Tubulin Post-Translational Modifications and Microtubule Dynamics
Abstract
1. Introduction
2. Polyamination and Cold-Stable Microtubules
3. Microtubule Acetylation
4. Methylation, Modification of the Minus End of a Subset of Mitotic Microtubules
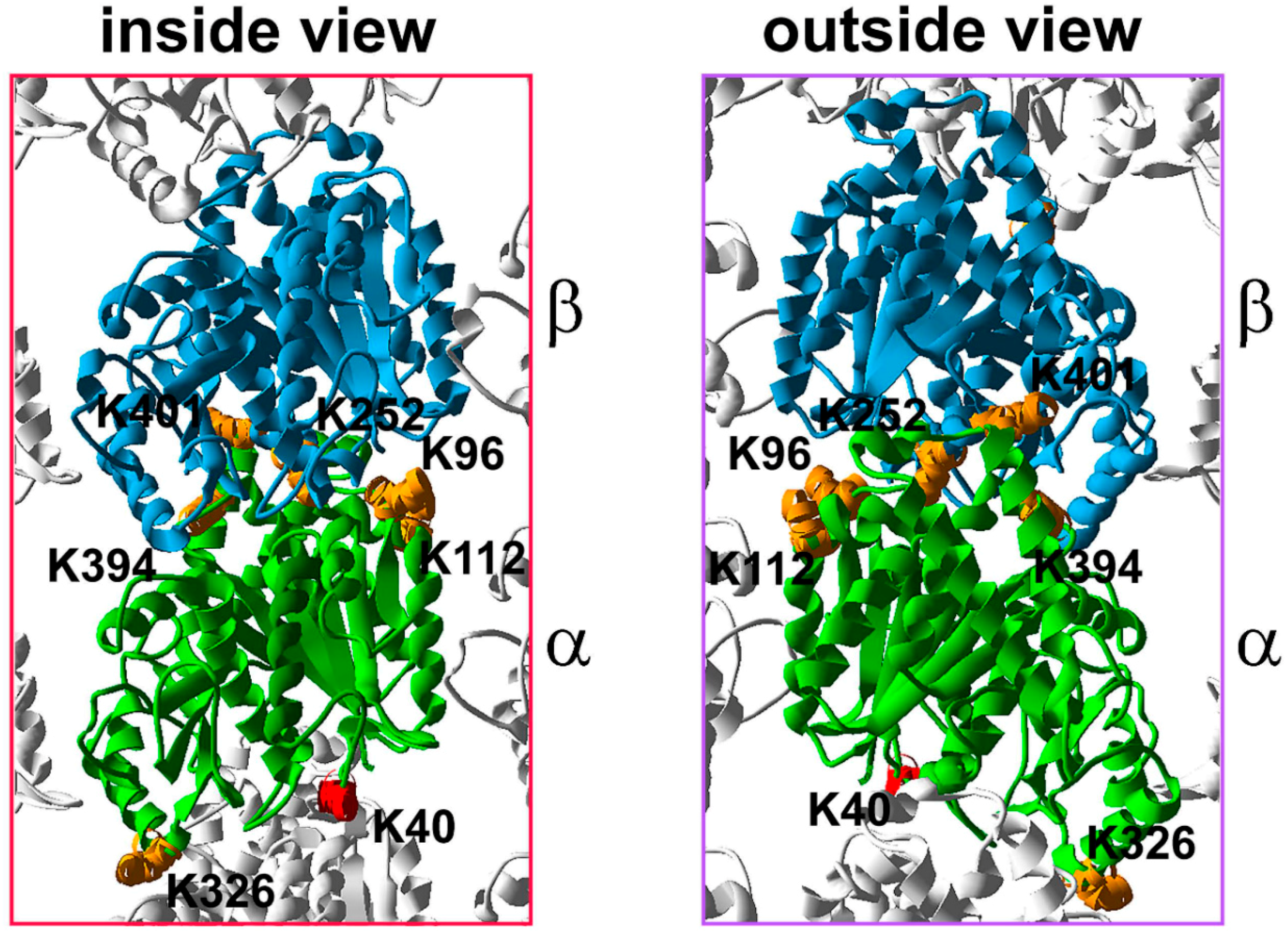
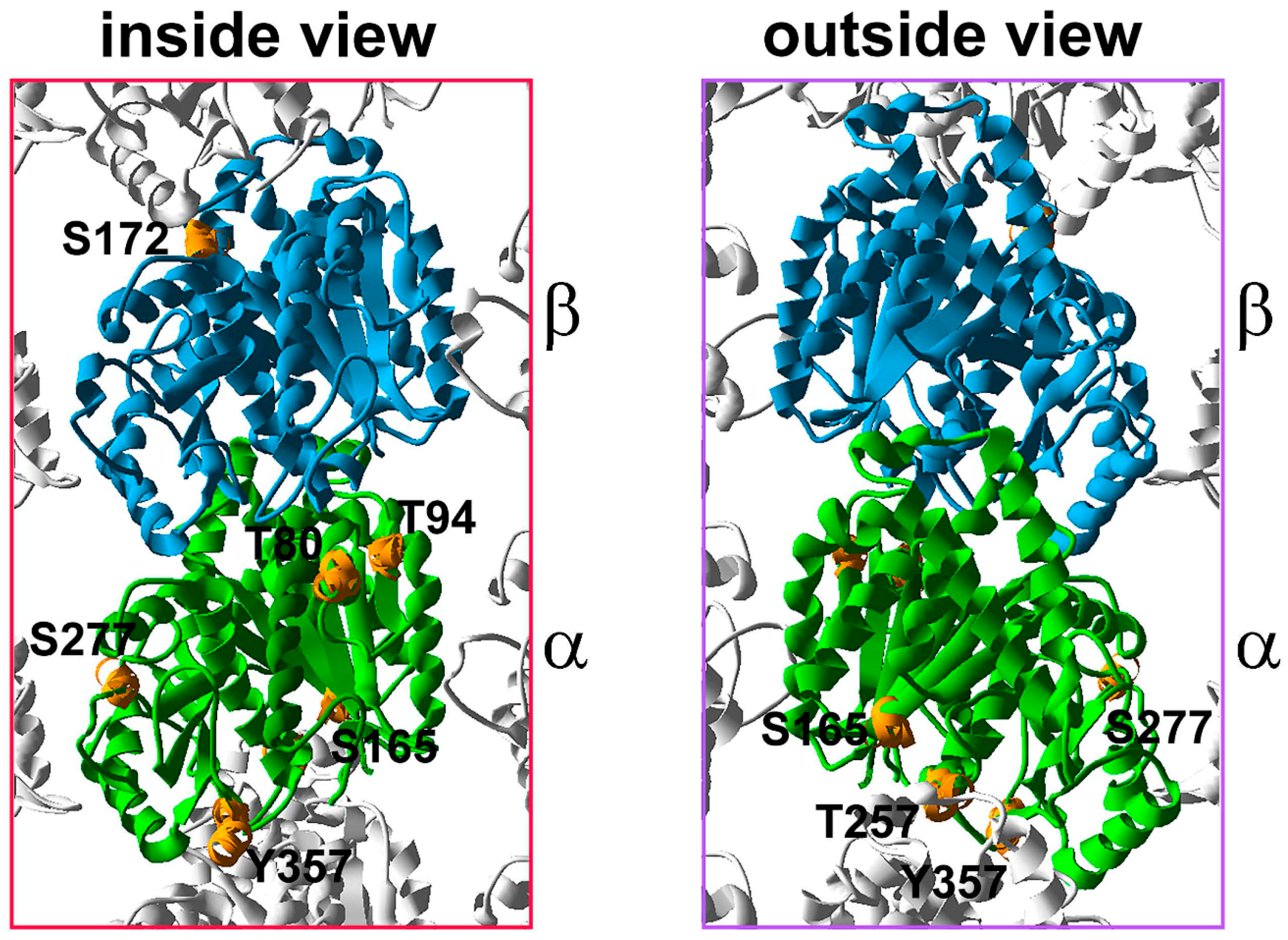
5. Tubulin Phosphorylation
6. Post-Translational Modifications Specific to the Tubulin Tail
6.1. Tyrosination, Detyrosination and Δ2 Modification
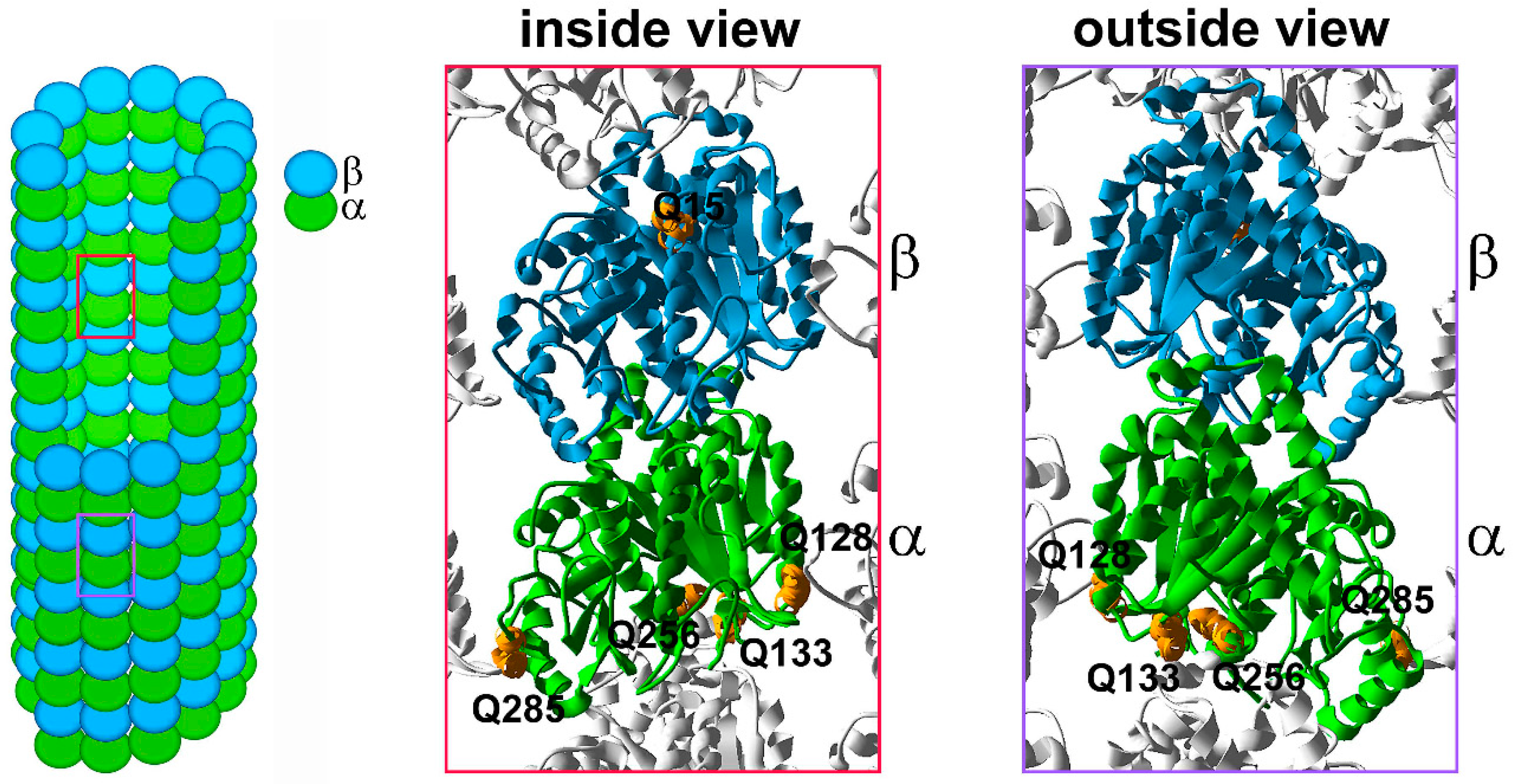
6.2. Glutamylation and Glycylation, Polymodifications of the Tubulin Tail
7. Other Modifications
Acknowledgments
Conflicts of Interest
References
- Cambray-Deakin, M.A.; Burgoyne, R.D. Acetylated and detyrosinated α-tubulins are co-localized in stable microtubules in rat meningeal fibroblasts. Cell Motil. Cytoskelet. 1987, 8, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Webster, D.R.; Borisy, G.G. Microtubules are acetylated in domains that turn over slowly. J. Cell Sci. 1989, 92, 57–65. [Google Scholar] [PubMed]
- Khawaja, S.; Gundersen, G.G.; Bulinski, J.C. Enhanced stability of microtubules enriched in detyrosinated tubulin is not a direct function of detyrosination level. J. Cell Biol. 1988, 106, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Verhey, K.J.; Gaertig, J. The tubulin code. Cell Cycle 2007, 6, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Fukushige, T.; Siddiqui, Z.K.; Chou, M.; Culotti, J.G.; Gogonea, C.B.; Siddiqui, S.S.; Hamelin, M. MEC-12, an α-tubulin required for touch sensitivity in C. elegans. J. Cell Sci. 1999, 112 Pt 3, 395–403. [Google Scholar] [PubMed]
- Brady, S.T.; Tytell, M.; Lasek, R.J. Axonal tubulin and axonal microtubules: Biochemical evidence for cold stability. J. Cell Biol. 1984, 99, 1716–1724. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Kirkpatrick, L.L.; Schilling, A.B.; Helseth, D.L.; Chabot, N.; Keillor, J.W.; Johnson, G.V.; Brady, S.T. Transglutaminase and polyamination of tubulin: Posttranslational modification for stabilizing axonal microtubules. Neuron 2013, 78, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E.; Whittaker, M.; Milligan, R.A.; Downing, K.H. High-resolution model of the microtubule. Cell 1999, 96, 79–88. [Google Scholar] [CrossRef]
- Inclán, Y.F.; Nogales, E. Structural models for the self-assembly and microtubule interactions of γ-, δ- and ε-tubulin. J. Cell Sci. 2001, 114 Pt 2, 413–422. [Google Scholar] [PubMed]
- LeDizet, M.; Piperno, G. Identification of an acetylation site of Chlamydomonas α-tubulin. Proc. Natl. Acad. Sci. USA 1987, 84, 5720–5724. [Google Scholar] [CrossRef] [PubMed]
- Akella, J.S.; Wloga, D.; Kim, J.; Starostina, N.G.; Lyons-Abbott, S.; Morrissette, N.S.; Dougan, S.T.; Kipreos, E.T.; Gaertig, J. MEC-17 is an α-tubulin acetyltransferase. Nature 2010, 467, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Shida, T.; Cueva, J.G.; Xu, Z.; Goodman, M.B.; Nachury, M.V. The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 2010, 107, 21517–21522. [Google Scholar] [CrossRef] [PubMed]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kwon, S.; Yamaguchi, T.; Cubizolles, F.; Rousseaux, S.; Kneissel, M.; Cao, C.; Li, N.; Cheng, H.L.; Chua, K.; et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 2008, 28, 1688–1701. [Google Scholar] [CrossRef] [PubMed]
- Soppina, V.; Herbstman, J.F.; Skiniotis, G.; Verhey, K.J. Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS ONE 2012, 7, e48204. [Google Scholar] [CrossRef] [PubMed]
- Howes, S.C.; Alushin, G.M.; Shida, T.; Nachury, M.V.; Nogales, E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol. Biol. Cell 2014, 25, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Coombes, C.; Yamamoto, A.; McClellan, M.; Reid, T.A.; Plooster, M.; Luxton, G.W.; Alper, J.; Howard, J.; Gardner, M.K. Mechanism of microtubule lumen entry for the α-tubulin acetyltransferase enzyme αTAT1. Proc. Natl. Acad. Sci. USA 2016, 113, E7176–E7184. [Google Scholar] [CrossRef] [PubMed]
- Ly, N.; Elkhatib, N.; Bresteau, E.; Piétrement, O.; Khaled, M.; Magiera, M.M.; Janke, C.; Le Cam, E.; Rutenberg, A.D.; Montagnac, G. αTAT1 controls longitudinal spreading of acetylation marks from open microtubules extremities. Sci. Rep. 2016, 6, 35624. [Google Scholar] [CrossRef] [PubMed]
- Szyk, A.; Deaconescu, A.M.; Spector, J.; Goodman, B.; Valenstein, M.L.; Ziolkowska, N.E.; Kormendi, V.; Grigorieff, N.; Roll-Mecak, A. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 2014, 157, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Gaertig, J.; Cruz, M.A.; Bowen, J.; Gu, L.; Pennock, D.G.; Gorovsky, M.A. Acetylation of lysine 40 in α-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 1995, 129, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- L’Hernault, S.W.; Rosenbaum, J.L. Chlamydomonas α-tubulin is posttranslationally modified in the flagella during flagellar assembly. J. Cell Biol. 1983, 97, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Piperno, G.; Fuller, M.T. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 1985, 101, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- LeDizet, M.; Piperno, G. Cytoplasmic microtubules containing acetylated α-tubulin in Chlamydomonas reinhardtii: Spatial arrangement and properties. J. Cell Biol. 1986, 103, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Piperno, G.; LeDizet, M.; Chang, X.J. Microtubules containing acetylated α-tubulin in mammalian cells in culture. J. Cell Biol. 1987, 104, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, A.; Shimazu, T.; Sumida, Y.; Saito, A.; Yoshimatsu, Y.; Seigneurin-Berny, D.; Osada, H.; Komatsu, Y.; Nishino, N.; Khochbin, S.; et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002, 21, 6820–6831. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.D.; Marmo, T.P.; Salam, A.A.; Che, S.; Finkelstein, E.; Kabarriti, R.; Xenias, H.S.; Mazitschek, R.; Hubbert, C.; Kawaguchi, Y.; et al. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 2007, 120 Pt 8, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Haggarty, S.J.; Koeller, K.M.; Wong, J.C.; Grozinger, C.M.; Schreiber, S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA 2003, 100, 4389–4394. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.; Ackerman, B.; Gundersen, G.G. Cell biology: Tubulin acetylation and cell motility. Nature 2003, 421, 230. [Google Scholar] [CrossRef] [PubMed]
- Kalebic, N.; Martinez, C.; Perlas, E.; Hublitz, P.; Bilbao-Cortes, D.; Fiedorczuk, K.; Andolfo, A.; Heppenstall, P.A. Tubulin acetyltransferase αTAT1 destabilizes microtubules independently of its acetylation activity. Mol. Cell. Biol. 2013, 33, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Kalebic, N.; Sorrentino, S.; Perlas, E.; Bolasco, G.; Martinez, C.; Heppenstall, P.A. αTAT1 is the major α-tubulin acetyltransferase in mice. Nat. Commun. 2013, 4, 1962. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Schaedel, L.; Portran, D.; Aguilar, A.; Gaillard, J.; Marinkovich, M.P.; Théry, M.; Nachury, M.V. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 2017, 356, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, Y.; Ballestrem, C.; Carramusa, L.; Mazitschek, R.; Khochbin, S.; Bershadsky, A. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J. Cell Sci. 2009, 122 Pt 19, 3531–3541. [Google Scholar] [CrossRef] [PubMed]
- Castro-Castro, A.; Janke, C.; Montagnac, G.; Paul-Gilloteaux, P.; Chavrier, P. ATAT1/MEC-17 acetyltransferase and HDAC6 deacetylase control a balance of acetylation of α-tubulin and cortactin and regulate MT1-MMP trafficking and breast tumor cell invasion. Eur. J. Cell Biol. 2012, 91, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.J. Tubulin acetylation: Responsible enzymes, biological functions and human diseases. Cell Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef] [PubMed]
- Ran, J.; Yang, Y.; Li, D.; Liu, M.; Zhou, J. Deacetylation of α-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly. Sci. Rep. 2015, 5, 12917. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Au, M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 1989, 243 (4894 Pt 1), 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Thomson, J.N. Organization of neuronal microtubules in the nematode Caenorhabditis elegans. J. Cell Biol. 1979, 82, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Topalidou, I.; Keller, C.; Kalebic, N.; Nguyen, K.C.; Somhegyi, H.; Politi, K.A.; Heppenstall, P.; Hall, D.H.; Chalfie, M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr. Biol. 2012, 22, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Cueva, J.G.; Hsin, J.; Huang, K.C.; Goodman, M.B. Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr. Biol. 2012, 22, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; Hilliard, M.A. Loss of MEC-17 leads to microtubule instability and axonal degeneration. Cell Rep. 2014, 6, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Savage, C.; Hamelin, M.; Culotti, J.G.; Coulson, A.; Albertson, D.G.; Chalfie, M. mec-7 is a β-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Gene. Dev. 1989, 3, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Maruta, H.; Greer, K.; Rosenbaum, J.L. The acetylation of α-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 1986, 103, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Portran, D.; Schaedel, L.; Xu, Z.; Théry, M.; Nachury, M.V. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell Biol. 2017, 19, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Baas, P.W. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J. Neurosci. 2010, 30, 7215–7226. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.X.; Xiong, Y.; Xiong, Z.; Wang, Q.; Zhang, Y.Q.; Jin, S. Microtubule-severing protein katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development 2014, 141, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Leo, L.; Yu, W.; D'Rozario, M.; Waddell, E.A.; Marenda, D.R.; Baird, M.A.; Davidson, M.W.; Zhou, B.; Wu, B.; Baker, L.; et al. Vertebrate fidgetin restrains axonal growth by severing labile domains of microtubules. Cell Rep. 2015, 12, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Valenstein, M.L.; Roll-Mecak, A. graded control of microtubule severing by tubulin glutamylation. Cell 2016, 164, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.W.; Hou, F.; Zhang, J.; Phu, L.; Loktev, A.V.; Kirkpatrick, D.S.; Jackson, P.K.; Zhao, Y.; Zou, H. A novel acetylation of β-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 2011, 22, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of α β-tubulin at 3.5 A resolution. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xiong, Y.; Ren, Y.; Zhang, L.; He, X.; Wang, X.; Liu, M.; Li, D.; Shui, W.; Zhou, J. Proteomic profiling and functional characterization of multiple post-translational modifications of tubulin. J. Proteome Res. 2015, 14, 3292–3304. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xiong, Y.; Li, S.; Ren, Y.; He, Q.; Gao, S.; Zhou, J.; Shui, W. New HDAC6-mediated deacetylation sites of tubulin in the mouse brain identified by quantitative mass spectrometry. Sci. Rep. 2015, 5, 16869. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Wandosell, F.; Serrano, L.; Avila, J. Phosphorylation of α-tubulin carboxyl-terminal tyrosine prevents its incorporation into microtubules. J. Biol. Chem. 1987, 262, 8268–8273. [Google Scholar] [PubMed]
- Matten, W.T.; Aubry, M.; West, J.; Maness, P.F. Tubulin is phosphorylated at tyrosine by pp60c-src in nerve growth cone membranes. J. Cell Biol. 1990, 111 (5 Pt 1), 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.D.; Furlong, M.T.; Asai, D.J.; Harrison, M.L.; Geahlen, R.L. Syk, activated by cross-linking the B-cell antigen receptor, localizes to the cytosol where it interacts with and phosphorylates α-tubulin on tyrosine. J. Biol. Chem. 1996, 271, 4755–4762. [Google Scholar] [PubMed]
- Fernandez, J.A.; Keshvara, L.M.; Peters, J.D.; Furlong, M.T.; Harrison, M.L.; Geahlen, R.L. Phosphorylation- and activation-independent association of the tyrosine kinase Syk and the tyrosine kinase substrates Cbl and Vav with tubulin in B-cells. J. Biol. Chem. 1999, 274, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Faruki, S.; Geahlen, R.L.; Asai, D.J. Syk-dependent phosphorylation of microtubules in activated B-lymphocytes. J. Cell Sci. 2000, 113 Pt 14, 2557–2565. [Google Scholar] [PubMed]
- Laurent, C.E.; Delfino, F.J.; Cheng, H.Y.; Smithgall, T.E. The human c-Fes tyrosine kinase binds tubulin and microtubules through separate domains and promotes microtubule assembly. Mol. Cell. Biol. 2004, 24, 9351–9358. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sayeski, P.P. Identification of tubulin as a substrate of Jak2 tyrosine kinase and its role in Jak2-dependent signaling. Biochemistry 2007, 46, 7153–7162. [Google Scholar] [CrossRef] [PubMed]
- Ori-McKenney, K.M.; McKenney, R.J.; Huang, H.H.; Li, T.; Meltzer, S.; Jan, L.Y.; Vale, R.D.; Wiita, A.P.; Jan, Y.N. Phosphorylation of β-tubulin by the down syndrome kinase, Minibrain/DYRK1a, regulates microtubule dynamics and dendrite morphogenesis. Neuron 2016, 90, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Fourest-Lieuvin, A.; Peris, L.; Gache, V.; Garcia-Saez, I.; Juillan-Binard, C.; Lantez, V.; Job, D. Microtubule regulation in mitosis: Tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol. Biol. Cell 2006, 17, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Caudron, F.; Denarier, E.; Thibout-Quintana, J.C.; Brocard, J.; Andrieux, A.; Fourest-Lieuvin, A. Mutation of Ser172 in yeast β tubulin induces defects in microtubule dynamics and cell division. PLoS ONE 2010, 5, e13553. [Google Scholar] [CrossRef] [PubMed]
- Ley, S.C.; Verbi, W.; Pappin, D.J.; Druker, B.; Davies, A.A.; Crumpton, M.J. Tyrosine phosphorylation of α tubulin in human T lymphocytes. Eur. J. Immunol. 1994, 24, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Abeyweera, T.P.; Chen, X.; Rotenberg, S.A. Phosphorylation of α6-tubulin by protein kinase Cα activates motility of human breast cells. J. Biol. Chem. 2009, 284, 17648–17656. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Tsimounis, A.; Chen, X.; Rotenberg, S.A. Phosphorylation of α-tubulin by protein kinase C stimulates microtubule dynamics in human breast cells. Cytoskeleton (Hoboken) 2014, 71, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Mitsopoulos, C.; Zihni, C.; Garg, R.; Ridley, A.J.; Morris, J.D. The prostate-derived sterile 20-like kinase (PSK) regulates microtubule organization and stability. J. Biol. Chem. 2003, 278, 18085–18091. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.C.; Tiu, S.Y.; Li, Q.; Qi, R.Z. Direct regulation of microtubule dynamics by protein kinase CK2. J. Biol. Chem. 2004, 279, 4433–4439. [Google Scholar] [CrossRef] [PubMed]
- Saoudi, Y.; Paintrand, I.; Multigner, L.; Job, D. Stabilization and bundling of subtilisin-treated microtubules induced by microtubule associated proteins. J. Cell Sci. 1995, 108 Pt 1, 357–367. [Google Scholar] [PubMed]
- Yu, I.; Garnham, C.P.; Roll-Mecak, A. Writing and Reading the Tubulin Code. J. Biol. Chem. 2015, 290, 17163–17172. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Brady, S.T. Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 2015, 25, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Joachimiak, E.; Louka, P.; Gaertig, J. Posttranslational modifications of tubulin and cilia. Cold Spring Harb. Perspect. Biol. 2016, 9, a028159. [Google Scholar] [CrossRef] [PubMed]
- Hallak, M.E.; Rodriguez, J.A.; Barra, H.S.; Caputto, R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977, 73, 147–150. [Google Scholar] [CrossRef]
- Kumar, N.; Flavin, M. Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. J. Biol. Chem. 1981, 256, 7678–7686. [Google Scholar] [PubMed]
- Barra, H.S.; Rodriguez, J.A.; Arce, C.A.; Caputto, R. A soluble preparation from rat brain that incorporates into its own proteins (14 C) arginine by a ribonuclease-sensitive system and (14 C) tyrosine by a ribonuclease-insensitive system. J. Neurochem. 1973, 20, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Arce, C.A.; Barra, H.S.; Rodriguez, J.A.; Caputto, R. Tentative identification of the amino acid that binds tyrosine as a single unit into a soluble brain protein. FEBS Lett. 1975, 50, 5–7. [Google Scholar] [CrossRef]
- Raybin, D.; Flavin, M. An enzyme tyrosylating α-tubulin and its role in microtubule assembly. Biochem. Biophys. Res. Commun. 1975, 65, 1088–1095. [Google Scholar] [CrossRef]
- Ersfeld, K.; Wehland, J.; Plessmann, U.; Dodemont, H.; Gerke, V.; Weber, K. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 1993, 120, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Szyk, A.; Deaconescu, A.M.; Piszczek, G.; Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 2011, 18, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Magiera, M.M.; Kuijpers, M.; Bargsten, K.; Frey, D.; Wieser, M.; Jaussi, R.; Hoogenraad, C.C.; Kammerer, R.A.; Janke, C.; Steinmetz, M.O. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J. Cell Biol. 2013, 200, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Paturle-Lafanechère, L.; Eddé, B.; Denoulet, P.; Van Dorsselaer, A.; Mazarguil, H.; Le Caer, J.P.; Wehland, J.; Job, D. Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry 1991, 30, 10523–10528. [Google Scholar] [CrossRef] [PubMed]
- Rogowski, K.; van Dijk, J.; Magiera, M.M.; Bosc, C.; Deloulme, J.C.; Bosson, A.; Peris, L.; Gold, N.D.; Lacroix, B.; Bosch Grau, M.; et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 2010, 143, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Berezniuk, I.; Vu, H.T.; Lyons, P.J.; Sironi, J.J.; Xiao, H.; Burd, B.; Setou, M.; Angeletti, R.H.; Ikegami, K.; Fricker, L.D. Cytosolic carboxypeptidase 1 is involved in processing α- and β-tubulin. J. Biol. Chem. 2012, 287, 6503–6517. [Google Scholar] [CrossRef] [PubMed]
- Berezniuk, I.; Lyons, P.J.; Sironi, J.J.; Xiao, H.; Setou, M.; Angeletti, R.H.; Ikegami, K.; Fricker, L.D. Cytosolic carboxypeptidase 5 removes α- and γ-linked glutamates from tubulin. J. Biol. Chem. 2013, 288, 30445–30453. [Google Scholar] [CrossRef] [PubMed]
- Kreitzer, G.; Liao, G.; Gundersen, G.G. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol. Biol. Cell 1999, 10, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Bulinski, J.C.; Gundersen, G.G. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays 1991, 13, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Webster, D.R.; Wehland, J.; Weber, K.; Borisy, G.G. Detyrosination of α tubulin does not stabilize microtubules in vivo. J. Cell Biol. 1990, 111, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Skoufias, D.A.; Wilson, L. Assembly and colchicine binding characteristics of tubulin with maximally tyrosinated and detyrosinated α-tubulins. Arch. Biochem. Biophys. 1998, 351, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Raybin, D.; Flavin, M. Modification of tubulin by tyrosination in cells and extracts and its effect on assembly in vitro. J. Cell Biol. 1977, 73, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Wehland, J.; Willingham, M.C.; Sandoval, I.V. A rat monoclonal antibody reacting specifically with the tyrosylated form of α-tubulin. I. Biochemical characterization, effects on microtubule polymerization in vitro and microtubule polymerization and organization in vivo. J. Cell Biol. 1983, 97, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.J.; Bulinski, J.C. Cellular microtubules heterogeneous in their content of microtubule-associated protein 4 (MAP4). Cell Motil. Cytoskelet. 1994, 27, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Feng, J.; Bo, W.; Wu, R.; Dong, Z.; Liu, Y.; Qiang, L.; Liu, M. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol. Biol. Cell 2017, 28, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Barnat, M.; Benassy, M.N.; Vincensini, L.; Soares, S.; Fassier, C.; Propst, F.; Andrieux, A.; von Boxberg, Y.; Nothias, F. The GSK3-MAP1B pathway controls neurites branching and microtubule dynamics. Mol. Cell. Neurosci. 2016, 72, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.; Hyman, A.A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007, 19, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Peris, L.; Wagenbach, M.; Lafanechere, L.; Brocard, J.; Moore, A.T.; Kozielski, F.; Job, D.; Wordeman, L.; Andrieux, A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 2009, 185, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, M.; Rice, L.M.; Vale, R.D. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 2014, 16, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Homma, N.; Takei, Y.; Tanaka, Y.; Nakata, T.; Terada, S.; Kikkawa, M.; Noda, Y.; Hirokawa, N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell 2003, 114, 229–239. [Google Scholar] [CrossRef]
- Peris, L.; Thery, M.; Fauré, J.; Saoudi, Y.; Lafanechère, L.; Chilton, J.K.; Gordon-Weeks, P.; Galjart, N.; Bornens, M.; Wordeman, L.; et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 2006, 174, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M.; Maesaki, R.; Kasa, M.; Watanabe, T.; Fukata, M.; Kaibuchi, K.; Hakoshima, T. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc. Natl. Acad. Sci. USA 2007, 104, 10346–10351. [Google Scholar] [CrossRef] [PubMed]
- Bieling, P.; Kandels-Lewis, S.; Telley, I.A.; van Dijk, J.; Janke, C.; Surrey, T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 2008, 183, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Nirschl, J.J.; Magiera, M.M.; Lazarus, J.E.; Janke, C.; Holzbaur, E.L. α-Tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep. 2016, 14, 2637–2652. [Google Scholar] [CrossRef] [PubMed]
- Eddé, B.; Rossier, J.; Le Caer, J.P.; Desbruyères, E.; Gros, F.; Denoulet, P. Posttranslational glutamylation of α-tubulin. Science 1990, 247, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Redeker, V.; Levilliers, N.; Schmitter, J.M.; Le Caer, J.P.; Rossier, J.; Adoutte, A.; Bré, M.H. Polyglycylation of tubulin: A posttranslational modification in axonemal microtubules. Science 1994, 266, 1688–1691. [Google Scholar] [CrossRef] [PubMed]
- Redeker, V.; Levilliers, N.; Vinolo, E.; Rossier, J.; Jaillard, D.; Burnette, D.; Gaertig, J.; Bré, M.H. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of α- and β-tubulin and affect the ciliary matrix in Tetrahymena. J. Biol. Chem. 2005, 280, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Sahab, Z.J.; Kirilyuk, A.; Zhang, L.; Khamis, Z.I.; Pompach, P.; Sung, Y.; Byers, S.W. Analysis of tubulin α-1A/1B C-terminal tail post-translational poly-glutamylation reveals novel modification sites. J. Proteome Res. 2012, 11, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Rogowski, K.; Wloga, D.; Regnard, C.; Kajava, A.V.; Strub, J.M.; Temurak, N.; van Dijk, J.; Boucher, D.; van Dorsselaer, A.; et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 2005, 308, 1758–1762. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, J.; Rogowski, K.; Miro, J.; Lacroix, B.; Eddé, B.; Janke, C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 2007, 26, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Rogowski, K.; Juge, F.; van Dijk, J.; Wloga, D.; Strub, J.M.; Levilliers, N.; Thomas, D.; Bré, M.H.; Van Dorsselaer, A.; Gaertig, J.; et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell 2009, 137, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Webster, D.M.; Rogowski, K.; Bré, M.H.; Levilliers, N.; Jerka-Dziadosz, M.; Janke, C.; Dougan, S.T.; Gaertig, J. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell 2009, 16, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Horigome, D.; Mukai, M.; Livnat, I.; MacGregor, G.R.; Setou, M. TTLL10 is a protein polyglycylase that can modify nucleosome assembly protein 1. FEBS Lett. 2008, 582, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Mukai, M.; Tsuchida, J.; Heier, R.L.; Macgregor, G.R.; Setou, M. TTLL7 is a mammalian β-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 2006, 281, 30707–30716. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Rogowski, K.; Sharma, N.; Van Dijk, J.; Janke, C.; Eddé, B.; Bré, M.H.; Levilliers, N.; Redeker, V.; Duan, J.; et al. Glutamylation on α-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot. Cell 2008, 7, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Mukai, M.; Ikegami, K.; Sugiura, Y.; Takeshita, K.; Nakagawa, A.; Setou, M. Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on β-tubulin through a random sequential pathway. Biochemistry 2009, 48, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Suryavanshi, S.; Eddé, B.; Fox, L.A.; Guerrero, S.; Hard, R.; Hennessey, T.; Kabi, A.; Malison, D.; Pennock, D.; Sale, W.S.; et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 2010, 20, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Dave, D.; Meagley, J.; Rogowski, K.; Jerka-Dziadosz, M.; Gaertig, J. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell 2010, 9, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Audebert, S.; Desbruyères, E.; Gruszczynski, C.; Koulakoff, A.; Gros, F.; Denoulet, P.; Eddé, B. Reversible polyglutamylation of α- and β-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell 1993, 4, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Bré, M.H.; Redeker, V.; Vinh, J.; Rossier, J.; Levilliers, N. Tubulin polyglycylation: Differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in paramecium. Mol. Biol. Cell 1998, 9, 2655–2665. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Kurabe, N.; Ikegami, K.; Tsutsumi, K.; Konishi, Y.; Kaplan, O.I.; Kunitomo, H.; Iino, Y.; Blacque, O.E.; Setou, M. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J. Biol. Chem. 2010, 285, 22936–22941. [Google Scholar] [CrossRef] [PubMed]
- Tort, O.; Tanco, S.; Rocha, C.; Bièche, I.; Seixas, C.; Bosc, C.; Andrieux, A.; Moutin, M.J.; Avilés, F.X.; Lorenzo, J.; Janke, C. The cytosolic carboxypeptidases CCP2 and CCP3 catalyze posttranslational removal of acidic amino acids. Mol. Biol. Cell 2014, 25, 3017–3027. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Bulinski, J.C. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat. Rev. Mol. Cell. Biol. 2011, 12, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Wloga, D.; Gaertig, J. Post-translational modifications of microtubules. J. Cell Sci. 2010, 123 Pt 20, 3447–3455. [Google Scholar] [CrossRef] [PubMed]
- Thazhath, R.; Liu, C.; Gaertig, J. Polyglycylation domain of β-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 2002, 4, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Thazhath, R.; Jerka-Dziadosz, M.; Duan, J.; Wloga, D.; Gorovsky, M.A.; Frankel, J.; Gaertig, J. Cell context-specific effects of the β-tubulin glycylation domain on assembly and size of microtubular organelles. Mol. Biol. Cell 2004, 15, 4136–4147. [Google Scholar] [CrossRef] [PubMed]
- Pathak, N.; Obara, T.; Mangos, S.; Liu, Y.; Drummond, I.A. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell 2007, 18, 4353–4364. [Google Scholar] [CrossRef] [PubMed]
- Pathak, N.; Austin, C.A.; Drummond, I.A. Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility. J. Biol. Chem. 2011, 286, 11685–11695. [Google Scholar] [CrossRef] [PubMed]
- Bosch Grau, M.; Gonzalez Curto, G.; Rocha, C.; Magiera, M.M.; Marques Sousa, P.; Giordano, T.; Spassky, N.; Janke, C. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 2013, 202, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.S.; He, Y.; Dougherty, E.J.; Jimenez-Movilla, M.; Avella, M.; Grullon, S.; Sharlin, D.S.; Guo, C.; Blackford, J.A., Jr.; Awasthi, S.; et al. Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility. J. Biol. Chem. 2013, 288, 15167–15180. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, R.; Piasecki, B.P.; Silva, M.; Phirke, P.; Nguyen, K.C.; Hall, D.H.; Swoboda, P.; Barr, M.M. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr. Biol. 2011, 21, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Boucher, D.; Larcher, J.C.; Gros, F.; Denoulet, P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry 1994, 33, 12471–12477. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Boucher, D.; Lazereg, S.; Pedrotti, B.; Islam, K.; Denoulet, P.; Larcher, J.C. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B and MAP2 by tubulin polyglutamylation. J. Biol. Chem. 2001, 276, 12839–12848. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A.; McNally, F.J. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 2010, 22, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A.; Vale, R.D. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 2008, 451, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; van Dijk, J.; Gold, N.D.; Guizetti, J.; Aldrian-Herrada, G.; Rogowski, K.; Gerlich, D.W.; Janke, C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 2010, 189, 945–954. [Google Scholar] [CrossRef] [PubMed]
- McNally, K.P.; Buster, D.; McNally, F.J. Katanin-mediated microtubule severing can be regulated by multiple mechanisms. Cell Motil. Cytoskelet. 2002, 53, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Yu, W.; Andreadis, A.; Luo, M.; Baas, P.W. Tau protects microtubules in the axon from severing by katanin. J. Neurosci. 2006, 26, 3120–3129. [Google Scholar] [CrossRef] [PubMed]
- Wightman, R.; Chomicki, G.; Kumar, M.; Carr, P.; Turner, S.R. SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr. Biol. 2013, 23, 1902–1907. [Google Scholar] [CrossRef] [PubMed]
- Alderson, N.L.; Wang, Y.; Blatnik, M.; Frizzell, N.; Walla, M.D.; Lyons, T.J.; Alt, N.; Carson, J.A.; Nagai, R.; Thorpe, S.R.; et al. S-(2-Succinyl)cysteine: A novel chemical modification of tissue proteins by a Krebs cycle intermediate. Arch. Biochem. Biophys. 2006, 450, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Piroli, G.G.; Manuel, A.M.; Walla, M.D.; Jepson, M.J.; Brock, J.W.; Rajesh, M.P.; Tanis, R.M.; Cotham, W.E.; Frizzell, N. Identification of protein succination as a novel modification of tubulin. Biochem. J. 2014, 462, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Kang, J.G.; Park, S.Y.; Lee, J.; Oh, Y.J.; Cho, J.W. O-GlcNAcylation of tubulin inhibits its polymerization. Amino Acids 2011, 40, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Natarajan, K.; Curiel, J.; Janke, C.; Liu, J. The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton (Hoboken) 2016, 73, 521–550. [Google Scholar] [CrossRef] [PubMed]
- Barisic, M.; Maiato, H. The tubulin code: A navigation system for chromosomes during mitosis. Trends Cell Biol. 2016, 26, 766–775. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wloga, D.; Joachimiak, E.; Fabczak, H. Tubulin Post-Translational Modifications and Microtubule Dynamics. Int. J. Mol. Sci. 2017, 18, 2207. https://doi.org/10.3390/ijms18102207
Wloga D, Joachimiak E, Fabczak H. Tubulin Post-Translational Modifications and Microtubule Dynamics. International Journal of Molecular Sciences. 2017; 18(10):2207. https://doi.org/10.3390/ijms18102207
Chicago/Turabian StyleWloga, Dorota, Ewa Joachimiak, and Hanna Fabczak. 2017. "Tubulin Post-Translational Modifications and Microtubule Dynamics" International Journal of Molecular Sciences 18, no. 10: 2207. https://doi.org/10.3390/ijms18102207
APA StyleWloga, D., Joachimiak, E., & Fabczak, H. (2017). Tubulin Post-Translational Modifications and Microtubule Dynamics. International Journal of Molecular Sciences, 18(10), 2207. https://doi.org/10.3390/ijms18102207





