, A.L.; Gasser, R.B.; Lightowlers, M.W. Antibody responses of patients with cystic hydatid disease to recombinant myophilin of Echinococcus granulosus. Acta Trop. 1996, 61, 307–314. [Google Scholar] [CrossRef]
- Peng, H.L.; Song, K.; Huang, C.Y.; Ye, S.; Song, H.G.; Hu, W.; Han, Z.G.; McManus, D.P.; Zhao, G.P.; Zhang, Q.H. Expression, immunolocalization and serodiagnostic value of a myophilin-like protein from Schistosoma japonicum. Exp. Parasitol. 2008, 119, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Sloane, B.F. Cathepsin B and cystatins: Evidence for a role in cancer progression. Semin. Cancer Biol. 1990, 1, 137–152. [Google Scholar] [PubMed]
- Tong, B.; Wan, B.; Wei, Z.; Wang, T.; Zhao, P.; Dou, Y.; Lv, Z.; Xia, Y.; Dai, Y. Role of cathepsin B in regulating migration and invasion of fibroblast-like synoviocytes into inflamed tissue from patients with rheumatoid arthritis. Clin. Exp. Immunol. 2014, 177, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.D.; Ham, B.; Mogridge, J.; Saftig, P.; Lin, S.; Kim, S.O. Cathepsin B-mediated autophagy flux facilitates the anthrax toxin receptor 2-mediated delivery of anthrax lethal factor into the cytoplasm. J. Biol. Chem. 2010, 285, 2120–2129. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.E.; Ho, C.C.; Yang, S.F.; Lin, S.H.; Yeh, K.T.; Lin, C.W.; Chen, M.K. Cathepsin B Expression and the Correlation with Clinical Aspects of Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Vivar, C.; Potter, M.C.; van Praag, H. All about running: Synaptic plasticity, growth factors and adult hippocampal neurogenesis. Curr. Top. Behav. Neurosci. 2012, 15, 189–210. [Google Scholar] [CrossRef]
- Houseweart, M.K.; Pennacchio, L.A.; Vilaythong, A.; Peters, C.; Noebels, J.L.; Myers, R.M. Cathepsin B but not cathepsins L or S contributes to the pathogenesis of Unverricht-Lundborg progressive myoclonus epilepsy (EPM1). J. Neurobiol. 2003, 56, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Kominami, E.; Ishido, K.; Muno, D.; Sato, N. The primary structure and tissue distribution of cathepsin C. Biol. Chem. 1992, 373, 367–373. [Google Scholar] [CrossRef]
- Dolenc, I.; Turk, B.; Pungerc, G.; Ritonja, A.; Turk, V. Oligomeric structure and substrate induced inhibition of human cathepsin C. J. Biol. Chem. 1995, 270, 21626–21631. [Google Scholar] [CrossRef] [PubMed]
- Muno, D.; Ishidoh, K.; Ueno, T.; Kominami, E. Processing and transport of the precursor of cathepsin C during its transfer into lysosomes. Arch. Biochem. Biophys. 1993, 306, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Wolters, P.J.; Raymond, W.W.; Blount, J.L.; Caughey, G.H. Regulated expression, processing, and secretion of dog mast cell dipeptidyl peptidase I. J. Biol. Chem. 1998, 273, 15514–15520. [Google Scholar] [CrossRef] [PubMed]
- Martínez, D.S.; Krzywinska, E.; Rathore, M.G.; Saumet, A.; Cornillon, A.; Lopez-Royuela, N.; Martínez-Lostao, L.; Ramirez-Labrada, A.; Lu, Z.Y.; Rossi, J.F.; et al. All-trans retinoic acid (ATRA) induces miR-23a expression, decreases CTSC expression and granzyme B activity leading to impaired NK cell cytotoxicity. Int. J. Biochem. Cell Biol. 2014, 49, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–428. [Google Scholar] [CrossRef] [PubMed]


| Group Name | Total Spectra | Spectra | Ratio Identified | Peptides | Proteins |
|---|---|---|---|---|---|
| All | 425,477 | 44,436 | 10.40% | 10,097 | 1904 |
| KEGG Pathway | Upregulated Proteins | Downregulated Proteins |
|---|---|---|
| Lysosome | putative inorganic phosphate cotransporter (Picot; Accession Number: O61369), actin (Accession Number: Q93129) | ganglioside GM2 activator (GM2A; Accession Number: Q8HXX6), cathepsin C (CTSC; Accession Number: A0A023PJH7), cathepsin B (CTSB; Accession Number: A1E295), palmitoyl-protein thioesterase 1 (PPT1; Accession Number: Q8HXW6), N-acetylglucosamine-6-sulfatase (GNS; Accession Number: Q8BFR4), α-N-acetylgalactosaminidase (Accession Number: Q90744) |
| Adherens junction | actin A1 (Accession Number: Q5BQE5), actin-2 (Accession Number: P26197), actin | kitasatospora griseola strain MF730-N6 RKJC_4 (Accession Number: A0A0D0PVQ3), β actin (ACTB; Accession Number: G8HY07), epidermal growth factor receptor (EGFR; Accession Number: P0CY46), glycerophosphodiester phosphodiesterase domain-containing protein 1 (GDPD1; Accession Number: Q8N9F7) |
| Bladder cancer | - | thymidine phosphorylase (Tymp; Accession Number: Q5FVR2), EGFR, GDPD1 |
| Apoptosis | actin A1, actin-2, actin, myophilin (Accession Number: Q24799) | CTSC, GDPD1, CTSB |
| Thyroid hormone signaling pathway | actin A1, actin-2, actin, solute carrier family 2 facilitated glucose transporter member 3 (SLC2A3; Accession Number: P47843) | ACTB, GDPD1 |
| Endometrial cancer | - | kitasatospora griseola strain MF730-N6 RKJC_4, EGFR, GDPD1 |
| Shigellosis | actin A1, actin-2, actin | profilin (Accession Number: F4XXT7), ACTB, GDPD1 |
| Regulation of actin cytoskeleton | actin A1, actin-2, actin | profilin, ACTB, EGFR, GDPD1, phosphatidylinositol 5-phosphate 4-kinase type-2 β (PIP4K2B; Accession Number: P78356), myosin regulatory light chain sqh (Accession Number: P40423) |
| Salmonella infection | actin A1, actin-2, actin | profilin, ACTB, GDPD1 |
| Viral myocarditis | actin A1, actin-2, actin, myosin heavy chain (MYH; Accession Number: P24733), MYH II (Accession Number: O96700) | ACTB |
| Hippo signaling pathway-fly | actin A1, actin-2, actin, protocadherin Fat 4 (FAT4; Accession Number: Q6V0I7) | ACTB |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; You, W.; Luo, X.; Ke, C. iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2017, 18, 2237. https://doi.org/10.3390/ijms18112237
Huang J, You W, Luo X, Ke C. iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. International Journal of Molecular Sciences. 2017; 18(11):2237. https://doi.org/10.3390/ijms18112237
Chicago/Turabian StyleHuang, Jianfang, Weiwei You, Xuan Luo, and Caihuan Ke. 2017. "iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai" International Journal of Molecular Sciences 18, no. 11: 2237. https://doi.org/10.3390/ijms18112237
APA StyleHuang, J., You, W., Luo, X., & Ke, C. (2017). iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. International Journal of Molecular Sciences, 18(11), 2237. https://doi.org/10.3390/ijms18112237




